Abstract
Cryptochromes (CRYs) are flavoproteins that are known as blue light photoreceptors in many organisms. Recently, genome sequences from a variety of algae became available. Functional characterizations of animal-like CRYs from Oestreococcus tauri, Chlamydomonas reinhardtii and Phaeodactylum tricornutum highlighted novel functions and properties. As arising from studies in fungi, certain algal CRYs of the “cryptochrome photolyase family” (PtCPF1, OtCPF1) have dual or even triple functions. They are involved in blue light perception and/or in the circadian clock and are able to repair DNA damages. On the other hand, the animal-like aCRY from C. reinhardtii is not only acting as sensory blue light- but also as sensory red light receptor thus expanding our current view of flavoproteins in general and CRYs in particular. The observed broad spectral response points to the neutral radical state of flavin, which is assumed to be the dark form in aCRY in contrast to the plant CRYs.
Keywords: Chlamydomonas reinhardtii, cryptochrome, blue light photoreceptor, red light photoreceptor, photolyase, flavoprotein
Algal Genomes and Cryptochromes
Light has major impact on algae, which are dominant producers of biomass on Earth and contribute significantly to global productivity and biogeochemical cycling.1 On the one hand, light serves as energy source for photosynthesis. On the other hand, light perception via specialized photoreceptors may (1) activate gene expression, (2) lead to behavioral responses and (3) entrain the circadian clock by light-dark cycles (reviewed in Hegemann).2 In the past years, several algal genomes including those of diatoms (e.g., Thalassiosira pseudonana, Phaeodactylum tricornutum) or green algae (e.g., Oestreococcus tauri, Chlamydomonas reinhardtii) have been sequenced (reviewed in Grossman).1 These sequences along with molecular tools available in the mentioned algae served as an efficient basis to screen for algal photoreceptors and to begin with their functional characterization. UV-, blue-, green- and red-light absorbing photoreceptors have been found in algae. Some of them such as the blue-light absorbing cryptochromes (CRYs) and phototropins (PHOTs) were already known from higher plants, others such as channelrhodopsins, channelenzymerhodopsins or aureochromes were found for the first time in algae (reviewed in Hegemann).2,3 In this mini-review, we will focus on CRYs. They represent not only a widespread group of blue-light absorbing photoreceptors in algae, but occur also in prokaryotes and many other eukaryotes including fungi, plants and animals. Their biological functions are not restricted to photoperception. Some of them are key components of the endogenous pacemaker of the circadian clock, others act as magnetoreceptors in migratory birds (reviewed in Chaves).4 CRYs are mainly divided in (1) animal type I and type II CRYs that are close to (6-4) photolyases, in (2) plant CRYs being close to cyclobutane pyrimidine dimer (CPD) photolyases and in (3) DASH CRYs (DASH; Drosophila, Arabidopsis, Synechocystis, Homo).4 CRYs are flavoproteins having a conserved photolyase homology region at their N-terminus. For a long time, it was thought that the plant and animal-type CRYs evolved from photolyases but lost the ability to catalyze light-dependent DNA repair. It was also thought that CRYs along with their flavin adenin dinucleotide (FAD) chromophore are typical blue-light absorbing receptors. Recent data about fungal and algal members belonging to the “cryptochrome photolyase family 1” (CPF1) and to animal-like CRYs have challenged these two “dogmas.”5-9
Algal CRYs with Dual and Triple Functions
In the marine diatom P. tricornutum, several CRYs of the different types are present. One of them, PtCPF1 was the first one to be functionally characterized. It is evolutionarily close to the animal-type CRYs and (6-4) photolyases. Therefore, it was examined if it has photolyase activity. PtCPF1 is indeed able to repair (6-4) photoproduct damages.8 Notably, PtCPF1 plays as well a significant role in blue-light regulated gene expression. Transcript levels of several genes were altered after blue light treatment of dark-adapted cells in transgenic lines overexpressing PtCPF1 as compared with wild type.8 The genes belong to functional different groups such as photosynthetic light harvesting, tetrapyrrole biosynthesis, carotenoid biosynthesis, nitrogen metabolism and others. These data suggest a dual function of this algal CRY as (6-4) photolyase and blue light receptor. PtCPF1 was also tested for transcriptional repressor activity in a heterologous system of mammalian COS cells. When expressed there, PtCPF1 inhibits CLOCK-BMAL-mediated transcription of a reporter.4 In the mammalian circadian clock, this repressor activity for the heteromer transcription factor COCK/BMAL is mediated by the CRY/PERIOD complex.10 Thus, PtCPF1 seems also to be involved in regulation of central clock function, at least within a heterologous system.
Several genes belonging to the CPF family were also found in the green primitive alga O. tauri.11 One of them, OtCPF1 is a close relative of PtCPF1. Two of the O. tauri CRYs showed DNA repair activity either of (6-4) photoproducts (OtCPF1) or of CPD-damaged double-stranded DNA (OtCPF2). Both CRYs revealed also transcriptional repressor activity toward CLOCK/BMAL when applied in the above-mentioned reporter assay system of COS cells. Moreover, it was shown that OtCPF1 also controls circadian rhythmicity in O. tauri.11 Thus, in a diatom as well as in a green alga, animal-like CRYs exist, which possess properties of mammalian CRYs as well as photolyase activity. Also in fungi dual activities with regard to photolyases and a role in development were found,5-7 suggesting that photolyases and CRYs have not always been functionally differentiated. Obviously, these properties co-exist in different kingdoms before their separation in higher plants and animals.
An Animal-Like CRY Acting as Functional Blue and Red Light Receptor
Another green alga of which the entire genome is known is the flagellated alga C. reinhardtii that has been used for many years as a model for photosynthesis, flagella formation, light perception and many other processes.12 It contains a plant CRY (known as CPH113) and an animal-type CRY, named aCRY, as well as two DASH-CRYs.9,14,15 aCRY is closely related to PtCPF1 and OtCPF1, but also to (6-4) photolyases and animal type II CRYs. For functional studies, an acry mutant was screened for by an insertional mutagenesis approach. The mutant had the resistance marker introduced within intron seven of the aCRY gene. It expresses aCRY to a reduced level (about 20%) compared with wild type.9 Transcript levels of various genes encoding proteins involved in chlorophyll and carotenoid biosynthesis, light-harvesting complexes, nitrogen metabolism, cell cycle and the circadian clock were investigated in dark adapted cultures of wild type and the acry mutant and after exposure to blue and red light. Some of the selected genes, including the one encoding the light harvesting complex protein LHCBM6 were already known to be induced by blue and red light and to be under control of the blue light receptor phototropin also present in C. reinhardtii.16 Transcript levels were compared for both blue and red light for all chosen transcripts. Surprisingly, most transcripts that were increased by blue light were also upregulated by red light. The transcripts encode the already mentioned LHCBM6 (light harvesting complex of photosynthesis) as well as proteins of chlorophyll biosynthesis (GSA, CHLD, POR), of carotenoid biosynthesis (PDS), of nitrogen metabolism (GLN1), of the cell cycle (CDKB1) and of the circadian clock.9 In the latter case, the transcript levels of the clock-relevant C1 and C3 subunits of the RNA-binding protein CHLAMY117 were strongly (up to 10–15-fold) increased. In contrast, transcript levels of the putative clock-relevant transcription factor ROC1518 were significantly downregulated, again under blue light and red light. Notably, the levels of most of these transcripts were significantly altered in the acry mutant, both under blue and red light conditions, suggesting that aCRY is indirectly or directly involved in the regulation of the transcripts under these light conditions.
In C. reinhardtii, certain processes are regulated by red light such as the light-induced phase resetting of the circadian clock19 or expression of certain genes as mentioned before.16 In some of the above-mentioned genes (LHCBM6 and GSA), a decrease in the level of the blue light receptor PHOT also reduced the red-light mediated effect to some extent albeit not as strong as the blue light effect. However, PHOT as well as the plant CRY from C. reinhardtii can be excluded as red light photoreceptors, because they do not absorb in the red region of the visible spectrum.16,20-22 It seems more likely that PHOT is part of a signaling network together with a red light photoreceptor and thus mediates its effect.16 Phytochrome is the most well known red light photoreceptor in plants. However, from the genome data, no indication exists that C. reinhardtii has a phytochrome.12,14 Since DCMU, an inhibitor of photosynthetic electron transport, did not have any significant effect on the blue- and red-light stimulated expression of the selected genes,16 this lack of effect suggests signaling via a red-light absorbing photoreceptor. Thus, aCRY emerged as a potential candidate. In the acry mutant, the red-light induced increase of many of the studied transcripts was strongly reduced compared with wild type, as already mentioned.9
In accordance with spectroscopic analysis of heterologously expressed aCRY, three redox states can be associated with aCRY (i.e., oxidized, neutral radical and anionic fully reduced FAD). Investigated spectra of the oxidized and fully reduced states of aCRY revealed that these forms are not responsible for the red light effects.9 In contrast, the spectrum of the neutral radical state obtained under reductive conditions had the appropriate spectral characteristics.9 It showed that aCRY is able to absorb also in the yellow (maximum at 585 nm) and red light range (maximum at 633 nm) of the visible spectrum, but not in the far-red range (700 nm). Notably, transcript levels of genes from cells induced under yellow and far-red light were congruent with the spectral properties of aCRY (Fig. 1). Thus, transcript levels of GLN1, PDS, GSA and the C3 subunit were significantly upregulated by yellow light in wild type and the levels were significantly reduced in the acry mutant under these light conditions. In contrast, little if any effect was visible under far-red light treatment. Thus, aCRY acts as a sensory yellow, red and blue light photoreceptor.
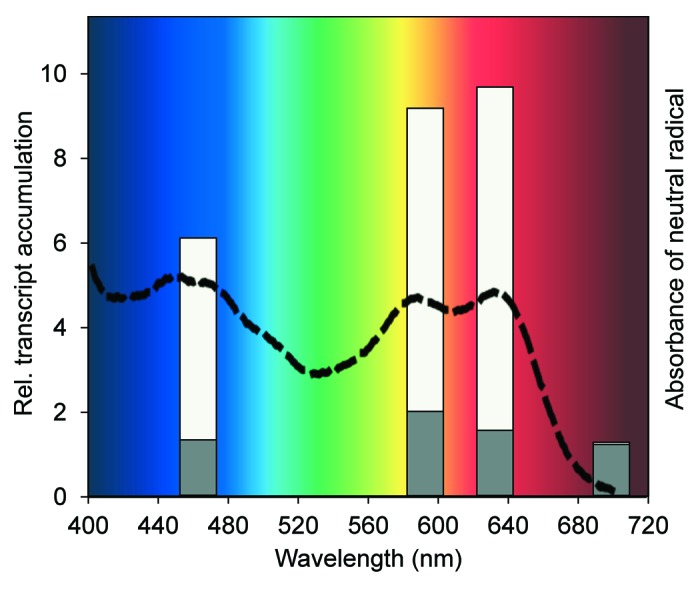
Figure 1. The absorption spectrum of the neutral radical of FAD in aCRY and in vivo responses of C. reinhardtii to light of different spectral qualities. Relative (rel.) transcript accumulation of GLN1 encoding glutamine synthetase 1 was quantified by RT-qPCR in wild type (white bars) and the acry mutant (gray bars; modified from Beel et al.).9 Cells were grown in a light/dark cycle. At the end of the light period, cells were maintained for 60 h in darkness before exposure for 120 min with either blue (465 nm), yellow (590 nm), red (635 nm) or far-red (700 nm) light. The changes in transcript levels following exposure of the cells are presented as fold change relative to RNA from dark-grown cells. For comparison, the absorption spectrum of the neutral radical of FAD in aCRY is shown (dashed line; modified from Beel et al.).9 aCRY was obtained with oxidized FAD from heterologous expression in E. coli. The spectrum of the radical was extracted after conversion with blue light.
Our data also indicate that C. reinhardtii may have another red-light absorbing photoreceptor besides aCRY. The reason for this hypothesis is based on the results obtained with the clock-relevant component ROC15, which is a putative transcription factor. Blue light and red light treatment of dark-adapted cells causes reduced ROC15 transcript levels in wild type.9 In the acry mutant, blue light treatment also reduces ROC15 levels, indicating that this decrease may not be mediated by aCRY. However under red light, ROC15 transcript levels are strongly increased. We assume that this increase in the mutant is due to another red light photoreceptor that is activated by the absence of aCRY. In agreement with such a postulation are the effects in the yellow range. There, ROC15 levels are also reduced in wild type, however they are not upregulated in the mutant as found after red light treatment. Thus, a different red light photoreceptor should exist in addition that does not react to yellow light.
Conclusions and Perspective
Blue light photoreceptors that are not able to absorb red light can still contribute to red light effects if associated with a functional network including red light receptors. The effects of a downregulated LOV-histidine kinase in O. tauri23 or of the downregulated PHOT in C. reinhardtii under red light have been attributed to such networking.16 Also, it is assumed that the impaired red light response in an Arabidopsis double mutant that is defective in both CRY1 and CRY2 is based on the physical interaction between CRYs and phytochromes.24 In all these cases with the two LOV-containing proteins and plant CRYs, the proteins are not able to absorb red light. In contrast, the neutral radical state of aCRY is able to absorb yellow and red light in addition to blue light, but not far-red light.9 The congruent activation of transcript levels under blue, yellow and red light, but not under far-red light reveals that aCRY is acting not only as a sensory blue light receptor. Thus, aCRY represents a long sought-after photoreceptor in C. reinhardtii that is responsive to red light. As mentioned above, there may still be another red light receptor in this organism.
In general, the spectroscopic properties of aCRY are different from so far known higher plant CRY photoperception. For plant CRYs, the dark form is proposed to contain an oxidized flavin,25,26 whereas the broad spectral responses of aCRY point to the neutral radical state in the dark.9 Thus, sensory flavoproteins may not only be categorized as potential blue light receptors in future. One has also to consider yellow and red light activation that are in agreement with the absorption spectrum of the neutral radical state of flavin.
Acknowledgments
Our work was supported by grants of Research Group FOR 1261 from the Deutsche Forschungsgemeinschaft (grants Mi373/11-1 and Mi373/12-1 to M.M. and Ko3580/1-1 to T.K.).
Glossary
Abbreviations:
- aCRY
animal-like cryptochrome
- CRY
cryptochrome
- CPF
cryptochrome photolyase family
- PHOT
phototropin
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22870
References
- 1.Grossman AR. In the grip of algal genomics. Adv Exp Med Biol. 2007;616:54–76. doi: 10.1007/978-0-387-75532-8_6. [DOI] [PubMed] [Google Scholar]
- 2.Hegemann P. Algal sensory photoreceptors. Annu Rev Plant Biol. 2008;59:167–89. doi: 10.1146/annurev.arplant.59.032607.092847. [DOI] [PubMed] [Google Scholar]
- 3.Luck M, Mathes T, Bruun S, Fudim R, Hagedorn R, Nguyen TM, et al. A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by UV and blue light. J Biol Chem. 2012 doi: 10.1074/jbc.M112.401604. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, et al. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–64. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 5.Berrocal-Tito GM, Esquivel-Naranjo EU, Horwitz BA, Herrera-Estrella A. Trichoderma atroviride PHR1, a fungal photolyase responsible for DNA repair, autoregulates its own photoinduction. Eukaryot Cell. 2007;6:1682–92. doi: 10.1128/EC.00208-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayram O, Biesemann C, Krappmann S, Galland P, Braus GH. More than a repair enzyme: Aspergillus nidulans photolyase-like CryA is a regulator of sexual development. Mol Biol Cell. 2008;19:3254–62. doi: 10.1091/mbc.E08-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bluhm BH, Dunkle LD. PHL1 of Cercospora zeae-maydis encodes a member of the photolyase/cryptochrome family involved in UV protection and fungal development. Fungal Genet Biol. 2008;45:1364–72. doi: 10.1016/j.fgb.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Coesel S, Mangogna M, Ishikawa T, Heijde M, Rogato A, Finazzi G, et al. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 2009;10:655–61. doi: 10.1038/embor.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beel B, Prager K, Spexard M, Sasso S, Weiss D, Müller N, et al. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell. 2012;24:2992–3008. doi: 10.1105/tpc.112.098947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 11.Heijde M, Zabulon G, Corellou F, Ishikawa T, Brazard J, Usman A, et al. Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant Cell Environ. 2010;33:1614–26. doi: 10.1111/j.1365-3040.2010.02168.x. [DOI] [PubMed] [Google Scholar]
- 12.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–51. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reisdorph NA, Small GD. The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiol. 2004;134:1546–54. doi: 10.1104/pp.103.031930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittag M, Kiaulehn S, Johnson CH. The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol. 2005;137:399–409. doi: 10.1104/pp.104.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulze T, Prager K, Dathe H, Kelm J, Kiessling P, Mittag M. How the green alga Chlamydomonas reinhardtii keeps time. Protoplasma. 2010;244:3–14. doi: 10.1007/s00709-010-0113-0. [DOI] [PubMed] [Google Scholar]
- 16.Im C-S, Eberhard S, Huang K, Beck CF, Grossman AR. Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J. 2006;48:1–16. doi: 10.1111/j.1365-313X.2006.02852.x. [DOI] [PubMed] [Google Scholar]
- 17.Iliev D, Voytsekh O, Schmidt E-M, Fiedler M, Nykytenko A, Mittag M. A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol. 2006;142:797–806. doi: 10.1104/pp.106.085944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M. A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev. 2008;22:918–30. doi: 10.1101/gad.1650408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo T, Johnson CH, Hastings JW. Action Spectrum for Resetting the Circadian Phototaxis Rhythm in the CW15 Strain of Chlamydomonas: I. Cells in Darkness. Plant Physiol. 1991;95:197–205. doi: 10.1104/pp.95.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kottke T, Heberle J, Hehn D, Dick B, Hegemann P. Phot-LOV1: photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophys J. 2003;84:1192–201. doi: 10.1016/S0006-3495(03)74933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo H, Kottke T, Hegemann P, Dick B. The phot LOV2 domain and its interaction with LOV1. Biophys J. 2005;89:402–12. doi: 10.1529/biophysj.104.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Immeln D, Schlesinger R, Heberle J, Kottke T. Blue light induces radical formation and autophosphorylation in the light-sensitive domain of Chlamydomonas cryptochrome. J Biol Chem. 2007;282:21720–8. doi: 10.1074/jbc.M700849200. [DOI] [PubMed] [Google Scholar]
- 23.Djouani-Tahri B, Christie JM, Sanchez-Ferandin S, Sanchez F, Bouget F-Y, Corellou F. A eukaryotic LOV-histidine kinase with circadian clock function in the picoalga Ostreococcus. Plant J. 2011;65:578–88. doi: 10.1111/j.1365-313X.2010.04444.x. [DOI] [PubMed] [Google Scholar]
- 24.Yang YJ, Zuo ZC, Zhao XY, Li X, Klejnot J, Li Y, et al. Blue-light-independent activity of Arabidopsis cryptochromes in the regulation of steady-state levels of protein and mRNA expression. Mol Plant. 2008;1:167–77. doi: 10.1093/mp/ssm018. [DOI] [PubMed] [Google Scholar]
- 25.Banerjee R, Schleicher E, Meier S, Viana RM, Pokorny R, Ahmad M, et al. The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J Biol Chem. 2007;282:14916–22. doi: 10.1074/jbc.M700616200. [DOI] [PubMed] [Google Scholar]
- 26.Bouly JP, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, Bakrim N, et al. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J Biol Chem. 2007;282:9383–91. doi: 10.1074/jbc.M609842200. [DOI] [PubMed] [Google Scholar]


