Abstract
We report the 3D structure of human β2 adrenergic receptor (AR) predicted by using the MembStruk first principles method. To validate this structure, we use the HierDock first principles method to predict the ligand-binding sites for epinephrine and norepinephrine and for eight other ligands, including agonists and antagonists to β2 AR and ligands not observed to bind to β2 AR. The binding sites agree well with available mutagenesis data, and the calculated relative binding energies correlate reasonably with measured binding affinities. In addition, we find characteristic differences in the predicted binding sites of known agonists and antagonists that allow us to infer the likely activity of other ligands. The predicted ligand-binding properties validate the methods used to predict the 3D structure and function. This validation is a successful step toward applying these procedures to predict the 3D structures and function of the other eight subtypes of ARs, which should enable the development of subtype-specific antagonists and agonists with reduced side effects.
The adrenergic receptors (ARs) are the class of G protein-coupled receptors (GPCR) responsible for mediating the effects of the catecholamines epinephrine and norepinephrine. There are currently nine known human ARs, partitioned into three subclasses: α1 (three subtypes located in vascular smooth muscle, the digestive tract, liver, and postsynaptically in the CNS), α2 (three subtypes located pre- and postsynaptically in the CNS, and in a wide variety of peripheral sites), and β (three subtypes located primarily in cardiac, vascular, and adipose tissues, respectively).
The members of this receptor class mediate a wide variety of physiological responses, including vasodilation and vasoconstriction, heart rate modulation, regulation of lipolysis, and blood clotting. These diverse and important functions make the adrenergic receptors a tempting pharmaceutical target, but attempts to create effective and specific drugs acting on these receptors have been slowed down by the lack of a 3D structure for any GPCR other than the bovine photoreceptor rhodopsin. The focus of this paper is the β2AR, which is targeted by agonist therapy in the treatment of asthma. Unfortunately, β2 agonists also exhibit crossreactivity with the other βARs, causing side effects such as increased heart rate and blood pressure (1). Three-dimensional models of the ARs would be extremely useful in the design of subtype-specific pharmaceutical compounds. In addition, the ARs have been thoroughly studied experimentally so that there are ample data for validating the structural predictions, which may in turn provide improved understanding for the superfamily of GPCRs.
We report here the predicted 3D structure of β2AR, which we use to predict detailed binding sites of agonists and antagonists to β2AR. This is an excellent case for validation because there is a wealth of experimental data on ligand-binding sites and mutational analysis with which to compare our results (2, 3).
We use the MembStruk computational method to predict the atomic level tertiary structure of GPCRs using only the primary sequence, and we use the HierDock method to predict the binding site and binding energy of ligands binding to the protein (4, 5). These methods have been validated for bovine rhodopsin (5, 6) where the predicted binding site is in good agreement with the experimental results. In addition, recent results for human D2 dopamine receptor (unpublished data) lead to predicted binding sites and energies for dopamine and D2 agonists and antagonists that are in good agreement with experimental data.
Materials and Methods
Structure Prediction of GPCRs: The MembStruk Method. The MembStruk method (version 3.5) for predicting the structure of transmembrane proteins consists of the following steps (5, 6). All energy and force evaluations use the DREIDING force field, CHARMM22 (7) charges for the protein, and QM charges for the ligands. Transmembrane (TM) prediction (TM2ndS). First, we predict the seven TM domains by using hydropathicity analysis combined with information from sequence alignments. The extent of each TM region is predicted by using sequence alignments of 14 input sequences having sequence identities ranging from 40% to over 90%. Then, we calculate the average hydrophobicity for every residue position over all these sequences in the multiple sequence alignment while averaging over a window size 12 to 20 residues. The baseline for this profile serves as the threshold value for determining the TM regions.
Position of maximum hydrophobicity. For each TM region, we identify lipid-accessible residues from the sequence alignments (less conserved residues) and from analysis of the hydrophobicity maxima in the sequence. This position of maximum hydrophobicity is used to estimate the relative translational orientation of the helices. We use the Eisenberg scale for hydrophobicity (8). Assembly of the helix bundle and optimization of the translational orientation of the helices. The helical axes are oriented according to the 7.5-Å electron density map of frog rhodopsin (9). The initial relative translational orientation of each helix is optimized on placing into the same fitting plane all of the hydrophobic centers obtained from step 2.
Optimization of helical bends and kinks. We construct canonical helices for the predicted TM segments and optimize the structures of the individual helices using energy minimization followed by fast torsional Newton–Euler inverse mass operator method (NEIMO) dynamics (10, 11) for 500 ps. This procedure optimizes the bends and kinks in each helix.
Monte Carlo optimization of rotational orientation of the helices. The initial rotational orientation of each helix (about its axis) is determined by setting the direction of the net hydrophobic moment of the middle one-third of each helix (about its hydrophobic center). Because molecular dynamics is not likely to surmount the barriers that might separate one good rotational state from another, we carry out a systematic search in which each helix is rotated through a grid of rotational angles, for each value of which the other six helices are optimized sequentially. This sampling allows the system to surmount rotational energy barriers to sample various possible rotational minima for each helix. This rotational optimization is carried out for each helix in turn (over the same grid) until there is no longer a change.
Optimization of the assembled helical bundle with an explicit lipid environment. The optimized helix bundle is further equilibrated by immersing the protein into a lipid bilayer and performing rigid body molecular dynamics (12, 13). The helix bundle surrounded by a lipid bilayer was optimized by using rigid body dynamics.
Loop building. We construct the full GPCR protein by adding interhelical loops and disulfide bridges to the helix bundle using whatif (14). In β2AR, the second intracellular loop (60 residues between TM5 and TM6) was truncated by 23 residues in the middle of the loop (residues 241–264) because such large loops are quite flexible, leading to a multiplicity of conformations with similar energies. Because this deletion is in the middle of a 60-residue loop region and because this loop is on the intracellular side, these changes should have little effect on the structure of the TM region. Currently, we omit modeling the amino and carboxyl terminal regions. Optimization of the final model. We optimize the final structure by using conjugate gradient minimization of all atoms in the structure but with lipid fixed.
Validation: Structure prediction for bovine rhodopsin. The only GPCR with an experimental 3D crystal structure is bovine rhodopsin (15, 16), making this the best structure for validating MembStruk. Trabanino et al. (6) showed that the TM regions of the MembStruk-predicted structure for rhodopsin agree with the crystal structure, to 2.85 Å CRMS (rms deviation in alpha carbon coordinates) for all of the main chain atoms. This good agreement with the crystal structure indicates that the MembStruk procedure predicts the helical regions reasonably well, without using any knowledge of the crystal structure.
Functional Prediction of GPCRs: The HierDock Protocol. The HierDock ligand-screening protocol follows a hierarchical strategy for examining ligand-binding conformations and calculating their binding energies. The steps are as follows.
Scanning. We carry out a coarse grain docking procedure to generate a set of conformations for ligand binding in the receptor. Here, we use dock 4.0 (17) to generate and score 1,000 configurations, of which 10% were selected by using a buried surface area cutoff of 85% and by using energy scoring from dock 4.0, for further analysis.
Ligand relaxation. The 100 best conformations selected for each ligand from step 1 are subjected to all-atom minimization, keeping the protein fixed but the ligand movable. The solvation of each of these 100 minimized structures was calculated by using the analytical volume generalized born (AVGB) continuum solvation method (18). The binding energies (BE) were calculated by using BE = [PE (ligand in solvent) - PE (ligand in protein)] where PE is the potential energy. Then, the 10 structures based both on binding energies and buried surface areas were selected from these 100 structures for the next step.
Complex relaxation. We optimize the structure of the receptor/ligand complex allowing the structure of the protein to accommodate the ligand. This relaxation is essential to identify the optimum conformations for the complex. The all-atom receptor/ligand energy minimization was performed on the 10 structures from the previous step. Using these optimized structures, we calculate the binding energy as the difference between the energy of the ligand in the protein and the energy of the ligand in water. The energy of the ligand in water is calculated by using DREIDING force field and the analytical volume generalized born (AVGB) continuum solvation method (18).
Side-chain optimization. We select, from the 10 structures from the previous step, the one with the maximum number of hydrogen bonds between ligand and protein. For this structure, we use the scream side-chain replacement program to reassign all side-chain rotamers for the residues within 4 Å in the binding pocket [scream uses a side-chain rotamer library (1,478 rotamers with a 1.0-Å resolution) with the all-atom DREIDING energy function to evaluate the energy for the ligand–protein complex].
Scanning the entire receptor for binding sites. To locate the binding site, we scan the entire protein for potential binding regions. The entire molecular surface of the predicted structure of β2AR is mapped, and spheres representing the empty volume of the receptor are generated by using the Sphgen program in the dock 4.0 suite of programs. The entire set of receptor spheres is divided into six overlapping regions, and epinephrine is used to scan for a putative binding site. This first pass is carried out by performing only the first 2 steps of the HierDock protocol described above.
Determination of binding site and binding energy for all ligands. In the second pass, we then take a 10 × 10 × 10-Å cube surrounding the putative binding site and perform all of the steps of the HierDock protocol as described above for all ligands in the study. Validation for function prediction protocol. The HierDock protocol has been validated by using it to predict the binding site for aminoacyl t-RNA synthetases (19, 20) and 37 other co-crystals of globular proteins (21, 22). We also validated HierDock for binding of a ligand to a GPCR by docking 11-cis-retinal to bovine rhodopsin (5, 6). The CRMS between crystal structure and the docked structure for cis-retinal is 0.6 Å, which is excellent considering that no information of the binding site was assumed. We have also used HierDock to predict binding to olfactory receptors and to other GPCRs (4, 5).
Results
Predicted Structure of Human β2AR. Our predicted β2AR structure shows the same general topology as bovine rhodopsin. There are kinks at the conserved prolines of TM2 and TM6 and minor bends in several other helices. The overall β2AR structure deviates from rhodopsin by 3.96 Å coordinate rms in the alpha carbons (CRMSα) of the TM region (8.49 Å CRMSα over the entire structure). We suspect that this 3D structure represents the inactive state of the receptor because it seems to predict binding of agonist and antagonist equally well.
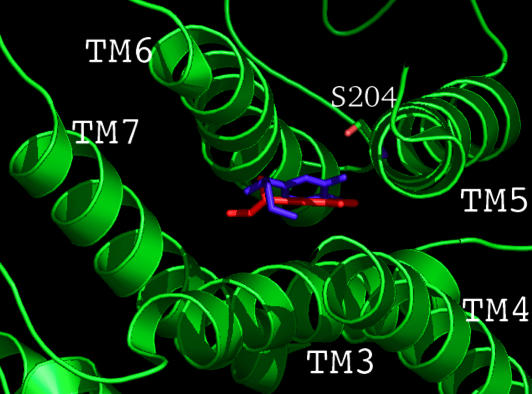
Predicted Binding Site of Epinephrine in β2AR. From the HierDock protocol described above, the predicted binding site of epinephrine is shown in Fig. 1 where epinephrine is nestled between TM3, 4, 5, and 6 as shown in the top view (Fig. 1B). This figure also shows the details of the 5.0-Å binding site of epinephrine, which suggests that the following residues are critical in binding the epinephrine.
Fig. 1.
Five-angstrom predicted binding site of epinephrine in the predicted structure for β2AR. Complete amino acids are shown in the side view (A) and top view (B) whereas only side chains are shown in (C). Protein model figures generated by using pymol software (31).
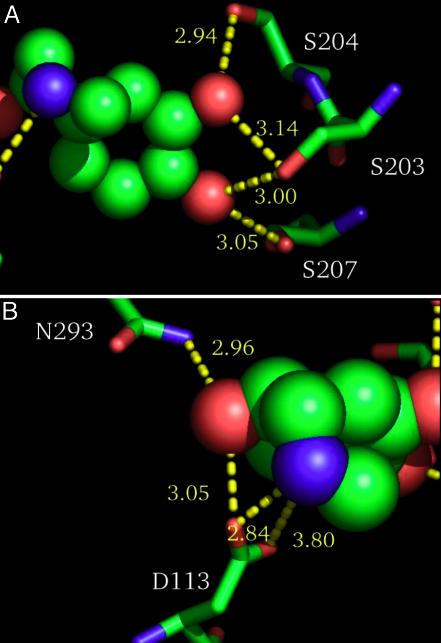
Asp-113 (TM3). Asp-113 forms a salt bridge with the amine (2.9 Å) of epinephrine and accepts a hydrogen bond from the alkyl OH group (3.0 Å). The Asp at this position is conserved across all biogenic amine receptors (whose endogenous ligands all include a similar amine), and numerous binding studies have shown that this residue interacts with the amine in epinephrine (2), validating our predicted binding site. The second oxygen of the carboxylate of Asp-113 is 3.8 Å from the amine, making the interaction with this part of the side chain much weaker.
Ser-203, -204, and -207 (TM5). We find that all three serines form a hydrogen-bonding network with the two-catechol OH groups of epinephrine as shown in Fig. 2. The four hydrogen bond distances are 3.1 Å (Ser-203-meta OH), 2.9 Å (Ser-204-meta OH), 3.0 Å (Ser-203-para OH), and 3.1 Å(Ser-207-para OH). Detailed mutational studies have shown that Ser-204 interacts specifically with the meta hydroxyl whereas Ser-207 interacts specifically with the para hydroxyl (2), validating our predicted binding site. In addition, a recent study indicates that Ser-203 (previously thought not to be involved in binding) is quite important to agonist binding, interacting with the meta OH of epinephrine (23). Our results indicate that Ser-203 interacts with both the meta and para OH groups to form the elegant hydrogen-bonding network shown in Fig. 2. The Ser-203-meta OH interaction is conserved across all agonists in our study whereas the Ser-203-para OH interaction was not, indicating that the meta interaction is probably functionally important whereas the network including the para OH may not be vital to agonist recognition. This conclusion agrees with mutation data indicating that binding of phenylephrine (epinephrine without the para OH) is not substantially affected by mutation of Ser-203 (23).
Fig. 2.
Important hydrogen bond contacts in the predicted binding site of epinephrine to human β2AR. (A) The hydrogen-bonded network formed between both catechol hydroxyl groups with all three highly conserved TM5 serines (S203, S204, and S207). (B) The hydrogen-bonded network involving the highly conserved TM3 aspartic acid and N293 with binding to the amine of the ligand.
Asn-293 (TM6). This residue donates a hydrogen bond (3.0 Å) to the alkyl OH of epinephrine, which is also coupled to Asp-113. Recently, point mutational studies by Shi et al. (24) showed that Asn-293 is important for binding, and it has been speculated to recognize the alkyl OH of epinephrine (24). A detailed picture of the ionic and hydrogen bond interactions between Asp-113, Asn-293, and the ligand is shown in Fig. 2B.
Ile-169 (TM4). This residue provides a hydrophobic interaction with the N-methyl group of epinephrine. It is established experimentally that epinephrine binds more strongly to β2AR than norepinephrine, and this methyl group is the only difference between these two ligands. Thus, it is plausible that Ile-169 is responsible for this selectivity
Val-117 (TM3) and Phe-290 (TM6). These residues provide hydrophobic interactions (favorable van der Waals interactions) with the ring of epinephrine. Phe-290 is in a hydrophobic region that also includes Phe-289 and Trp-286, collectively termed the WXXFF motif. This motif is conserved throughout the biogenic amine receptors (24). In our structure, only Phe-290 seems to be directly involved in binding the ligand, but the other two residues may act to position it properly while contributing to the hydrophobicity of this region of the binding pocket.
Binding Sites of Other Ligands. Predicted binding sites of all other ligands in the study are shown in Fig. 7 (which is published as supporting information on the PNAS web site). The predicted binding site of each ligand is described below; the structures of these ligands are shown in Fig. 3.
Fig. 3.
Structures of all ligands docked to the β2AR in this study.
Norepinephrine (native ligand). The predicted binding site is broadly similar to that of epinephrine, with two exceptions. Ile-169 interacts with the ring of norepinephrine because there is no N-methyl group in norepinephrine; this change allows the tail of norepinephrine to have a slightly different shape than in epinephrine. Additionally, in binding to norepinephrine, Asp-113 interacts only with the amine group (not with the hydroxyl group in the alkyl chain of norepinephrine), but both oxygens of the carboxylate of Asp-113 are strongly involved (distances of 2.9 Å and 3.2 Å). Fig. 4 compares the binding conformations of epinephrine and norepinephrine, along with the location of Ser-204. We see that epinephrine has a stronger interaction with this serine than norepinephrine, but a counterclockwise rotation of helix 5 by ≈20° could improve the interaction with both ligands. It is possible that our predicted structure has small errors in rotational orientation of the helices, or that this potential for rotation is indicative of domain-level movements that occur during activation, as will be discussed below.
Fig. 4.
Comparison of the predicted binding sites of epinephrine (blue) and norepinephrine (red) to the β2AR. Epinephrine is tilted toward S204 (making a strong contact) whereas norepinephrine is tilted slightly away (making a weak contact).
Isoproterenol (synthetic agonist). All interactions found in the binding of epinephrine are also found in the binding site for isoproterenol. However, isoproterenol has a larger alkyl group than epinephrine, which interacts with additional residues. Thus, Trp-109 has a direct hydrophobic interaction with this isopropyl tail whereas Ile-169 seems to interact more strongly. Additionally, Trp-173 (EC2) seems to close over the top of the binding site to interact with the isopropyl group of isoproterenol.
Salbutamol (β2AR-specific agonist). Salbutamol seems to form interactions similar to those of epinephrine and isoproterenol. However, there is sufficient space in the binding cavity for the t-butyl group of salbutamol to fit in the same location as the methyl of epinephrine, interacting with Ile-169, His-172, and Trp-173. The bulky hydrophobic tails of these three ligands cause them to bind in a slightly different conformation than norepinephrine and dopamine, which may contribute to βAR subtype specificities and differences in affinity between adrenergic and dopamine receptors.
Dopamine (native ligand for dopaminergic receptors; weak agonist for β2AR). The predicted binding site of dopamine seems broadly similar to that of norepinephrine, with two important exceptions. First, the catechol OHs of dopamine seem to interact only with Ser-203 and Ser-207 (Ser-204 is over 4.8 Å away). This result may be due to the lack of a hydrogen bond with Asn-293, which would otherwise pull the ligand in the direction of Ser-204. Second, because there is no alkyl OH, Asn-293 does not interact with this part of the ligand and instead seems to interact weakly with the amine.
Propranolol (synthetic antagonist). Propranolol binds in the same pocket as the other ligands, but in a substantially different conformation. Most notable is that the ligand hydroxyl group forms a bifurcated hydrogen bond with Ser-203 and Ser-204, but there is no interaction with Ser-207. In addition, it seems that the Cβ of Ser-203 is also involved in a hydrophobic interaction with the backbone and ring system of propranolol. The amine of propranolol seems to interact with Asp-113 (as do the other ligands), but here the aromatic ring system of the ligand sits much closer to TM6. As a result the aromatic ring interacts strongly with Phe-289, Phe-290, and Ile-294. The isopropyl tail seems to be in the same conformation as for isoproterenol, interacting with Trp-109. Ile-169 seems to be involved in a hydrophobic interaction with the alkyl backbone of the ligand.
Butoxamine (β2-selective antagonist). Butoxamine binds in a conformation almost identical to that of propranolol, with an aromatic system interacting with TM6 and a t-butyl group on the amine that binds in exactly the same position as the same portion of salbutamol. Oxygens on butoxamine are involved in hydrogen-bonding interactions with Ser-203 and Ser-204, but there is no interaction between butoxamine and Ser-207. Although there is no interaction with Asn-293 in our initially predicted structure, we find that a 180° rotation of the asparagine side chain allows a strong interaction between the asparagine amine and a methyl ether oxygen of butoxamine. This interaction improves the binding energy by ≈2.5 kcal/mol. The N-isopropyl group seems to make the same interactions as the hydrophobic tails of salbutamol and isoproterenol.
Metoprolol (β1-selective antagonist). Metoprolol binds to the β2AR in a conformation that allows its amine group to interact with Asp-113, while allowing a pair of oxygens in the ligand to interact with Ser-203 and Ser-204. A third oxygen in metoprolol interacts with the amine of Trp-109 whereas hydrophobic portions of the ligand have strong interactions with Phe-290 and Ile-169. Although metoprolol is marketed as a specific β1AR blocker, its side effect profile indicates it could have some antagonistic crossreactivity with β2AR, as will be discussed below. Xamoterol (β1-selective agonist) and Atenolol (β1-selective antagonist). The predicted binding sites are shown in Fig. 7, but our calculations showed that they do not bind appreciably to β2AR.
Discussion
Difference in Agonist and Antagonist Binding Sites. Of the eight most strongly bound molecules in our calculations, some are known to act as agonists whereas others act as antagonists. We find that the binding sites of the known agonists and antagonists described above show consistently different patterns in the binding site. Thus, the agonists epinephrine, norepinephrine, salbutamol, and isoproterenol all form hydrogen bonds with both Ser-204 and Ser-207, and the antagonists that bind strongly to the receptor form hydrogen bonds only with Ser-203 (or Ser-203 and Ser-204 for butoxamine), but never Ser-207. This difference between the binding of epinephrine and butoxamine is shown in Fig. 5. Both agonists and antagonists have a strong bond to the Asp-113 of TM3. Based on these results, we suggest that an agonist must couple strongly to TM5 with both Ser-204 and Ser-207 while also bonding to TM3 through Asp-113. On the other hand, the antagonists need to recognize the same size (to block the agonist) but lead to a more flexible coupling of TM3 and TM5 so that they do not induce the transition to the activated state.
Fig. 5.
Critical hydrogen-bonding contacts in the binding of butoxamine (A) and epinephrine (B)to β2AR. Note that butoxamine does not contact S207 (6 Å away) whereas epinephrine has a strong hydrogen bond (3-Å contact).
Difference in Agonist-Binding Sites. Among the agonists in this study, we find two slightly different binding conformations. Isoproterenol, salbutamol, and epinephrine (the stronger agonists for β2AR) bind in a conformation that allows them to form a strong hydrogen-bonding network with all three of the conserved TM5 serines (as shown in Fig. 2). In contrast, the weaker agonists norepinephrine and dopamine form strong contacts only with Ser-203 and Ser-207, lying rather far from Ser-204. On the other hand, for both cases, only a small rotation of TM5 would bring Ser-204 into an excellent hydrophilic interaction with the ligand, while also improving the hydrogen bond angles to the other two residues. These results suggest that β2AR activation may be mediated by a rotation of TM5 to optimize hydrogen bonding with agonist ligands. A small counterclockwise (as viewed from the extracellular side) rotation of TM5 would greatly improve the interactions of norepinephrine and dopamine with Ser-204 (and also Ser-207 for dopamine). It would also improve the interaction of salbutamol with Ser-207 and that of isoproterenol with Ser-204. Although the hydrogen-bonding interactions with epinephrine seem optimal in the docked conformation, it is so close to Ser-203 that it has a minor van der Waals clash with the Cβ hydrogens of this side chain; a slight rotation of TM5 could relieve this clash while also allowing some improvement in the hydrogen bond angles to Ser-203. The underlying trend behind these interactions is that all of the agonists seem to bind with their catechol hydroxyls much closer to the beta carbon position of Ser-203 than those of Ser-204 or Ser-207, even in cases (such as epinephrine) where hydrogen bond distances to Ser-204 and Ser-207 are optimal in these structures. This conformation is possible because the rotamers of Ser-204 and Ser-207 are fully extended toward the ligand whereas that of Ser-203 is more parallel with the ring of the ligand (perpendicular to the cytoplasmic plane). This feature makes it possible for interactions with all agonists to be improved by a rotation of TM5 to improve the hydrogen bonding network that they form with the serines of TM5. This TM5 rotation will improve the binding to ligand with little or no motion of the ligand, allowing the binding interactions with TM3 and TM6 to be preserved through this rotation (we believe that anchoring interactions with TM3 and TM6 may be what causes the ligand to fall into a slightly nonoptimal conformation with respect to its TM5 contacts). Thus, we postulate that all β adrenergic agonists bind strongly to the three conserved serines in TM5 and exert activating effects by inducing a rotation of TM5 that may have broad-reaching effects elsewhere in the protein. In contrast, antagonists do not form this network of interactions with the TM5 serines, making them incapable of inducing the conformational changes leading to activation.
Recent experimental studies indicate that activation of the β2AR involves a movement of TM6 relative to TM5 and TM3 (25), causing disruption of an ionic lock between the cytoplasmic ends of TM3 and TM6 (26). Detailed interhelical-linkage studies on the β2AR have shown that modifications that maintain close contact between the cytoplasmic ends of TM3 and TM6 completely block receptor activation (27). These results, combined with Ghanouni's fluorescence experiments (25), have led to the conclusion that, on activation, TM6 either rotates slightly counterclockwise (as viewed from the extracellular region) or tilts its intracellular end toward TM5. It is possible that a small rotation of TM5 is the critical step in causing the protein to transition to the activated state. Clearly, the linkage of TM5 and TM6 by the IL-3 loop must play some key role in this. A recent study on bacteriorhodopsin showed that a potential-based change in the conformation of the IL-3 loop may be directly responsible for an outward tilting of TM6 occurring at the end of the M segment of the photocycle (28). Although the function of bacteriorhodopsin is quite different from that of the β2AR, this experiment illustrates that manipulating the conformation of IL-3 in a seven-TM protein can cause a meaningful spatial movement of TM6. A similar mechanism may be involved in activation of the β2AR (and potentially other GPCRs as well), with the agonist-induced rotation of TM5 causing the IL-3 loop to change conformations, and thus leading to a translation of TM6 and a full transition to the activated state. More detailed studies will be required to determine the exact relationship between these activation events.
Binding Energies and Interactions of the Various Ligands. Classification as agonist or antagonist. The analysis of hydrogen-bonding contacts described above to classify each binding ligand as either an agonist or antagonist leads to the classifications shown in Fig. 6. This criterion correctly classifies all ligands as either agonists or antagonists. We classify dopamine as a weak agonist because it makes the appropriate serine contacts, but its contact to S204 is quite weak, and thus dopamine may not be as effective in inducing the activating conformational change (described above) as the other agonists in this study.
Fig. 6.
Predicted binding energies and contact distances (hydrogen-bonding and ionic) for the ligands of Fig. 4 to β2AR (all distances are heavy atom–heavy atom). A more positive energy signifies stronger binding. Based on the TM5 criteria described in the text, ligands were classified as either agonists or antagonists; their experimentally recognized effects are also indicated. Hydrogen bond lengths and electrostatic interactions between the receptor are also shown for each of five key residues of the protein; strong interactions (contacts <3.5 Å) are shown in bold. An x indicates no interaction. The hydrogen bond in butoxamine marked with an asterisk indicates a contact that is made only with the side chain of N293 flipped 180° from the simulated conformation, as described in the text.
Comparison of predicted binding energies and effects with experimental data. The binding energies for all ligands in this study are shown in Fig. 6. These results are in qualitative agreement with experiment. Thus, it is known that isoproterenol exhibits the strongest binding affinity among the common agonists to β2AR and that epinephrine binds more strongly than norepinephrine (2), all in agreement with our calculations. We also find that salbutamol (known to be a β2-specific agonist) binds quite strongly, on par with other strong agonists for this receptor. With only one exception, the results for the agonists, and their relative ordering, are perfectly in accord with experimental affinities (29). Only for dopamine is there a deviation between our results and the known relative affinities for β2AR. Dopamine is known to bind less strongly than norepinephrine, but we predict it to bind more strongly. We do not have an explanation for this result because we find an almost identical binding mode.
We should emphasize that the binding energies reported here come from energy minimization, corresponding to the binding enthalpy at 0 K (except that we do not include zero point energy). The calculations do include solvation effects (based on the properties of water at 300 K), but they do not include explicit entropic terms or the temperature corrections in the enthalpy. Nevertheless, these results provide valuable information about the active site, including an interpretation of difference between agonists or antagonists, and indeed the calculated binding energies show a very good correlation with experimental dissociation constants, particularly when agonists and antagonists are analyzed separately. A graph comparing our calculated binding energies with experimental energies is included in Fig. 8 (which is published as supporting information on the PNAS web site).
Our results suggest that both propranolol and butoxamine act as antagonists for this receptor. Butoxamine is known to be a selective antagonist for β2AR whereas propranolol is an unselective β antagonist. The other three ligands considered here (metoprolol, atenolol, and xamoterol) were designed to be specific for β1AR. Of these ligands, we find that only metoprolol binds appreciably to β2AR, and, because it does not interact with Ser-207, we expect metoprolol to act as an antagonist. This finding is consistent with the side effect profile for metoprolol, which indicates the occurrence of breathing problems consistent with β2AR antagonism (30).
Full analysis of the subtype selectivity of these compounds (agonists and antagonists) must await completion of similar studies for β1AR, but the current results are promising, both because salbutamol (a strong selective β2 agonist) was found to be strongly active and because two β1-specific compounds were found not to bind.
Conclusion
We find that the predicted 3D structure of the β2AR leads to predicted binding sites of various ligands that are in excellent agreement with all available experimental data. The predicted docking conformations for epinephrine and all other ligands with well characterized binding sites match very closely to experimental results. Most notably, all experimentally determined hydrophilic interactions are predicted successfully in our computational models.
The predicted binding site and energetics of other ligands to the predicted β2AR structure also lead to good agreement with experiment. Moreover, this analysis leads to a classification of ligands as either agonists or antagonists, based on whether or not they interact with both of the highly conserved TM5 serines of the receptor: Any ligand that binds but does not contact both serines will act as an antagonist whereas those ligands binding to both serines will act as agonists. This correlation allows us to successfully classify all ligands in the study as agonists or antagonists (or nonbinding), and it may prove useful in predicting the effects of novel ligands to β2AR or other GPCRs. We hope that detailed analyses of the reasons behind this correlation may aid in finding a detailed description of how receptor activation occurs, both in the β2AR and in other receptors in this family.
This study of β2AR, and other successful recent studies in other GPCRs, suggests that these ab initio predictions of the 3D structure and function may be useful for other GPCRs. We anticipate that these methods may also be useful for predicting the function of less well characterized GPCRs, such as lipid and peptide receptors, and may be useful in quickly assaying potential crossreactivity effects of new compounds. Carrying out similar studies for the binding site of each ligand to the other eight adrenergic receptors should help develop an understanding of the origins of subtype specificities in the ARs.
Supplementary Material
Acknowledgments
This research was supported partially by National Institutes of Health Grants BRGRO1-GM625523, R29AI40567, and HD36385. The computational facilities were provided by a Shared University Research grant from IBM and Defense University Research Instrumentation Program grants from the Army Research Office (ARO) and the Office of Naval Research (ONR). The facilities of the Materials and Process Simulation Center are also supported by the Department of Energy, the National Science Foundation, the Multidisciplinary University Research Initiative (MURI)-ARO, MURI-ONR, General Motors, ChevronTexaco, Seiko-Epson, the Beckman Institute, and Asahi Kasei.
Abbreviations: AR, adrenergic receptor; GPCR, G protein-coupled receptor; TM, transmembrane; CRMS, rms deviation in alpha carbon coordinates.
References
- 1.Giroux, M. & Ferrieres, J. (2003) J. Asthma 40, 41-48. [DOI] [PubMed] [Google Scholar]
- 2.Strader, C. D. (1994) Annu. Rev. Biochem. 63, 101-132. [DOI] [PubMed] [Google Scholar]
- 3.Lotvall, J. (2002) Pulm. Pharmacol. Ther. 15, 497-501. [DOI] [PubMed] [Google Scholar]
- 4.Floriano, W. B., Vaidehi, N., Goddard, W. A., III, Singer, M. S. & Shepherd, G. M. (2000) Proc. Natl. Acad. Sci. USA 97, 10712-10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaidehi, N., Floriano, W. B., Trabanino, R. J., Hall, S. E., Freddolino, P., Choi, E. J., Zamanakos, G. & Goddard, W. A., III (2002) Proc. Natl. Acad. Sci. USA 99, 12622-12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trabanino, R. J., Hall, S. E., Vaidehi, N., Floriano, W. B., Kam, V. & Goddard, W. A., III (2004) Biophys. J., in press. [DOI] [PMC free article] [PubMed]
- 7.MacKerell, A. D., Bashford, D., Bellott, M., Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Guo, H. & Ha, S. (1998) J. Phys. Chem. 102, 3586-3616. [DOI] [PubMed] [Google Scholar]
- 8.Eisenberg, D., Weiss, R. M. & Terwilliger, T. C. (1984) Proc. Natl. Acad. Sci. USA 8, 140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schertler, G. F. X. (1998) Eye 12, 504-510. [DOI] [PubMed] [Google Scholar]
- 10.Jain, A., Vaidehi, N. & Rodriguez, G. (1993) J. Comp. Phys. 106, 258-268. [Google Scholar]
- 11.Vaidehi, N., Jain, A. & Goddard, W. A., III (1996) J. Phys Chem. 100, 10508-10517. [Google Scholar]
- 12.Ding, H. Q., Karasawa, N. & Goddard, W. A., III (1992) J. Chem. Phys. 97, 4309-4315. [Google Scholar]
- 13.Lim, K.-T., Brunett, S., Iotov, M., McClurg, R. B., Vaidehi, N., Dasgupta, S., Taylor, S., Goddard, W. A., III (1997) J. Comput. Chem. 18, 501-521. [Google Scholar]
- 14.Vriend, G. (1990) J. Mol. Graphics 8, 52-56. [DOI] [PubMed] [Google Scholar]
- 15.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T. & Stenkamp, R. E. (2000) Science 289, 739-745. [DOI] [PubMed] [Google Scholar]
- 16.Teller, D. C., Okada, T., Behnke, C. A., Palczewski, K. & Stenkamp, R. E. (2001) Biochemistry 40, 7761-7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing, T. A. & Kuntz, I. D. (1997) J. Comput. Chem. 18, 1175-1189. [Google Scholar]
- 18.Zamanakos, G. (2001) Ph.D. thesis (California Institute of Technology, Pasadena, CA).
- 19.Wang, P., Vaidehi, N., Tirrell, D. A. & Goddard, W. A., III (2002) J. Am. Chem. Soc. 124, 14442-14449. [DOI] [PubMed] [Google Scholar]
- 20.Kekenes-Huskey, P. M., Vaidehi, N., Floriano, W. B. & Goddard, W. A., III (2003) J. Phys. Chem. B 107, 11549-11557. [Google Scholar]
- 21.Datta, D., Vaidehi, N. & Goddard, W. A., III (2002) Proc. Natl. Acad. Sci. USA 99, 2636-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Floriano, W. B., Vaidehi, N., Zamanakos, G. & Goddard, W. A., III (2004) J. Med. Chem. 47, 56-71. [DOI] [PubMed] [Google Scholar]
- 23.Liapakis, G., Ballesteros, J. A., Papachristou, S., Chan, W. X., Chen, X. & Javitch, J. A. (2000) J. Biol. Chem. 275, 37779-37788. [DOI] [PubMed] [Google Scholar]
- 24.Shi, L. & Javitch, J. A. (2002) Annu. Rev. Phamacol. Toxicol. 42, 437-467. [DOI] [PubMed] [Google Scholar]
- 25.Ghanouni, P., Steenhuis, J. J., Farrens, D. L. & Kobilka, B. K. (2001) Proc. Natl. Acad. Sci. USA 98, 5997-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballesteros, J. A., Anne, D. J., Liapakis, G., Rasmussen, S. G. F., Shi, L., Gether, U. & Javitch, J. A. (2001) J. Biol. Chem. 276, 29171-29177. [DOI] [PubMed] [Google Scholar]
- 27.Sheikh, S. P., Vilardarga, J.-P., Baranski, T. J., Lichtarge, O., Iiri, T., Meng, E. C., Nissenson, R. A. & Bourne, H. R. (1999) J. Biol. Chem., 274, 17033-17041. [DOI] [PubMed] [Google Scholar]
- 28.Alexiev, U., Rimke, I. & Pohlmann, T. (2003) J. Mol. Biol. 328, 705-719. [DOI] [PubMed] [Google Scholar]
- 29.Seeman, P. (1993) Receptor Tables: Drug Dissociation Constants for Neuroreceptors and Transporters (SZ Research, Toronto), Vol. 2.
- 30.Pritchett, A. M. & Redfield, M. M. (2002) MM Mayo Clinic Proc. 77, 839-846. [DOI] [PubMed] [Google Scholar]
- 31.DeLano, W. L. (2003) The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.