Abstract
The rhizobial FixL/FixJ system, a paradigm of heme-based oxygen sensors, belongs to the ubiquitous two-component signal transduction system. Oxygen-free (deoxy) FixL is autophosphorylated at an invariant histidine residue by using ATP and catalyzes the concomitant phosphoryl transfer to FixJ, but oxygen binding to the FixL heme moiety inactivates the kinase activity. Here we demonstrate that ADP acts as an allosteric effector, reducing the oxygen-binding affinity of the sensor domain in FixL when it is produced from ATP in the kinase reaction. The addition of ADP to a solution of purified wild-type FixL resulted in an ≈4- to 5-fold decrease in oxygen-binding affinity in the presence of FixJ. In contrast, phosphorylation-deficient mutants, in which the well conserved ATP-binding catalytic site of the kinase domain is impaired, showed no such allosteric effect. This discovery casts light on the significance of homodimerization of two-component histidine kinases; ADP, generated in the phosphorylation reaction in one subunit of the homodimer, enhances the histidine kinase activity of the other, analogous to a two-cylinder reciprocating engine by reducing the ligand-binding affinity.
Two-component signal transduction systems composed of sensory histidine kinases and their cognate response regulators are widely distributed in bacteria, fungi, and plants, and are responsible for cellular adaptations to various environmental stimuli or stresses (1). The rhizobial FixL/FixJ two-component system directs the expression of nitrogen fixation genes at low oxygen tensions in plant root nodules (2). FixL is an orthodox sensory histidine kinase consisting of a heme-based sensor domain as an oxygen-binding site and a histidine kinase domain, which are aligned in a single polypeptide (3). Deoxy FixL is autophosphorylated at a conserved histidine residue by using ATP, and the phosphoryl group is then transferred to FixJ; but the kinase activity is down-regulated when molecular oxygen binds to the heme moiety of FixL (4–6). Unphosphorylated FixL preferably exists in the form of a quaternary complex of FixL2/FixJ2 (7) like the EnvZ2/OmpR2 complex (8), but on oxygen-regulated phosphorylation, phospho-FixJ is liberated to bind to the promoter regions of the nifA and fixK genes (9, 10). Hence, FixL/FixJ is referred to as a biological direct oxygen sensor system (11, 12). These in vitro reconstitution studies on oxygen binding by FixL, its oxygen-linked autophosphorylation, the phosphorylation-induced dimerization of FixJ, and its DNA binding clearly demonstrated that the FixL/FixJ system typifies a molecular adaptation to extracellular stimuli.
The paradigm for molecular adaptation to an extracellular stimulus involves ligand recognition in the receptor and coupled chemical reactions on the part of the transmitter. Intracellular chemical signals are, in turn, mediated via several proteins, leading to the regulation of temporal and spatial gene expression. In many cases, therefore, the signaling flows are precisely regulated by positive and negative feedback loops. A positive feedback, for example, can account for the stabilization, amplification, or prolongation of the outputs in the course of a signaling cascade. In fact, the feedback loops have been reported in two-component systems with some transcriptional factors looping downstream of the signaling pathways (13–15). However, the issue of whether ligand recognition or the ligand-binding affinity of sensory histidine kinases is directly modulated by such a feedback loop remains unclear, because few ligand-binding studies for other two-component sensory histidine kinases have been reported because of the experimental difficulties of measuring their binding affinities. To examine this possibility, taking advantage of the fact that the oxy and deoxy forms of FixL can be spectroscopically distinguished as the ligand-bound and -free forms, we determined the oxygen-binding affinity from oxygen equilibrium curves (OECs) under various conditions by using highly purified FixL and FixJ proteins and discovered an allosteric effect by ADP, instead of the action of phosphorylated FixJ, on the oxygen-binding affinity of FixL, which possibly facilitates the deoxygenation-linked phosphorylation reactions.
Materials and Methods
Expression and Purification of Recombinant FixL and FixJ. The expression and purification of the His6-tag FixL and FixJ derived from Sinorhizobium meliloti have been described (6, 16). FixL mutants, N403Q, N403E, N403C, and H285V, were constructed by PCR-based nucleotide changes, AAC to CAG, AAC to GAA, AAC to TGC, and CAC to GTT, respectively. These mutations were confirmed by DNA sequencing.
Visible Spectra and OEC Measurements. Visible spectra and OECs were obtained by using a spectrophotometer (UV2500, Shimadzu, Kyoto) equipped with an oxygenation/deoxygenation cell at 25°C (17). Oxygen concentrations were monitored with a polarographic oxygen meter (DOL-40, Denki Kagaku Kogyo, Tokyo). FixL and FixJ were conventionally diluted in a standard kinase buffer (50 mM Tris·HCl pH 7.8/50 mM KCl/0.2 mM MnCl2/0.2 mM ATP or ADP) (6) supplemented with 10 mM DTT at final concentrations of 5 and 10 μM, respectively. When necessary, FixJ, MnCl2, and adenine nucleotides were added at the final concentrations indicated in the figures and tables. Kd values were obtained from the OECs at 417 nm by hyperbolic curve fitting (Hill's coefficient values of ≈0.9–1.2).
Stopped-Flow Kinetic Measurement. Oxygen association/dissociation rate constants were determined by using a stopped-flow rapid mixing apparatus (Unisoku, Osaka). The generation of oxy FixL was monitored at 420 nm after mixing the deoxy FixL solution in the N2-saturated anoxic buffer and an equal volume of the kinase buffer equilibrated with dissolved oxygen at concentrations of 245, 490, 735, 980, and 1,225 μM. Oxy FixL is thought to be generated by a pseudo-first-order reaction (v = kobs[FixL], kobs = kon[O2] + koff) when [FixL] is much less than [O2]. As the actual time courses for the change in absorbance at 420 nm exhibited biphasic kinetics, similar to the binding of carbon monoxide (18), the observed rate constants (kobs) and amplitudes (ΔA) of these phases were analyzed by fitting into a two-term exponential function of ΔAtotal = ΔAfast exp(-kobs,fastt) + ΔAslow exp(-kobs,slowt). The plots of kobs for the two phases versus [O2] exhibited a linear relationship (data not shown), giving the kon and koff values for each phase from the equation kobs = kon[O2] + koff.
Results
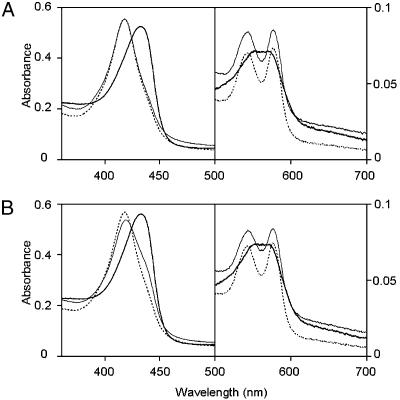
Effect of ADP on the Oxygen-Binding Affinity of FixL. FixL binds molecular oxygen reversibly as monitored by changes in the electronic absorption spectrum on conversion between the oxy (dotted and thin lines) and deoxy forms in the absence of ATP (thick line in Fig. 1A), which is responsible for the switching of the autophosphorylation of FixL and the subsequent phosphoryl transfer to FixJ. If the absorption spectra under air-saturated conditions are different from each other before and after the phosphorylation reactions, this would suggest that a feedback regulation affecting the oxygen-binding affinity is present in the FixL/FixJ system. Indeed, the spectral profile (thin line) of the reoxygenated FixL after the anaerobic kinase reactions with ATP was found to be different from that for the initial oxy FixL at the same oxygen tension (dotted line in Fig. 1B). A decrease in the peak at 417 nm, corresponding to the oxy form, and the appearance of a shoulder at 434 nm that is attributed to the deoxy form (19) under air-saturated conditions indicate a decrease in the oxygen-binding affinity of FixL. This observation suggests that some effectors that reduce the oxygen-binding affinity are generated during the phosphorylation reactions.
Fig. 1.
Changes in the electronic absorption spectrum on conversion between the oxy and deoxy forms of FixL responsible for the switching of the kinase activity of FixL. Spectral changes for FixL without (A) and with (B) ATP in a kinase buffer containing FixJ were measured in air-saturated and nitrogen gas-saturated conditions at 25°C. The concentrations of FixL and FixJ were 5 and 10 μM, respectively. MnCl2 and ATP were added at final concentrations of 0.2 mM. DTT was added at a concentration of 10 mM to prepare ferrous FixL. The concentration of dissolved oxygen in the air-saturated solutions was 170 μM because of the autoxidation of FixL in the presence of DTT. The oxygen concentration at anaerobic conditions was reduced to 6 μM under a stream of nitrogen gas for 20 min, and reoxygenation was performed by flushing with air for 25 min. Broken lines, initial oxy form; solid lines, deoxy form; hair lines, reoxygenated form.
To identify the effector molecules, the Kd values for FixL were determined from the OECs in the presence of various adenine nucleotides (Table 1). It should be noted that the OEC measurement was initiated after the anaerobic treatment of each sample under a stream of nitrogen gas for 20–30 min, permitting the kinase reactions to proceed. FixL exhibited Kd values of ≈50 μM in the presence and absence of FixJ. These values are in a reasonable range to sense the physiological changes in oxygen tension between the aerobic (21% oxygen content) and microaerobic (<1% oxygen content) growth conditions, and are consistent with the previously reported value (19). The addition of ATP reduced the oxygen-binding affinity by ≈3-fold in the presence of FixJ, consistent with the data shown in Fig. 1B. Nonhydrolyzable ATP analogues, β,γ-methyleneadenosine 5′-triphosphate (AMP-PCP) and β,γ-imidoadenosine 5′-triphosphate (AMP-PNP), had no significant effect (Table 1), suggesting that the binding of ATP is not directly responsible for the reduction in the affinity. Surprisingly, the addition of ADP resulted in a 4-fold reduction in the oxygen-binding affinity (Fig. 2A and Table 1). Because the removal of ATP/ADP from the postphosphorylation solution by size exclusion column chromatography abolished the decrease in oxygen-binding affinity of the FixL/phospho-FixJ solution, the observed changes are not due to the production of phospho-FixJ or chemical modifications of FixL (data not shown).
Table 1. Effect of adenine nucleotides on the oxygen-binding affinity of FixL.
|
Kd, μM
|
||
|---|---|---|
| FixL | –FixJ | +FixJ |
| None | 50 ± 10 | 56 ± 8 |
| +ATP | 47 ± 6 | 176 ± 7 |
| +AMP-PCP | nt | 55 ± 1 |
| +AMP-PNP | nt | 46 ± 9 |
| +ADP | 118 ± 30 | 218 ± 27 |
Kd values of FixL were determined from the OECs at 25°C. The concentrations of FixL and FixJ in the kinase buffer were 5 and 10 μM, respectively. MnCl2 and adenine nucleotides were added at the final concentrations of 0.2 mM. nt, not tested.
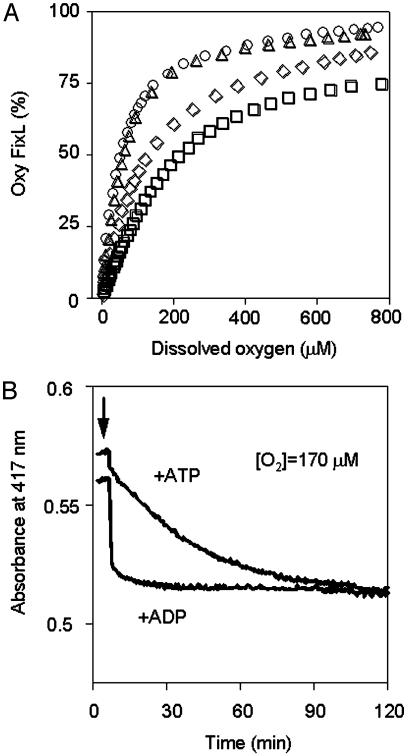
Fig. 2.
Allosteric effect of ADP on the oxygen-binding affinity of FixL. (A) OECs of FixL. Circles, FixL; triangles, FixL + FixJ; diamonds, FixL + ADP; squares, FixL + FixJ + ADP. The OEC measurement was initiated after anaerobic treatment of each sample under a stream of nitrogen gas for 20–30 min, and the absorbance changes at 417 nm corresponding to the absorption peak of oxy FixL were recorded while the oxygen tension was raised by adding air and, later, oxygen. (B) Time courses for the change in absorbance of FixL induced by ATP and ADP in air-saturated conditions. Absorbance changes of the Soret peak for oxy FixL at 417 nm were recorded. The protein concentrations and buffer compositions were the same as in Fig. 1. The arrows indicate the addition of ATP or ADP (final concentration, 0.2 mM). Dissolved oxygen was maintained at 170 μM under a stream of air by monitoring with an oxygen electrode.
Fig. 2B confirms that the binding of ADP directly lowers the oxygen-binding affinity. Under aerobic conditions, the oxygen-binding affinity immediately drops as a result of added ADP. In contrast, the effect of ATP is slow because ADP gradually accumulates via the phosphorylation reactions of the residual deoxy form of FixL (15–20% of total FixL) in the air-saturated solution in the presence of FixJ. It should be noted that neither GTP nor GDP exerted any detectable allosteric effect (data not shown).
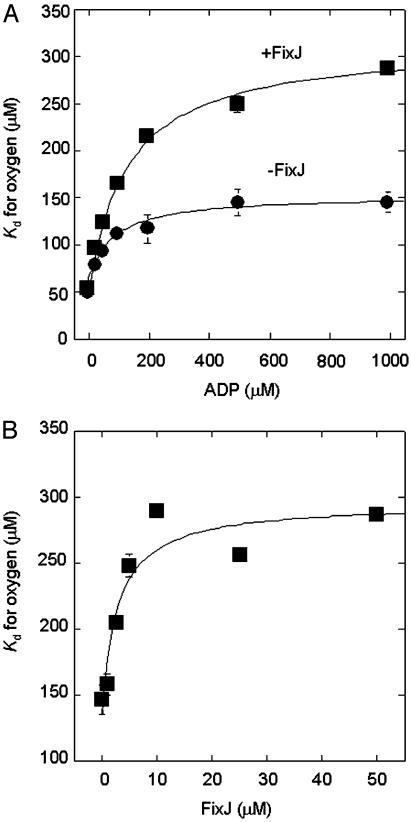
The maximal reduction in affinity requires rather high concentrations of ADP and is strengthened by FixJ (Fig. 3A). Because FixJ alone does not affect the affinity (Table 1), FixJ binding may modify the configuration of the bound ADP or the local protein structure of the ADP-binding site. The addition of FixJ is effective at low concentrations, and its apparent Kd for affecting the oxygen-binding affinity was estimated to be 3 μM (Fig. 3B). Because this value coincides with the Km of FixJ phosphorylation (20), the FixJ-binding sites in FixL appear to be shared with each other. Finally, based on these findings, we conclude that ADP, but not phospho-FixJ or ATP, acts as an allosteric effector reducing the oxygen-binding affinity in the FixL/FixJ system.
Fig. 3.
ADP and FixJ dependence of the oxygen-binding affinity of FixL. (A) ADP-dependent Kd change. The concentrations of FixL, FixJ, and MnCl2 were 5 μM, 10 μM, and 1 mM, respectively. (B) FixJ-dependent Kd change. The concentrations of FixL, ADP, and MnCl2 were 5 μM, 1 mM, and 1 mM, respectively.
Effect of ADP on the Oxygen Association/Dissociation Rate Constants of FixL. Kinetic analyses using a stopped-flow apparatus indicate biphasic oxygen-binding profiles, where a fraction of the fast component is predominant, similar to the carbon monoxide binding previously reported (18). The addition of ADP significantly alters the koff values of the fast phase component (Table 2), suggesting that the ADP effect might affect the protein portion in the vicinity of the heme. However, the perturbation of the heme environment appears to be subtle, in that the Fe-His stretching band of deoxy FixL derived from the heme-H194 bond, and the Fe-O2 stretching band of oxy FixL in the presence of FixJ were changed only slightly by the addition of ADP, as evidenced by Raman spectroscopic resonance measurements (data not shown).
Table 2. Oxygen-binding parameters for FixL in the presence of FixJ at 25°C.
| Fast phase
|
Slow phase
|
|||
|---|---|---|---|---|
| ADP, μM | kon, M-1·s-1 | koff, s-1 | kon, M-1·s-1 | koff, s-1 |
| 0 | (4.3 ± 0.2) × 104 | 13 ± 1 | (5.2 ± 0.6) × 103 | 2.3 ± 0.3 |
| 200 | (4.0 ± 0.2) × 104 | 25 ± 1 | (3.0 ± 0.5) × 103 | 2.5 ± 0.2 |
| 1,000 | (4.1 ± 0.3) × 104 | 40 ± 1 | (2.3 ± 0.9) × 103 | 5.0 ± 0.4 |
The concentrations of FixL and FixJ in the kinase buffer were 5 and 10 μM, respectively. MnCl2 was added at a final concentration of 1 mM.
ADP Binds to the ATP-Binding Catalytic Site in the Kinase Domain. The ATP-binding catalytic domain, consisting of the N and G boxes, is well conserved in the kinase domain of sensory histidine kinases (1, 21). The invariant asparagine residues in the N boxes, N347 of osmosensor histidine kinase EnvZ, N409 of chemotactic sensor kinase CheA, and N389 of phosphate-regulating kinase PhoQ, which are equivalent to N403 of FixL, are required for ATP binding as well as for kinase activity (22–25). Because no other binding motif for adenine nucleotides is present, it would appear that ADP binds to the catalytic site of the kinase domain of FixL.
To verify the ADP-binding site, the oxygen-binding affinities of the N403 mutants of FixL, which have no histidine kinase activity (H. Nakamura, unpublished observation), were determined in the presence and absence of ADP. The observed Kd values indicate that the allosteric effect by ADP was completely absent in the N403Q, N403E, and N403C mutants (Table 3). In contrast, H285V, another autophosphorylation-deficient mutant caused by the disruption of the conserved phosphorylation site in the H box (refs. 5 and 22; H. Nakamura, unpublished observation), retained the allosteric effect (Table 3). In this mutant, the allosteric effect by ADP, i.e., the appearance of the shoulder at 430 nm in the air-saturated conditions like Fig. 1B, was eliminated by further addition of an equal amount of ATP, suggesting that ATP competitively binds to the catalytic site with a higher affinity (data not shown). These results unambiguously demonstrate that N403 also participates in ADP binding. The small difference in oxygen-binding affinity of the N403C and H285V mutants compared with that of wild type may be due to a minor mutational effect.
Table 3. Oxygen-binding affinities of FixL and its mutants.
|
Kd, μM
|
||
|---|---|---|
| FixL | –ADP | +ADP |
| Wild type | 56 ± 8 | 218 ± 27 |
| N403Q | 42 ± 1 | 40 ± 7 |
| N403E | 47 ± 2 | 45 ± 1 |
| N403C | 79 ± 2 | 73 ± 2 |
| H285V | 69 ± 1 | 173 ± 21 |
Kd values of FixLs were determined in the presence of FixJ from the OECs at 25°C. The concentrations of FixL and FixJ in the kinase buffer were 5 and 10 μM, respectively. MnCl2 and ADP were added at final concentrations of 0.2 mM.
Discussion
The observation of a decrease in the oxygen-binding affinity of FixL after phosphorylation prompted us to investigate the existence of a positive feedback loop, because a decrease in binding affinity is accompanied by an increase in the fraction of the kinase-active deoxy form. At the initial stage of the present study, phospho-FixJ had been expected to be the most likely candidate for the allosteric effector. However, ADP was identified as the responsible molecule when it was observed to bind to the catalytic site of the kinase domain. In addition, the FixJ-binding site for strengthening the ADP allosteric effect would likely be identical to that for the phosphoryl transfer reaction. Taking these results into account, the significance and mechanisms of the ADP allosteric effect on the oxygen-regulated kinase reaction are discussed.
The nature of the action of ADP is apparently that of positive feedback in which the overall kinase activity is up-regulated with respect to reducing the oxygen-binding affinity. Because ADP occupies the catalytic site of the kinase domain when the allosteric effect is exerted, however, kinase activity would be expected to be inhibited. This paradox is understandable, considering that FixL forms a homodimer (3) that contains two nucleotide-binding sites per dimer complex. Indeed, homodimer formation is essential for the molecular function of sensory histidine kinases in that the catalytic domain of one polypeptide trans-phosphorylates the histidine residue of the partner molecule (26–29). In the case of homodimeric FixL, the in situ ADP generated by the phosphorylation reaction appears to be responsible for the allosteric effect. It is also evident that, to ensure a turnover of phosphorylation reactions, the generated ADP should be simultaneously liberated from FixL when phospho-FixJ is dissociated to bind to the fixK and nifA promoters, similar to CheY (30) and OmpR (31).
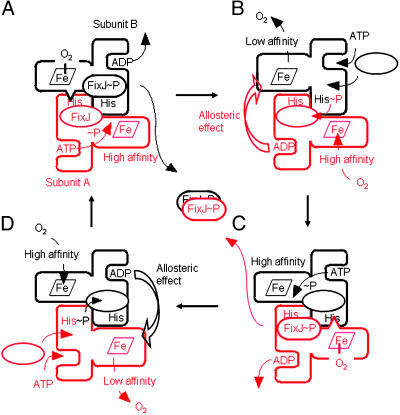
Given the extent of our current understanding of the ADP allosteric effect, a reciprocating model of the in situ ADP-dependent acceleration of the kinase reaction is proposed (Fig. 4). Although several reciprocating models have been proposed for other homodimeric enzymes (32–35), the present model is unique from the standpoint that ADP, a product of protein phosphorylation reactions, enhances deoxygenation-linked autophosphorylation alternately in each subunit (Fig. 4, B to C and D to A), leading to the enhanced production of phospho-FixJ. It should be noted that the gross autophosphorylation activity is regulated by oxygen binding at the high affinity hemes (Fig. 4, subunit A in A and subunit B in C), influencing the content of the active FixL fraction undergoing the turnover cycle.
Fig. 4.
Two-cylinder reciprocating engine model of FixL/FixJ phosphorylation reactions. Oxygen-free FixL exerts an autophosphorylation reaction in a turnover cycle. FixL, like other histidine kinases, is thought to be transautophosphorylated in the homodimer form. Although it is currently unknown whether the oxygen-bound sensor domain in one subunit down-regulates the histidine-phosphorylation of the same polypeptide (cis-acting repression) or that of the partner (trans-acting repression), the latter transacting mode is assumed in the present model, as shown by wedges in A and C. It should be noted that, in the trans-acting mode, the oxygen-free sensor domain would unmask the histidine in the other subunit or promote an ATP-phosphoryl transfer reaction in the same subunit. To follow the reactions, one cycle in subunit A is colored red. (A) ATP is catalyzed at the nucleotide-binding site of the kinase domain because of deoxygenation of the sensor domain in subunit A, and the phosphoryl group is transferred to the histidine in subunit B. (B) ADP produced at the binding site reduces the oxygen-binding affinity of subunit B in a trans-acting manner (larger sensor domain of subunit B). The phosphoryl group of subunit B is transferred to FixJ. (C) Phospho-FixJ and ADP are then released from subunit A. In turn, the ATP-phosphoryl transfer reaction in subunit B is enhanced because of the allosteric effect in B. (D) FixJ and ATP are reloaded into subunit A. The oxygen-binding affinity of subunit A is decreased by the ADP that is produced in subunit B, leading to the ATP-phosphoryl transfer reaction in A. On overall phosphoryl transferring reactions, two phospho-FixJ are liberated from FixL to form a regulatory active dimer (center). Oxygen binding at the high affinity hemes (subunit A in A and subunit B in C) determines the content of kinase-active FixL in the turnover cycle, and the allosteric effect facilitates the velocity of the transitions from B to C and from D to A, resulting in the amplification of the phospho-FixJ production. If the oxygen-bound sensor domain inhibits the histidine-phosphorylation of the same subunit (cis-acting repression), the model implies that the ADP allosteric effect would be exerted in the sensor domain of the same subunit (cis-acting manner).
As mentioned above, homodimerization and trans-autophosphorylation across individual subunits are essential for the two-component histidine kinases. The sensor domains also consist of two subunits, even though they exist separately from the kinase polypeptides. It is interesting to note that homodimeric chemoreceptors of Tar and Tsr exhibit strong cooperativities of ligand binding (36, 37), whereas the CheA histidine kinase dimer solely isolated catalyzes noncooperative autophosphorylation (28). Thus, the regulatory linkage between cooperative ligand binding and noncooperative autophosphorylation should be studied by the reconstitution of the entire chemotactic complexes of Tar or Tsr/CheW/CheA. In contrast, each FixL subunit noncooperatively binds oxygen unlike chemoreceptors (ref. 19 and this study), but the generation of low affinity FixL in the turnover cycle seems to exert efficient autophosphorylation and phosphoryl transfer to FixJ, although it is difficult to experimentally evaluate the ADP effect on FixJ phosphorylation. Therefore, the present study proposes an alternate regulatory mode of two-component signal transduction systems that accounts for the amplification of the output signal by homodimeric histidine kinases instead of cooperative ligand binding.
A large number of two-component signal transduction systems have emerged as the result of sequence determinations of the whole genomes of prokaryotes and eukaryotes. In particular, the orthodox sensory histidine kinases contain both the sensor and the kinase domain in single polypeptides (1), and they may share a common regulatory mechanism with respect to ligand sensing and phosphorylation signaling. The present study, which could be completed because of the experimental advantage that the oxygen-binding affinity of FixL can be spectroscopically determined, demonstrates that ADP is an allosteric effector for the modulation of the ligand-binding affinity of sensory histidine kinases coupled with phosphorylation reactions, and provides insight into the physiological significance of homodimer formation and trans-phosphorylation reactions. It should be elucidated whether the change in ligand-binding affinity of the sensory histidine kinase discovered here is true for other two-component systems.
Acknowledgments
We thank Profs. Takehiko Shibata and Takeshi Nishino for their encouragement throughout the work. This work was supported by the Bioarchitect Research Program, the Body (Bio-Organism) Defense Network Research Program of RIKEN, and Grant-in-Aid for Scientific Research of Priority Areas on Metal Sensors 12147210 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: OEC, oxygen equilibrium curve.
References
- 1.Stock, A. M., Robinson, V. L. & Goudreau, P. N. (2000) Annu. Rev. Biochem. 69, 183-215. [DOI] [PubMed] [Google Scholar]
- 2.David, M., Daveran, M.-L., Batut, J., Dedieu, A., Domergue, O., Ghai, J., Hertig, C., Boistard, P. & Kahn, D. (1988) Cell 54, 671-683. [DOI] [PubMed] [Google Scholar]
- 3.Gilles-Gonzalez, M. A., Ditta, G. S. & Helinski, D. R. (1991) Nature 350, 170-172. [DOI] [PubMed] [Google Scholar]
- 4.Gilles-Gonzalez, M. A. & Gonzalez, G. (1993) J. Biol. Chem. 268, 16293-16297. [PubMed] [Google Scholar]
- 5.Monson, E. K., Ditta, G. S. & Helinski, D. R. (1995) J. Biol. Chem. 270, 5243-5250. [DOI] [PubMed] [Google Scholar]
- 6.Saito, K., Ito, E., Hosono, K., Nakamura, K., Imai, K., Iizuka, T., Shiro Y. & Nakamura, H. (2003) Mol. Microbiol. 48, 373-383. [DOI] [PubMed] [Google Scholar]
- 7.Miyatake, H., Kanai, M., Adachi, S., Nakamura, H., Tamura, K., Tanida, H., Tsuchiya, T., Iizuka, T. & Shiro, Y. (1999) Acta Crystallogr. D 55, 1215-1218. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida, T., Qin, L. & Inouye, M. (2002) Mol. Microbiol. 46, 1273-1282. [DOI] [PubMed] [Google Scholar]
- 9.Da Re, S., Bertagnoli, S., Fourment, J., Reyrat, J.-M. & Kahn, D. (1994) Nucleic Acids Res. 22, 1555-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galinier, A., Garnerone, A.-M., Reyrat, J.-M., Kahn, D., Batut, J. & Boistard, P. (1994) J. Biol. Chem. 269, 23784-23789. [PubMed] [Google Scholar]
- 11.Delgado-Nixon, V. M., Gonzalez, G. & Gilles-Gonzalez, M.-A. (2000) Biochemistry 39, 2685-2691. [DOI] [PubMed] [Google Scholar]
- 12.Akimoto, S., Tanaka, A., Nakamura, K., Shiro, Y. & Nakamura, H. (2003) Biochem. Biophys. Res. Commun. 304, 136-142. [DOI] [PubMed] [Google Scholar]
- 13.Raivio, T. L., Popkin, D. L. & Silhavy, T. J. (1999) J. Bacteriol. 181, 5263-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prigent-Combaret, C., Brombacher, E., Vidal, O., Ambert, A., Lejeune, P., Landini, P. & Dorel, C. (2001) J. Bacteriol. 183, 7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato, A., Latifi, T. & Groisman, E. A. (2003) Proc. Natl. Acad. Sci. USA 100, 4706-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura, H., Saito, K., Ito, E., Tamura, K., Tsuchiya, T., Nishigaki, K., Shiro, Y. & Iizuka, T. (1998) Biochem. Biophys. Res. Commun. 247, 427-431. [DOI] [PubMed] [Google Scholar]
- 17.Imai, K. (1981) Methods Enzymol. 76, 438-449. [DOI] [PubMed] [Google Scholar]
- 18.Rodgers, K. R., Tang, L., Lukat-Rodgers, G. S. & Wengenack, N. L. (2001) Biochemistry 40, 12932-12942. [DOI] [PubMed] [Google Scholar]
- 19.Gilles-Gonzalez, M. A., Gonzalez, G., Perutz, M. F., Kiger, L., Marden, M. C. & Poyart, C. (1994) Biochemistry 33, 8067-8073. [DOI] [PubMed] [Google Scholar]
- 20.Tuckerman, J. R., Gonzalez, G., Dioum, E. M. & Gilles-Gonzalez, M.-A. (2002) Biochemistry 41, 6170-6177. [DOI] [PubMed] [Google Scholar]
- 21.Hoch, J. A. & Silhavy, T. J. (1995) Two-Component Signal Transduction (Am. Soc. Microbiol., Washington, DC).
- 22.Yang, Y. & Inouye, M. (1993) J. Mol. Biol. 231, 335-342. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka, T., Saha, S. K., Tomomori, C., Ishima, R., Liu, D., Tong, K. I., Park, H., Dutta, R., Qin, L., Swindells, M. B., et al. (1998) Nature 396, 88-92. [DOI] [PubMed] [Google Scholar]
- 24.Bilwes, A. M., Quezada, C. M., Croal, L. R., Crane, B. R. & Simon, M. I. (2001) Nat. Struct. Biol. 8, 353-360. [DOI] [PubMed] [Google Scholar]
- 25.Marina, A., Mott, C., Auyzenberg, A., Hendrickson, W. A. & Waldburger, C. D. (2001) J. Biol. Chem. 276, 41182-41190. [DOI] [PubMed] [Google Scholar]
- 26.Yang, Y. & Inouye, M. (1991) Proc. Natl. Acad. Sci. USA 88, 11057-11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ninfa, E. G., Atkinson, M. R., Kamberov, E. S. & Ninfa, A. J. (1993) J. Bacteriol. 175, 7024-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surette, M. G., Levit, M., Liu, Y., Lukat, G., Ninfa, E. G., Ninfa, A. & Stock, J. B. (1996) J. Biol. Chem. 271, 939-945. [DOI] [PubMed] [Google Scholar]
- 29.Cai, S.-J. & Inouye, M. (2003) J. Mol. Biol. 329, 495-503. [DOI] [PubMed] [Google Scholar]
- 30.Schuster, S. C., Swanson, R. V., Alex, L. A., Bourret, R. B. & Simon, M. I. (1993) Nature 365, 343-347. [DOI] [PubMed] [Google Scholar]
- 31.Mattison, K. & Kenney, L. J. (2002) J. Biol. Chem. 277, 11143-11148. [DOI] [PubMed] [Google Scholar]
- 32.Harada, K. &Wolfe, R. G. (1968) J. Biol. Chem. 243, 4131-4137. [PubMed] [Google Scholar]
- 33.Senior, A. E., Al-Shawi, M. K. & Urbatsch, I. L. (1995) FEBS Lett. 377, 285-289. [DOI] [PubMed] [Google Scholar]
- 34.Kovina, M. V. & Kochetov, G. A. (1998) FEBS Lett. 440, 81-84. [DOI] [PubMed] [Google Scholar]
- 35.Jones, P. M. & George, A. M. (2002) Proc. Natl. Acad. Sci. USA 99, 12639-12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biemann, H.-P. & Koshland, D. E., Jr. (1994) Biochemistry 33, 629-634. [DOI] [PubMed] [Google Scholar]
- 37.Lin, L.-N., Li, J., Brandts, J. F. & Weis, R. M. (1994) Biochemistry 33, 6564-6570. [DOI] [PubMed] [Google Scholar]