Abstract
Yeast tRNA ligase (Trl1) is an essential enzyme that converts cleaved tRNA half-molecules into spliced tRNAs containing a 2′-PO4, 3′-5′ phosphodiester at the splice junction. Trl1 also catalyzes splicing of HAC1 mRNA during the unfolded protein response. Trl1 performs three reactions: the 2′,3′-cyclic phosphate of the proximal RNA fragment is hydrolyzed to a 3′-OH, 2′-PO4 by a cyclic phosphodiesterase; the 5′-OH of the distal RNA fragment is phosphorylated by a GTP-dependent polynucleotide kinase; and the 3′-OH, 2′-PO4, and 5′-PO4 ends are then sealed by an ATP-dependent RNA ligase. The removal of the 2′-PO4 at the splice junction is catalyzed by the essential enzyme Tpt1, which transfers the RNA 2′-PO4 to NAD+ to form ADP-ribose 1′′-2′′-cyclic phosphate. Here, we show that the bacteriophage T4 enzymes RNA ligase 1 and polynucleotide kinase/phosphatase can fulfill the tRNA and HAC1 mRNA splicing functions of yeast Trl1 in vivo and bypass the requirement for Tpt1. These results attest to the portability of RNA-repair systems, notwithstanding the significant differences in the specificities, mechanisms, and reaction intermediates of the individual yeast and T4 enzymes responsible for the RNA healing and sealing steps. We surmise that Tpt1 and its unique metabolite ADP-ribose 1′′-2′′-cyclic phosphate do not play essential roles in yeast independent of the tRNA-splicing reaction. Our finding that one-sixth of spliced HAC1 mRNAs in yeast cells containing the T4 RNA-repair system suffered deletion of a single nucleotide at the 3′ end of the splice-donor site suggests a model whereby the yeast RNA-repair system evolved a requirement for the 2′-PO4 for RNA ligation to suppress inappropriate RNA recombination.
RNA ligases participate in repair, splicing, and editing pathways that either reseal broken RNAs or alter their primary structure. Bacteriophage T4 RNA ligase 1 (Rnl1) is the founding member of this class of enzymes (1), which includes yeast tRNA ligase and trypanosome RNA-editing ligases (2, 3). The function of Rnl1 in vivo is to repair a break in the anticodon loop of Escherichia coli tRNALys triggered by phage activation of a host-encoded anticodon nuclease PrrC (4, 5). Depletion of tRNALys by PrrC blocks phage protein synthesis and arrests the infection before it can spread. However, the bacteriophage T4 enzymes polynucleotide kinase/phosphatase (Pnkp) and Rnl1 repair the broken tRNAs and thereby thwart the host defense mechanism.
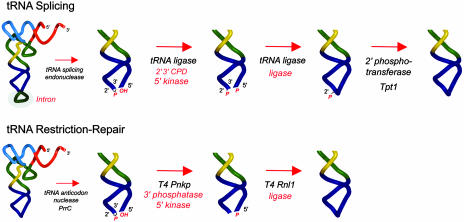
The enzymatic steps in bacteriophage tRNA restriction/repair are broadly similar to those of yeast tRNA splicing, the process whereby introns are removed seamlessly from the tRNA anticodon loop (2). The incision steps in both cases result in the formation of 2′,3′-cyclic phosphate and 5′-OH termini. tRNA splicing requires two breaks in the backbone of the pre-tRNA to excise the intron, whereas tRNA restriction involves a single break in the mature tRNA (Fig. 1). The 2′,3′-cyclic phosphate and 5′-OH ends are not substrates for T4 RNA ligase or yeast tRNA ligase, which seal only 3′-OH and 5′-PO4 RNA termini. Thus, the broken ends must be “healed” before they can be sealed. Healing entails two steps: (i) hydrolysis of the 2′,3′-cyclic phosphate to form a 3′-OH and (ii) phosphorylation of the 5′-OH to form a 5′-PO4 end. The end-healing and strand-sealing steps of the phage-encoded tRNA repair pathway are performed by two separate enzymes (Pnkp and Rnl1), whereas a single enzyme Trl1 performs these steps in yeast tRNA splicing.
Fig. 1.
Yeast tRNA-splicing and phage tRNA-restriction-repair pathways. In tRNA splicing, the pre-tRNA is cleaved at the exon–intron junctions in the anticodon loop by a tRNA-splicing endonuclease, which leaves a 2′,3′-cyclic phosphate end on the proximal half-molecule and a 5′-OH on the distal half-molecule. The ends are then remodeled and sealed by tRNA ligase (Trl1), a multifunctional protein with 2′,3′-CPD, 5′ kinase, and ligase activities. The residual 2′-PO4 at the splice junction is then removed by the NAD+-dependent 2′ phosphotransferase Tpt1. In tRNA-restriction repair, the mature tRNALys is cleaved in the anticodon loop by PrrC, which leaves 2′,3′-cyclic phosphate and 5′-OH ends. The ends are then healed by T4 Pnkp, which removes the phosphate at the 3′ end and phosphorylates the 5′ terminus. T4 Rnl1 then seals the 3′-OH and 5′-PO4 termini to form a standard 3′-5′ phosphodiester linkage and thus restore tRNALys function in protein synthesis.
T4 Pnkp remodels both ends of the broken tRNA as follows: (i) it converts the 2′,3′-cyclic phosphate to a 3′-PO4 and then hydrolyzes the 3′-PO4 to a 3′-OH, and (ii) it transfers the γ-phosphate from ATP to the 5′-OH end to form a 5′-PO4 (6–9). T4 Rnl1 then joins the 3′-OH and 5′-PO4 RNA ends by means of three nucleotidyl transfer steps (10). Step 1 is the reaction of RNA ligase with ATP to form a covalent ligase-(lysyl-N)-AMP intermediate (11). In step 2, the AMP is transferred to a 5′-PO4 RNA end to form an RNA-adenylate intermediate, AppRNA (12). In step 3, attack by an RNA 3′-OH on RNA-adenylate releases AMP and forms a standard 3′-5′ phosphodiester linkage.
The healing phase of yeast tRNA splicing involves a different ensemble of intermediates and an additional repair reaction not seen in the T4 reaction pathway. Yeast Trl1 performs three reactions: (i) the 2′,3′-cyclic phosphate of the proximal tRNA half-molecule is hydrolyzed to a 3′-OH, 2′-PO4 by a cyclic phosphodiesterase (CPD); (ii) the 5′-OH of the distal half-molecule is phosphorylated by a GTP-dependent polynucleotide kinase; and (iii) the 3′-OH, 2′-PO4 and 5′-PO4 ends are sealed by an ATP-dependent RNA ligase to form a spliced tRNA containing an unconventional 2′-PO4, 3′-5′ phosphodiester at the splice junction (Fig. 1 and refs. 13–16). The final step in yeast tRNA splicing is the removal of the 2′-PO4 at the splice junction by the 2′ phosphotransferase Tpt1, which catalyzes the transfer of the tRNA 2′-PO4 to NAD+ to form ADP-ribose 1′′-2′′-cyclic phosphate (ref. 17 and Fig. 1). Trl1 and Tpt1 are both essential for yeast viability (18, 19). In the case of Trl1, each of the component catalytic modules is essential for cell growth and single missense mutations in the active sites of the CPD, kinase, and ligase domains are lethal in vivo (20). Phizicky and coworkers (21) have shown that the failure of Tpt1 to remove the 2′-PO4 from the splice junction precludes subsequent tRNA modification reactions in the anticodon loop.
Here, we ask whether the differences in the phage tRNA-restriction and yeast tRNA-splicing pathways can instruct us regarding the evolution of RNA-repair mechanisms. An intriguing question is why tRNA splicing in yeast is biochemically more complex than tRNA restriction/repair. Is the formation of an unconventional 2′-PO4, 3′-5′ phosphodiester by Trl1 somehow essential for tRNA splicing in vivo? What is the rationale for exploiting NAD+ as a cofactor for removal of the 2′ phosphate from the splice junction; i.e., does the formation of ADP-ribose 1′′-2′′-cyclic phosphate play a critical role per se in cell growth? In addition to its role in tRNA splicing, Trl1 is responsible for nonspliceosomal splicing of mRNA during the unfolded protein response pathway triggered by stress on the endoplasmic reticulum (22, 23). Is the complexity of the yeast tRNA splicing pathway somehow related to its dual role in mRNA splicing?
To address these issues, we have tested the “portability” of the divergent RNA repair pathways; i.e., whether the healing or sealing components of the tRNA splicing system can be replaced in vivo by their bacteriophage analogs. We find that whereas the yeast ligase domain and T4 Rnl1 are functionally interchangeable as isolated sealing modules, the respective healing enzymes are not. T4 Pnkp could substitute for the yeast kinase and CPD modules only in tandem with T4 Rnl1, which implies that the yeast RNA ligase requires a 2′-PO4 terminus for its strand-sealing activity in vivo. We find that Tpt1 is completely dispensable in yeast cells in which Trl1 was replaced by Rnl1 and Pnkp. This result shows that: (i) the sole essential function of Tpt1 is to dephosphorylate the 2′-PO4 at splice junctions formed by Trl1 and (ii) the formation of ADP-ribose 1′′-2′′-cyclic phosphate by Tpt1 is not essential for growth. Moreover, we show that splicing of HAC1 mRNA during the unfolded protein response occurs in trl1Δ tpt1Δ cells that express Rnl1 and Pnkp, albeit with decreased “fidelity,” insofar as one-sixth of the population of spliced HAC1 mRNAs suffered deletion of a single nucleotide at the 3′ end of the splice-donor site. We discuss a model for the evolution of the complexity of the yeast RNA-repair system as a means to suppress inappropriate RNA recombination.
Materials and Methods
Yeast Expression Plasmids. CEN plasmids for expression of either native Trl1, the N-terminal ligase domain Trl1(1–388), and the C-terminal kinase-CPD domain Trl1(389–827) in yeast under the transcriptional control of the natural TRL1 promoter were described (20). A CEN TRP1 plasmid expressing T4 Rnl1 and T4 Pnkp was constructed as follows. First, NdeI–BamHI fragments containing the T4 Rnl1 and T4 Pnkp ORFs were excised from pET-Rnl1 and pET-Pnkp plasmids (24, 25) and inserted singly between the NdeI and BamHI sites of yeast shuttle vector pYX1 (CEN TRP1) to yield pYX-RNL1 and pYX-PNKP. In these vectors, the T4 genes are under the transcriptional control of the Saccharomyces cerevisiae TPI1 promoter. (The T4 coding sequences in the pYX vectors are flanked at the 5′ end by an in-frame leader encoding a hexahistidine tag; an untranslated BamHI–HindIII fragment from pET16b is present 3′ of the T4 gene.) Then, an NgoMIV fragment containing the TPI1-PNKP expression cassette was excised from pYX-PNKP and inserted into an XmaI site in pYX-RNL1 to generate the dual-expression plasmid pYX-RNL1/PNKP.
Complementation of trl1Δ by Plasmid Shuffle. Yeast trl1Δstrain YRS1 (MATα ura3–1 ade2–1 trp1–1 his3–11,15 leu2–3,11 can1–100 trl1::kanMX p360-TRL1) was transformed with CEN plasmids encoding either the Trl1 ligase and kinase-CPD domains, or T4 Rnl1 and T4 Pnkp. Transformants were selected on appropriate drop-out media. Two individual colonies were patched on drop-out agar medium and cells from each patch were then streaked on agar containing 0.75 mg/ml 5-fluoroorotic acid (5-FOA). The plates were incubated at 18, 25, 30 and 37°C. Combinations of Trl1 domains or T4 Rnl1 and Pnkp that did not allow formation of 5-FOA-resistant colonies after 7 days at any of the temperatures tested were scored as lethal (- growth). Other combinations supported 5-FOA-resistant colony formation within 4 days. Two individual colonies from each streak were picked from the 5-FOA plate, patched on yeast extract/peptone/dextrose (YPD) medium and then tested for growth on YPD agar at 25, 30, and 37°C. Strains that formed colonies of size indistinguishable from the wild-type TRL1 strain at all temperatures were scored as +++.
Deletion of TPT1 in S. cerevisiae. A 1.3-kbp DNA fragment containing the TPT1 ORF plus 505 base pairs of upstream (5′) and 105 base pairs of downstream (3′) chromosomal DNA was amplified by PCR from total yeast DNA using primers that introduced an EcoRI site at the 5′ end and a BamHI site at the 3′ end. The PCR product was digested with EcoRI and BamHI and then inserted into yeast expression vectors pSE360 (CEN URA3) and pSE358 (CEN TRP1) to yield p360-TPT1 and p358-TPT1, respectively. The TPT1 ORF was sequenced completely in both plasmids to confirm that no coding changes were introduced during amplification and cloning. A haploid derivative of yeast strain W303 (MATa ura3–1 ade2–1 trp1–1 his3–11,15 leu2–3,11–2 can1–100) was transformed with p360-TPT1. The chromosomal TPT1 gene was disrupted in this strain by transformation with a linear LEU2 cassette flanked by 1.0- and 0.9-kbp tracts of yeast genomic DNA corresponding to the sequences 5′ and 3′ of the chromosomal TPT1 gene, respectively. The tpt1Δ strain YBS501, in which the TPT1 ORF was deleted and replaced by LEU2, depended for viability on the CEN URA3 TPT1 plasmid. Whereas YBS501 was unable to form colonies on agar medium containing 5-FOA, a drug that selects against the URA3 gene, transformation of YBS501 with a CEN TRP1 TPT1 plasmid permitted growth on medium containing 5-FOA.
Construction of a trl1Δ tpt1Δ Strain Expressing T4 RNA-Repair Enzymes. TPT1 was disrupted by using the LEU2 cassette in the trl1Δ diploid strain YRS2 (MATa/MATα, ura3–1/ura3–1 ade2–1/ade2–1 trp1–1/trp1–1 his3–11,15/his3–11,15 leu2–3,11–2/leu2–3,11–2 can1–100/can1–100 trl1::kanMX trl1::kanMX) containing plasmids p360-TRL1 (CEN URA3 TRL1) and pYX-RNL1/PNKP (CEN TRP1 RNL1 PNKP). Leu+ Trp+ Ura+ diploids were screened by Southern blotting to confirm correct integration of the LEU2 cassette at one of the two chromosomal TPT1 loci. The TPT1 tpt1::LEU2 trl1::kanMX trl1::kanMXdiploid was sporulated and tetrads were dissected to obtain haploid progeny. All Leu+ haploids lacking Tpt1 were also Trp+, signifying that they contained the T4 RNA-repair enzyme system. Moreover, the LEU2 TRP1 haploids grew on medium containing 5-FOA. All Ura+ Trp- haploids containing the plasmid-borne yeast tRNA ligase gene (and lacking the T4 enzymes) were also Leu-, meaning they contained the intact chromosomal copy of TPT1. These results show that Tpt1 is essential when tRNA splicing is catalyzed by yeast Trl1, but dispensable when the tRNA splicing is catalyzed by T4 Rnl1 and Pnkp.
HAC1 mRNA Splicing in the Unfolded Protein Response. Cultures (200 ml) of yeast trl1Δ p358-TRL1, trl1Δ pYX-RNL1/PNKP, and Δtrl1 Δtpt1 pYX-RNL1/PNKP strains were grown at 30°C in YPD broth until the OD600 reached 0.6. The cultures were split into two 100-ml aliquots: one of which was adjusted to 1.5 μg/ml tunicamycin (Sigma). Cells were harvested 1 h later by centrifugation and cell pellets were stored at -80°C. Total RNA was isolated by the hot phenol method (26). Aliquots of RNA (30 μg, calculated on the basis of OD260) from control and tunicamycin-treated cells were resolved by electrophoresis through a formaldehyde/1.6% agarose gel. The RNA was then transferred to a Hybond membrane (Amersham Pharmacia). Radiolabeled HAC1 and ACT1 probes were prepared by using a random priming kit according to the instructions of the vendor (Boehringer). Hybridization was performed as described (26). Hybridized probe was visualized by autoradiography of the membrane. The strength of the hybridization signal was quantitated by scanning the membrane with a PhosphorImager.
The spliced HAC1 transcript was also detected by RT-PCR amplification as follows. Total yeast RNA (30 μg) was mixed with 20 pmol of an HAC1-specific antisense DNA primer, 5′-d(CATGAAGTGATGAAGAAATC). The mixture was heated for 3 min at 90°C and then cooled to 37°C over 1 h. Primer extension was performed with AMV reverse transcriptase (Life Sciences) in reaction buffer containing 50 mM Tris·HCl (pH 8.3), 10 mM DTT, 60 mM NaCl, 6 mM Mg acetate, and 0.4 mM dNTPs. After incubation for 5 min at 37°C and then for 30 min at 42°C, the cDNA was recovered by phenol extraction and ethanol precipitation. The reverse transcription product was amplified by 40 cycles of PCR with Herculase polymerase (Stratagene) in a reaction primed by the HAC1 antisense oligonucleotide and a HAC1 sense-strand oligonucleotide 5′-d(GACTACATCTGCAGTATC). The PCR products were analyzed by electrophoresis through a 1.2% agarose gel containing ethidium bromide. Gel-purified PCR fragments were cloned into a pCR-Blunt II-TOPO vector (Invitrogen) and transformed into E. coli. Plasmid DNAs were isolated from individual kanamycin-resistant transformants and the HAC1 insert was sequenced.
Results
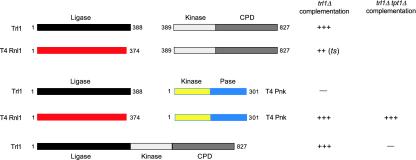
T4 Rnl1 Can Substitute in Vivo for the Ligase Domain of Yeast Trl1. The 827-aa Trl1 polypeptide consists of an N-terminal adenylyltransferase/ligase domain, a central polynucleotide kinase domain, and a C-terminal CPD domain. All three domain modules are essential in vivo, although they need not be linked in the same polypeptide to sustain yeast cell growth (20). For example, complementation of a lethal trl1Δ mutation by the plasmid shuffle method can be achieved by expressing the ligase domain Trl1(1–388) and the kinase-CPD domain Trl1(389–827) as unlinked polypeptides (Fig. 2).
Fig. 2.
The T4 RNA-repair system can replace yeast tRNA ligase in vivo and bypass the requirement for Tpt1. The N-terminal adenylyltransferase/ligase domain is black and the C-terminal kinase-CPD domains of Trl1 are white/gray. T4 Rnl1 is red. T4 Pnkp is yellow (N-terminal kinase) and blue (C-terminal phosphatase). CEN plasmids expressing the indicated components were tested by plasmid shuffle for complementation of the yeast trl1Δ strain and, where indicated, the trl1Δ tpt1Δ strain. Complementation was scored as described in Materials and Methods.
Amino acid sequence comparisons and available mutational data suggest that the N-terminal domain of Trl1 resembles T4 Rnl1, at least with respect to the peptide motifs that comprise the adenylyltransferase active site (20, 25). To test whether the phage RNA ligase could substitute in vivo for the yeast RNA ligase domain, we expressed the 374-aa Rnl1 protein in yeast under the control of a constitutive yeast promoter on a single-copy CEN plasmid. Coexpression of T4 Rnl1 and the yeast kinase-CPD module supported growth of trl1Δ cells on YPD agar at 25 and 30°C, with the colony size slightly smaller than that of isogenic TRL1 cells (scored as ++ growth in Fig. 2). However, the trl1Δ RNL1 TRL1(389–827) strain did not grow at 37°C (denoted by ts in Fig. 2). Expression of T4 Rnl1 alone (i.e., without the Trl1 kinase-CPD domain) was unable to complement trl1Δ in the plasmid shuffle assay (data not shown). We surmise that T4 Rnl1 is capable of sealing the ends of the tRNA half-molecules that had been processed by Trl1(389–827).
T4 Pnkp Can Replace the Kinase-CPD Domain of Trl1 Only in Tandem with T4 Rnl1. The central kinase domain of Trl1 resembles the N-terminal kinase domain of T4 Pnkp with respect to the presence of a Walker A-box motif, GxGK(S/T), and a second motif, RxxxR, located ≈100 amino acids downstream of the A-box. These two motifs form the nucleoside triphosphate-binding pocket of the kinase active site (24, 27–29). In contrast, the structures and mechanisms of the 3′ end-processing components are completely different in the T4 and yeast RNA-repair systems. The C-terminal CPD domain of yeast Trl1 resembles the 2H phosphoesterase enzyme superfamily, which is defined by two copies of a histidine-containing motif HφTφ (where φ is a hydrophobic residue; ref. 30). Mutations of the defining histidines in Trl1 are either lethal or confer a ts growth defect in vivo (20). The 3′ phosphatase domain of T4 Pnk belongs to the DxDxT family of phosphotransferases that act through a covalent acyl-phosphoenzyme intermediate in which the phosphate is linked to the first aspartate in the DxDxT motif (27, 29, 31).
To test whether T4 Pnkp could substitute in vivo for the Trl1 kinase-CPD domain, we expressed the 301-aa Pnkp protein in yeast under the control of a constitutive yeast promoter on a CEN plasmid. Coexpression of T4 Pnkp and the N-terminal ligase domain Trl1(1–388) failed to complement growth of trl1Δ cells at 25, 30, or 37°C (scored as - growth in Fig. 2). However, T4 Pnkp did complement trl1Δ when expressed in tandem with T4 Rnl1. Complete replacement of Trl1 by the two-component phage RNA-repair system restored wild-type growth of trl1Δ cells on YPD agar at 25, 30, and 37°C (Fig. 2). Thus, although Pnkp is apparently capable of healing the ends of the incised tRNA half-molecules in vivo, the resulting 3′-OH and 5′-PO4 termini can be sealed by T4 Rnl1, but not by the yeast ligase domain Trl1(1–388).
Because the kinase modules of the phage and yeast end-healing enzymes are structurally similar and the reaction products identical (i.e., a 5′-PO4 RNA end), we ascribe the fact that the Trl1(1–388) functions only in tandem with Trl1(388–821) to the distinctive 3′-OH, 2′-PO4 end configuration generated by yeast CPD versus the 3′-OH, 2′-OH end produced by Pnkp. In other words, the yeast tRNA ligase requires the 2′-PO4 terminus to seal tRNAs in vivo. This inference is consistent with the in vitro requirements for a 2′-PO4 for RNA joining by purified Trl1 (14) and by the functionally homologous RNA ligase/kinase/CPD enzyme isolated from wheat germ (32, 33). We find that purified recombinant Trl1(1–388) is incapable of sealing an RNA substrate with 5′-PO4 and 3′-OH/2′-OH termini that are ligated quantitatively by purified recombinant T4 Rnl1 (data not shown). The ability of T4 Rnl1 to function in yeast tRNA splicing in tandem with Trl1(389–827) implies that the phage ligase is able to seal the 3′-OH, 2′-PO4 termini formed in vivo by the yeast CPD enzyme.
The T4 RNA-Repair System Bypasses the in Vivo Requirement for the 2′ Phosphotransferase Tpt1. Tpt1 catalyzes the transfer of the tRNA 2′-PO4 to NAD+ to form ADP-ribose 1′′-2′′-cyclic phosphate and nicotinamide (17, 34). The mechanism entails two component steps. First, NAD+ reacts with the tRNA 2′ phosphate to expel nicotinamide and generate a 2′ phospho-ADP-ribosylated RNA intermediate. Then, transesterification of the ADP-ribose 2′-O to the tRNA 2′-phosphate displaces the tRNA product and generates ADP-ribose 1′′-2′′-cyclic phosphate (34). Tpt1 exemplifies a family of structurally homologous proteins found in eukaryal, archaeal, and bacterial proteomes (35). Tpt1 is the only activity in yeast cells capable of efficiently removing the 2′ phosphate from tRNA splice junctions; the 2′ phosphate at the splice junction is comparatively resistant to hydrolysis by generic phosphatases such as calf alkaline phosphatase (36). Because Tpt1 homologs are found in species that have no intron-containing tRNAs, it has been suggested that members of this enzyme family may dephosphorylate other substrates in vivo through a shared mechanism of phosphoryl transfer to NAD+ to form ADP-ribose 1′′-2′′-cyclic phosphate (35). One speculation is that the ADP-ribose 1′′-2′′-cyclic phosphate product of the Tpt1 reaction may serve as a signaling molecule as part of a metabolic or regulatory pathway, as do other nucleotide cyclic phosphates (19). In addition, Spinelli et al. (35) have considered the prospect that Ttp1 may play an essential noncatalytic function, akin to the in vivo requirement for T4 Rnl1 for attachment of the tail fiber during phage morphogenesis (37).
To address these issues, we gauged the requirements for yeast Tpt1 in vivo as a function of the source of the tRNA splicing machinery. An initial gene disruption experiment confirmed that a tpt1Δ mutation was lethal in a haploid TRL1 strain (Fig. 2 and Materials and Methods). To see whether the requirement for Tpt1 could be bypassed by expressing the T4 enzymes, we then disrupted one chromosomal copy of TPT1 with a LEU2 cassette in a trl1Δ trl1Δ diploid strain containing URA3 TRL1 and TRP1 RNL1/PNKP plasmids. The TPT1 tpt1::LEU2 trl1::kanMX trl1::kanMX diploid was sporulated and asci were dissected to obtain haploid progeny. The instructive finding was that all Leu+ haploids lacking Tpt1 were also Trp+, signifying that they all contained the T4 RNA-repair enzyme system. All of the LEU2 TRP1 haploids grew on medium containing 5-FOA; i.e., they easily lost the TRL1 plasmid. On the other hand, all Ura+ Trp- haploids containing TRL1 plasmid (and lacking the T4 enzymes) were also Leu-; i.e., they contained the intact chromosomal copy of TPT1. We found that trl1Δ tpt1Δ cells containing the T4 RNA-repair system as the only source of tRNA-splicing activity grew as well as the parental wild-type TRL1 TPT1 yeast strain on YPD agar at all temperatures tested (scored as +++ growth in Fig. 2). The trl1Δ tpt1Δ RNL1/PNKP strain also grew as well as the parental wild-type strain on minimal agar medium containing either glucose, galactose, or raffinose as the carbon sources, and on YP agar medium containing either sucrose or glycerol as the carbon sources (data not shown).
These results show that: (i) Tpt1 is essential when tRNA splicing is catalyzed by yeast Trl1, but dispensable when the tRNA splicing is catalyzed by T4 Rnl1 and Pnkp; (ii) there is not a strict requirement for the formation and removal of a 2′ phosphate at the tRNA splice junction; (iii) Tpt1 plays no essential role in vivo (at least not for growth under laboratory conditions tested) other than removal of the 2′ phosphates generated by Trl1; and (iv) there is no essential growth role in vivo (under laboratory conditions tested) for the ADP-ribose 1′′-2′′-cyclic phosphate product of the Tpt1 reaction.
T4 RNA-Repair Enzymes Can Splice HAC1 mRNA During the Unfolded Protein Response. Trl1 is responsible for nonspliceosomal splicing of HAC1 mRNA in the yeast unfolded protein response pathway (22, 23). Endoplasmic reticulum stress-induced cleavage of the HAC1 mRNA by endonuclease Ire1 excises a 252-nt intron and leaves 2′,3′-cyclic phosphate and 5′-OH termini at the cleavage sites. After healing and sealing by Trl1, the HAC1 translation reading frame is altered so as to replace the C-terminal 10 amino acids of the uninduced form of Hac1 (encoded in the intron) with a different 18-aa segment (encoded in the second exon). The induced form of Hac1 is a transcription factor that efficiently activates certain nuclear genes in response to endoplasmic reticulum stress (38, 39). Excision of the intron from HAC1 mRNA also relieves a block to HAC1 translation caused by long-range base pairing between the intron and the 5′-UTR (40).
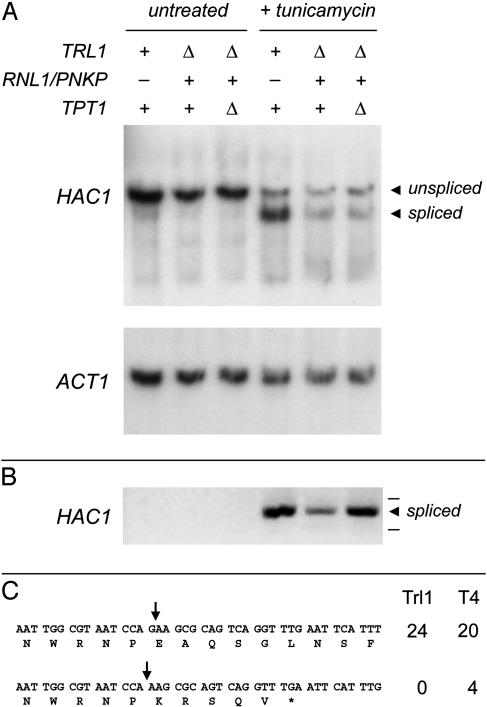
Is Trl1 uniquely required for mRNA splicing in the unfolded protein response? To answer this question, we assessed the splicing of HAC1 mRNA in response to tunicamycin (an inducer of the unfolded protein response) in yeast cells expressing either Trl1 or T4 Rnl1/Pnkp as their source of tRNA-splicing enzymes. Total RNA isolated from yeast cells treated for 1 h with 1.5 μg/ml tunicamycin and from mock-treated controls was analyzed by Northern blotting. HAC1 transcripts were detected by hybridization to a 32P-labeled HAC1 DNA probe (Fig. 3A). As expected, the larger unspliced HAC1 transcript predominated in control cells, whereas the shorter spliced HAC1 transcript was the major species present in tunicamycin-treated cells, comprising 74% of the total (spliced plus unspliced) HAC1 mRNA. The instructive findings were that HAC1 splicing was also triggered by tunicamycin in trl1Δ cells containing the T4 RNA-repair system, with or without the 2′ phosphotransferase Tpt1 (Fig. 3A). Spliced mRNA comprised 68% and 63% of the total HAC1 transcript in tunicamycin-treated trl1Δ RNL1/PNKP TPT1 and trl1Δ RNL1/PNKP tpt1Δ strains, respectively.
Fig. 3.
Splicing of HAC1 mRNA in the unfolded protein response. (A) The genotypes of the yeast strains with respect to the tRNA-splicing enzymes are indicated above the lanes. Total RNA isolated from uninduced and tunicamycin-induced yeast cultures was resolved by agarose gel electrophoresis, transferred to a membrane, and probed for HAC1 and ACT1 mRNAs, as indicated. The hybridized 32P-labeled probes were detected by autoradiography. The spliced and unspliced HAC1 mRNAs are indicated by the arrowheads at right. (B) Spliced HAC1 mRNA was amplified by RT-PCR. The DNA products were analyzed by agarose gel electrophoresis, stained with ethidium bromide, and visualized by short-wave UV transillumination. An inverse image of the stained gel is shown. The amplified spliced HAC1 cDNA (indicated by the arrowhead at right) migrated between the 600- and 400-bp linear duplex DNA markers (-, above and below the arrowhead). (C) Individual cDNA clones of spliced HAC1 mRNAs derived from tunicamycin-treated cell containing either yeast tRNA ligase (Trl1; n = 24) or T4 Rnl1 and Pnkp (T4; n = 24) were sequenced. The coding strand sequence flanking the correctly spliced junction of Ire1-incised HAC1 mRNA is shown above the sequence of the minority population (4 of 24) of incorrectly spliced HAC1 mRNAs detected in RNL1 PNKP cells.
RT-PCR amplification of HAC1 from the samples of total yeast RNA was performed with DNA primers flanking the intron. The unspliced HAC1 transcript is poorly copied by reverse transcriptase, because the extensive secondary structure of the HAC1 intron is an impediment to primer extension. However, the spliced HAC1 mRNA can be copied by reverse transcriptase and the cDNA subsequently amplified by PCR. A 534-nt PCR product corresponding to the spliced form of the HAC1 transcript was detected by RT-PCR amplification of RNA from tunicamycin-treated TRL1 TPT1, trl1Δ RNL1/PNKP TPT1 and trl1Δ RNL1/PNKP tpt1Δ cells, but not from RNA isolated from uninduced controls (Fig. 3B). These results show that the T4 RNA-repair enzymes can substitute for Trl1 in both tRNA and mRNA splicing in vivo.
To survey the fidelity of the HAC1 splicing reaction in vivo, the RT-PCR-amplified spliced HAC1 cDNAs were gel-purified, ligated into a bacterial plasmid vector, and transformed into E. coli. Plasmid DNAs were isolated from individual transformants and the HAC1 insert was sequenced. All of the 24 individual spliced cDNAs amplified from tunicamycin-treated wild-type TRL1 cells were accurately spliced; i.e., the splice junction (arrow in Fig. 3C) corresponds to the termini generated by Ire1 endonuclease incision of the unspliced HAC1 mRNA at the proximal exon–intron splice donor (5′-CCAG↓) and distal intron–exon splice acceptor (↓AAGC-3′) sites (23). The notable finding was that one-sixth of the surveyed population (4 of 24) of spliced HAC1 mRNAs from cells containing the T4 RNA-repair system suffered loss of a single guanylate nucleotide at the 3′ end of the splice-donor site (Fig. 3C). Because none of the splicing events mediated by the T4 system resulted in alteration of the splice-acceptor site or any other nucleotides in the vicinity of the splice junction (Fig. 3C), we surmise that the T4-mediated mRNA splicing reaction in yeast allows for exo-nucleolytic trimming of the splice-donor 3′ terminus before the end-sealing step. Simple explanations for the loss of fidelity during HAC1 splicing by the phage enzymes are as follows: (i) end-trimming is suppressed in TRL1 cells by the presence of a 2′-PO4 at the splice-donor 3′ terminus, which is not present when the end-healing steps are performed by T4 Pnkp and/or (ii) that any 3′ end-trimming that does occur in TRL1 cells will result in a 3′ terminus lacking a 2′-PO4, which is not a substrate for the end-sealing reaction of Trl1.
Discussion
The ability of bacteriophage enzymes Rnl1 and Pnkp to fulfill the tRNA and mRNA splicing functions of yeast Trl1 and bypass the essential yeast 2′-phosphotransferase Tpt1 attests to portability of RNA-repair systems in vivo, notwithstanding the significant differences in the specificities, mechanisms, and reaction intermediates of the individual enzymes responsible for the RNA-healing and -sealing steps. As noted above, our findings imply that Tpt1 and its unique metabolite, ADP-ribose 1′′-2′′-cyclic phosphate, do not play essential roles independent of the tRNA-splicing reaction.
Why then does yeast employ a more complex system of RNA repair than phage T4? In particular, what advantage might accrue to yeast cells by the specific requirement for a 2′-PO4 end for RNA ligation in vivo? We speculate that this requirement evolved to: (i) limit potential RNA joining events to the RNA 3′-OH, 2′-PO4 ends formed in vivo by the CPD activity of Trl1 and (ii) exclude “illegitimate” RNA-joining reactions between 5′-PO4 RNA termini (generated directly by endonucleolytic cleavage or 5′ exoribonucleolytic decay, or indirectly by phosphorylation of cleaved 5′-OH termini by the Pnk activity of Trl1) and the vast number of unmodified 3′-OH, 2′-OH ends present in the cell. The issue is not simply wasteful RNA ligation, but rather, the avoidance of producing recombinant mRNA molecules coding for potentially deleterious proteins. Yeast cells have distinct pathways of mRNA decay by 5′ to 3′ and 3′ to 5′ exoribonucleases (ref. 41 and references therein). If Trl1 were capable of efficiently sealing 3′-OH, 2′-OH ends, then the 3′-OH-terminated 5′ coding segment of one transcript (containing a 5′ cap) could, in principle, be illegitimately spliced to the 5′-PO4-terminated 3′ coding segment [and poly(A) or oligo(A) tail] of another transcript. The hybrid mRNA could thereby yield an aberrant protein product. Indeed, the illegitimate HAC1 splicing event seen here in 4 of 24 cDNAs entailed deletion of a single nucleotide at the proximal splice-donor site, resulting in a translational frameshift that appends a foreign peptide sequence to the C terminus of the Hac1 protein in lieu of the peptide encoded by the distal exon (Fig. 3C). Such events in the general RNA population, even if infrequent, are potentially toxic and better avoided; e.g., by incorporation of a CPD component into the tRNA-splicing machinery and imposing on the ligase component a requirement for a 2′-PO4 end.
If, as the adage goes, what's good for the goose is good for the gander, why does the phage T4 RNA-repair system exploit a different 3′ end-healing mechanism and impose no special requirements at the ligatable 3′ end? A simple explanation is that the phage is under no pressure to suppress illegitimate RNA-joining reactions in the bacterial host cell, because the bacterium will soon suffer a lytic death at the hands of the phage. The priority for the phage is to reverse the “altruistic” triggering of host cell death by tRNA restriction (a primitive form of apoptosis) just long enough to exploit the cellular translation machinery for its own replication.
Whereas the fungal tRNA-splicing system may have evolved to ensure fidelity of RNA splicing, the fact that a seemingly less stringent heterologous RNA-repair system can support yeast viability under laboratory conditions provides a means to identify, by functional complementation, RNA-splicing and -repair enzymes from other organisms. For example, the RNA ligase responsible for metazoan tRNA splicing has not been cloned and there are no obvious homologs of yeast tRNA ligase in available metazoan proteomes. Although the metazoan unfolded protein response, like that of yeast, involves induced mRNA cleavage and unconventional mRNA splicing (42, 43), the identity of the RNA ligase component of the metazoan endoplasmic reticulum stress response is also unknown. The unfolded protein response is implicated in human disease (44), thereby making the missing RNA ligase a potential therapeutic target.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CPD, cyclic phosphodiesterase; 5-FOA, 5-fluoroorotic acid; YPD, yeast extract/peptone/dextrose.
References
- 1.Silber, R., Malathi, V. G. & Hurwitz, J. (1972) Proc. Natl. Acad. Sci. USA 69, 3009-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abelson, J., Trotta, C. R. & Li, H. (1998) J. Biol. Chem. 273, 12685-12688. [DOI] [PubMed] [Google Scholar]
- 3.Simpson, L., Sbicego, S. & Aphasizhev, R. (2003) RNA 9, 265-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amitsur, M., Levitz, R. & Kaufman, G. (1987) EMBO J. 6, 2499-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufmann, G. (2000) Trends Biochem. Sci. 25, 70-74. [DOI] [PubMed] [Google Scholar]
- 6.Richardson, C. C. (1965) Proc. Natl. Acad. Sci. USA 54, 158-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novogrodsky, A. & Hurwitz, J. (1966) J. Biol. Chem. 241, 2923-2932. [PubMed] [Google Scholar]
- 8.Novogrodsky, A., Tal., M., Traub, A. & Hurwitz, J. (1966) J. Biol. Chem. 241, 2933-2943. [PubMed] [Google Scholar]
- 9.Cameron, V. & Uhlenbeck, O. C. (1977) Biochemistry 16, 5120-5126. [DOI] [PubMed] [Google Scholar]
- 10.Uhlenbeck, O. C. & Gumport, R. I. (1982) in The Enzymes, ed. Boyer, P. (Academic, New York), Vol. 15, pp. 31-58. [Google Scholar]
- 11.Cranston, J. W., Silber, R., Malathi, V. G. & Hurwitz, J. (1974) J. Biol. Chem. 249, 7447-7456. [PubMed] [Google Scholar]
- 12.Sugino, A., Snopek, T. J. & Cozarelli, N. R. (1978) J. Biol. Chem. 252, 1732-1738. [PubMed] [Google Scholar]
- 13.Greer, C. L., Peebles, C. L., Gegenheimer, P. & Abelson J. (1983) Cell 32, 537-546. [DOI] [PubMed] [Google Scholar]
- 14.Phizicky, E. M., Schwartz, R. C. & Abelson, J. (1986) J. Biol. Chem. 261, 2978-2986. [PubMed] [Google Scholar]
- 15.Apostol, B. L., Westaway, S. K., Abelson, J. & Greer, C. L. (1991) J. Biol. Chem. 266, 7445-7455. [PubMed] [Google Scholar]
- 16.Westaway, S. K., Belford, H. G., Apostol, B. L., Abelson, J. & Greer, C. L. (1993) J. Biol. Chem. 268, 2435-2443. [PubMed] [Google Scholar]
- 17.Culver, G. M., McCraith, S. M., Zillman, M., Kierzek, R., Michaud, N., LaReau, R. D., Turner, D. H. & Phizicky, E. M. (1993) Science 261, 206-208. [DOI] [PubMed] [Google Scholar]
- 18.Phizicky, E. M., Consaul, S. A., Nehrke, K. W. & Abelson, J. (1992) J. Biol. Chem. 267, 4577-4582. [PubMed] [Google Scholar]
- 19.Culver, G. M., McCraith, S. M., Consaul, S. A., Stanford, D. R. & Phizicky, E. M. (1997) J. Biol. Chem. 272, 13203-13210. [DOI] [PubMed] [Google Scholar]
- 20.Sawaya, R., Schwer, B. & Shuman, S. (2003) J. Biol. Chem. 278, 43928-43938. [DOI] [PubMed] [Google Scholar]
- 21.Spinelli, S. L., Consaul, S. A. & Phizicky, E. M. (1997) RNA 3, 1388-1400. [PMC free article] [PubMed] [Google Scholar]
- 22.Sidrauski, C., Cox, J. S. & Walter, P. (1996) Cell 87, 405-413. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez, T. N., Sidrauski, C., Dörfler, S. & Walter, P. (1999) EMBO J. 18, 3119-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, L. K. & Shuman, S. (2001) J. Biol. Chem. 276, 26868-26874. [DOI] [PubMed] [Google Scholar]
- 25.Wang, L. K., Ho, C. K., Pei, Y. & Shuman, S. (2003) J. Biol. Chem. 278, 29454-29462. [DOI] [PubMed] [Google Scholar]
- 26.Herrick, D., Parker, R. & Jacobsen, A. (1990) Mol. Cell. Biol. 10, 2269-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, L. K. & Shuman, S. (2002) Nucleic Acids Res. 30, 1073-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, L. K., Lima, C. D. & Shuman, S. (2002) EMBO J. 21, 3873-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galburt, E. A., Pelletier, J., Wilson, G. & Stoddard, B. L. (2002) Structure (London) 10, 1249-1260. [DOI] [PubMed] [Google Scholar]
- 30.Mazumder, R., Iyer, L., Vasudevan, S. & Aravind, L. (2002) Nucleic Acids Res. 30, 5229-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collet, J. F., Stroobant, V., Pirard, M., Delpierre, G. & Van Schaftingen, E. (1998) J. Biol. Chem. 273, 14107-14112. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz, R. C., Greer, C. L., Gegenheimer, P. & Abelson, J. (1983) J. Biol. Chem. 258, 8374-8383. [PubMed] [Google Scholar]
- 33.Pick, L., Furneaux, H. & Hurwitz, J. (1986) J. Biol. Chem. 261, 6694-6704. [PubMed] [Google Scholar]
- 34.Spinelli, S. L., Kierzek, R., Turner, D. H. & Phizicky, E. M. (1999) J. Biol. Chem. 274, 2637-2644. [DOI] [PubMed] [Google Scholar]
- 35.Spinelli, S. L., Malik, H. S., Consaul, S. A. & Phizicky, E. M. (1998) Proc. Natl. Acad. Sci. USA 95, 14136-14141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gegenheimer, P., Gabius, H. J., Peebles, C. L. & Abelson, J. (1983) J. Biol. Chem. 258, 8365-8373. [PubMed] [Google Scholar]
- 37.Snopek, T. J., Wood, W. B., Conley, M. P., Chen, P. & Cozzarelli, N. R. (1977) Proc. Natl. Acad. Sci. USA 74, 3355-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox, J. S. & Walter, P. (1996) Cell 87, 391-404. [DOI] [PubMed] [Google Scholar]
- 39.Mori, K., Ogawa, N., Kawahara, T., Yanagi, H. & Yura, T. (2000) Proc. Natl. Acad. Sci. USA 97, 4660-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rüesgegger, U., Leber, J. H. & Walter, P. (2001) Cell 107, 103-114. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell, P. & Tollervey, D. (2003) Mol. Cell 11, 1405-1413. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida, H., Matsui, T., Yamamato, A., Okada, T. & Mori, K. (2001) Cell 107, 881-891. [DOI] [PubMed] [Google Scholar]
- 43.Calfon, M., Zeng, H., Urano, F., Till, J. H., Hubbard, S. R., Harding, H. P., Clark, S. G. & Ron, D. (2002) Nature 415, 92-96. [DOI] [PubMed] [Google Scholar]
- 44.Kaufman, R. J. (2002) J. Clin. Invest. 110, 1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]