Abstract
The mitochondrial enzyme succinate dehydrogenase (SDH) consists of four subunits, a flavoprotein (SDH1), an iron-sulfur (Fe-S) protein (SDH2) and two integral membrane subunits (SDH3/SDH4). In mammals and yeast, an assembly factor termed SDHAF2/SDH5 is required for accumulation of flavinylated SDH1. In Arabidopsis, we have recently reported the characterization of an unknown function protein with low sequence similarity to SDHAF2 that is needed for assembly and activity of SDH and also for normal root elongation.1 In this short communication, we have reviewed the sequence diversity and conservation of SDHAF2 across kingdoms based on phylogenetic analysis. Given that flavinylation of SDH is dependent on the SDH1:SDHAF2 interaction, we have also discussed the conservation of the C-terminal tail of SDH1, which is required for this interaction process. In combination, we provide comparative evidence for a conserved role of SDHAF2 as an assembly factor from animals to plants.
Keywords: succinate dehydrogenase, plant mitochondria, flavinylation
Text
Succinate dehydrogenase (SDH, Complex II), oxidases succinate to fumarate and has a central role in respiration as a component of both the TCA cycle and the electron transport chain. The SDH complex contains four subunits: a flavoprotein (SDH1) that contains a bound FAD cofactor, an iron-sulfur (Fe-S) protein (SDH2) containing three Fe-S clusters and two integral membrane proteins (SDH3 and SDH4) that form the b- type cytochrome by binding heme.2 At the amino acid sequence level,SDH1 and SDH2 are highly conserved while SDH3 and SDH4 have very significantly diverged in plants, fungi and mammals.3,4
The enzymatic function of SDH is dependent on covalent attachment of FAD to the 70 kDa SDH1 protein. A non-enzymatically catalyzed covalent attachment of FAD to a histidine residue of SDH1 is required to allow electron donation from succinate for its oxidation to fumarate.5 This flavinylation reaction is known to be stimulated by citrate acid cycle intermediates, including succinate.6 An assembly factor (SDHAF2) that is required for the flavinylation of SDH1 was recently identified in humans through mutants causing disease.7 SDHAF2 is proposed to function by binding to SDH1 and allowing FAD entry, but it is unclear exactly how or if it facilitates the covalent flavinylation reaction.7 In plants, Arabidopsis At5g51040 is a functional ortholog of human SDHAF2 but only has low similarity at the amino acid sequence level. Knockdown of At5g51040 results in decreased SDH enzyme activity and loss of flavinylated SDH1.1 We have proposed that At5g51040 functions in a similar manner to human SDHAF2, i.e., via SDH1 interaction and facilitation of FAD entry.1 Herewith, we use phylogenetic analysis in support of this proposal. We show that SDHAF2-like proteins across fungi, plants and animals show great diversity at the amino acid sequence level, but that a glycine residue experimentally shown to be needed for SDH1:SDHAF2 physical interaction is conserved across all the species analyzed. Furthermore, we use sequence analysis to show the conservation of key residues in the C-terminal tail of SDH1,which are required for SDH flavinylation, based on recent mutation studies in yeast.8
SDHAF2 is Divergent at the Sequence Level but a Short Conserved Region Contains a Glycine Residue Required for SDH1 Interaction
We have showed that Arabidopsis SDHAF2 (At5g51040) has only 30% identity to human SDHAF2 (NP_060311) in a specific 96 AA region (position 69 to 165 in At5g51040) and 32% identity to yeast SDHAF2 (B3LIY9) in approximately the same region.1 A very recent study has indicated that, in the bacteria genus Serratia, SdhE (G4V4G2) is required for flavinylation of SDH.9 Arabidopsis SDHAF2 (At5g51040) has only 26% sequence identity (from position 82 to 134) to Serratia SdhE (BLAST E-value of 3 × e−09). To further analyze the conservation of SDHAF2, we aligned the sequences of SDHAF2-like proteins in plants, green algae, yeast, mammals and bacteria SdhE (Fig. 1A). The mammalian sequences clearly separate from green alga and plants, while and bacterial SdhE is quite different from all the eukaryotes. Among the plants, monocots and eudicots can be separated into two groups and appear to share a common ancestor, the moss Physcomitrella patens (Fig. 1A).

Figure 1. Diversity and conservation of SDHAF2-like protein sequences. (A) Phylogenetic tree analysis of SDHAF2-like protein sequences. The protein accession numbers of SDHAF2-like proteins used for analysis are: Homo sapiens, NP_060311; Mus musculus, NP_079609; Saccharomyces cerevisiae, B3LIY9; Serratia ATCC 39006, G4V4G2; Physcomitrella patens, XP_001752564; Ostreococcus tauri, XP_003074399; Chlamydomonas reinhardtii, XP_001694005; Glycine max, NP_001236603; Medicago truncatula, XP_003602736; Ricinuscommunis, XP_002511076; Arabidopsis thaliana, At5g51040; Vitis Vinifera, XP_003635192; Hordeum vulgare, BAJ90590; Brachypodium distachyon, XP_003577516; Oryza sativa, Os11 g32480; Zea mays, ACG26201; Sorghum bicolor, XP_002449598. (B) Sequence alignment of conserved regions containing a glycine residue (red arrow) required for flavinylation.
In humans, mutation of glycine 78 in SDHAF2 to arginine (G78R) is a cause of paraganglioma-pheochromocytoma.7 Further analysis of this mutation in human and yeast SDHAF2 indicate that glycine at position 78 is essential for SDHAF2:SDH1 interaction and accumulation of flavinylated SDH1.7 Alignment of amino acid sequences in this region across different species show that this glycine is extremely conserved, even in the bacterial Serratia sequence (Fig. 1B). Mutation of this glycine in Serratia SdhE also inhibited accumulation of flavinylated SDH1.9 Therefore, despite the great diversity evident in SDHAF2 sequence (Fig. 1A), the conserved glycine indicated in Figure 1B is likely to be essential for SDHAF2:SDH1 interaction and accumulation of flavinylated SDH1 across prokaryotes and eukaryotes.
The C-terminal Tail of SDH1 Required for SDHAF2 Binding and Flavinylation is Conserved
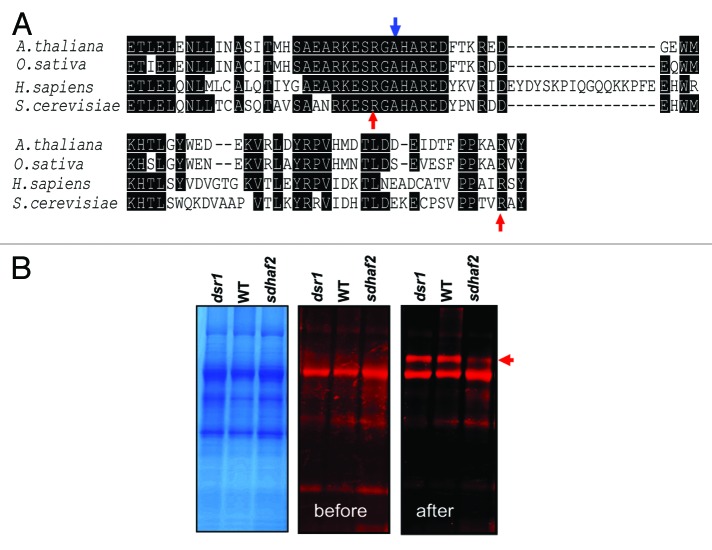
FAD insertion into SDH1 requires SDH1:SDHAF2 physical interaction.7 Recent evidence in yeast indicates that the C-terminal tail of SDH1 is important for both flavinylation of SDH1 and for this SDH1:SDHAF2 interaction.8 In yeast, two C-terminal Arg residues (R582 and R638; indicated by red arrows in Fig. 2A) which are distant from the FAD binding site are essential for SDH1 flavinylation and influence the binding and stability of SDHAF2.8 Kim et al.8 proposed that the Arg residues could have roles: (1) directly in SDHAF2 association, (2) in the recruitment and/or guidance of FAD and/or (3) in allowing succinate to the substrate site to catalyze the covalent flavinylation reaction. The SDH1 sequence is quite conserved across different species at the protein sequence level but still many differences are evident across kingdoms.3,4 Sequence alignment of SDH1 C-terminal tails showed that both Arg residues in yeast are also conseved in human, Arabidopsis and rice (red arrows, Fig. 2A). In human, mutation of Arg 589 (top red arrow) to Trp (R589W) is associated with paraganglioma,10 presumably due to reduction in SDH flavinylation and SDH activity.
Figure 2. C-terminal tail sequences of SDH1 required for flavinylation. (A) Sequence alignment of C-terminal tail of SDH1 in plants, human and yeast. Red arrows indicate arginine residues required for flavinylation in yeast. The blue arrow indicate the mutation site of Arabidopsis SDH1–1 (At5g66760) indsr1. The protein sequences used for alignments are: Arabidopsis thaliana (At5g66760), Oryza sativa (Os07 g04240); Homo sapiens, P31040; Saccharomyces cerevisiae, Q00711. (B) Flavinylated SDH1is indicated by the red arrow in Arabidopsis mitochondrial protein extracts from dsr1, WT and sdhaf2. The protein bound-flavin assay was conducted as described in Huang et al.1 After SDS-PAGE separation of mitochondria proteins, the protein bands with bound-flavin before and after 10% acetic acid treatment for 30 min were scanned using a Typhoon Trio laser imager (GE Healthcare) with excitation at 488 nm and emission at 526 nm. The protein bands containing bound-FAD are indicated by the red arrow.
The conserved C-terminal Arg residues required for SDHAF2 binding and flavinylation and very close to residues required for SDH1 activity and ATP activation in plants
In Arabidopsis, we also recently reported a mutant (dsr1) affecting SDH1–1 (A580T, indicated by blue arrow in Fig. 2A).11 As shown in Figure 2A, this alanine residue is only one amino acid away from the conserved R589 (red arrow) which is essential for FAD insertion. This Arabidopsis dsr1mutation (A580T) reduces SDH enzymatic activity by ~70% and eliminates ATP activation of the enzyme.11 However, SDH1 flavinylation in dsr1 is very similar to that of WT, while sdhaf2 has less SDH1 and less FAD-SDH111,1 (Fig. 2B). These results are consistent with our previous hypothesis that dsr1mutation in SDH1–1 affects the succinate binding pocket not FAD binding.11 These data show that this C-terminal region of SDH1 is a hotspot for modulating, but not eliminating, SDH activity by altering the substrate binding and flavinylation status of the enzyme.
Conclusions
In summary, we have recently reported that Arabidopsis At5g51040 is a SDHAF2-like protein and is required for optimal SDH function in plants.1 Direct evidence for SDH1:SDHAF2 interaction and its role in flavinylation are currently derived from studies of yeast and mammals. However, sequence analysis suggests a common mechanism of flavinylation of SDH via SDH1:SDHAF2 interaction is likely to be shared between animals, yeast and plants via the selective conservation of amino acid residues required for flavinylation in SDHAF2 and SDH1.
Acknowledgments
This research was funded by support from the ARC Centre of Excellence in in Plant Energy Biology (CE0561495) and The University of Western Australia and Agilent Technologies Australia support for the Centre of Comparative Analysis of Biomolecular Networks to A.H.M. A.H.M. was funded as an ARC Australian Future Fellow (FT110100242).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22815
References
- 1.Huang S, Taylor NL, Ströher E, Fenske R, Harvey Millar A. Succinate dehydrogenase assembly factor 2 is needed for assembly and activity of mitochondrial complex II and for normal root elongation in Arabidopsis. Plant J. 2012 doi: 10.1111/tpj.12041. In press. [DOI] [PubMed] [Google Scholar]
- 2.Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, et al. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–57. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 3.Burger G, Lang BF, Reith M, Gray MW. Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distant eukaryotes. Proc Natl Acad Sci U S A. 1996;93:2328–32. doi: 10.1073/pnas.93.6.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang S, Taylor NL, Narsai R, Eubel H, Whelan J, Millar AH. Functional and composition differences between mitochondrial complex II in Arabidopsis and rice are correlated with the complex genetic history of the enzyme. Plant Mol Biol. 2010;72:331–42. doi: 10.1007/s11103-009-9573-z. [DOI] [PubMed] [Google Scholar]
- 5.Tomasiak TM, Maklashina E, Cecchini G, Iverson TM. A threonine on the active site loop controls transition state formation in Escherichia coli respiratory complex II. J Biol Chem. 2008;283:15460–8. doi: 10.1074/jbc.M801372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson KM, Lemire BD. Covalent attachment of FAD to the yeast succinate dehydrogenase flavoprotein requires import into mitochondria, presequence removal, and folding. J Biol Chem. 1996;271:4055–60. doi: 10.1074/jbc.271.8.4055. [DOI] [PubMed] [Google Scholar]
- 7.Hao H-X, Khalimonchuk O, Schraders M, Dephoure N, Bayley J-P, Kunst H, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–42. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HJ, Jeong M-Y, Na U, Winge DR. Flavinylation and assembly of succinate dehydrogenase are dependent on the C-terminal tail of the flavoprotein subunit. J Biol Chem. 2012 doi: 10.1074/jbc.M112.405704. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNeil MB, Clulow JS, Wilf NM, Salmond GPC, Fineran PC. SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria. J Biol Chem. 2012;287:18418–28. doi: 10.1074/jbc.M111.293803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnichon N, Brière JJ, Libé R, Vescovo L, Rivière J, Tissier F, et al. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19:3011–20. doi: 10.1093/hmg/ddq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, et al. Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc Natl Acad Sci U S A. 2011;108:10768–73. doi: 10.1073/pnas.1016060108. [DOI] [PMC free article] [PubMed] [Google Scholar]