Abstract
β-Secretase [also known as the β-site amyloid precursor protein-cleaving enzyme 1 (BACE1)] is an enzyme involved in the production of Aβ-amyloid plaques in the brains of patients with Alzheimer's disease. The enhanced production of this enzyme occurs without corresponding changes in BACE1 mRNA levels. The complex 5′ leader of BACE1 mRNA contains three upstream ORFs (uORFs) preceding the BACE1 initiation codon. In this study, we investigated how this 5′ leader affects translation efficiency as a first step in understanding the enhanced production of the enzyme in the disease. Using reporter constructs in transfected mammalian cell lines and cell-free lysates, we showed that the translation mediated by the BACE1 5′ leader is cap-dependent and inhibited by cis-acting segments contained within the 5′ leader. Disruption of the uORFs had no effect on translation in B104 cells, which was surprising because the first two AUGs reside in contexts able to function as initiation codons. Possible mechanisms to explain how ribosomes bypass the uORFs, including reinitiation, leaky scanning, and internal initiation of translation were found to be inconsistent with the data. The data are most consistent with a model in which ribosomes shunt uORF-containing segments of the 5′ leader as the ribosomes move from the 5′ end of the mRNA to the initiation codon. In PC12 cells, however, the second uORF appears to be translated. We hypothesize that the translation efficiency of the BACE1 initiation codon may be increased in patients with Alzheimer's disease by molecular mechanisms that enhance shunting or increase the relative accessibility the BACE1 initiation codon.
Alzheimer's disease is a progressive neurodegenerative disease characterized by neuritic plaques composed of short, insoluble β-pleated peptides (Aβ) that are proteolytic cleavage products of an integral membrane protein known as the amyloid precursor protein (1, 2). β-Secretase enzyme, also known as the β-site amyloid precursor protein-cleaving enzyme (BACE), is a membrane-bound aspartic protease that catalyzes the first step of amyloid precursor protein cleavage (3). The gene encoding β-secretase, BACE1, has been identified (3–7) and validated by using various criteria (see refs. 8–12).
In the brains of patients with Alzheimer's disease, β-secretase protein levels are as much as 2.7-fold higher than in control brains (13–15), and the overexpression of this enzyme is sufficient to increase Aβ levels in transgenic mice (reviewed in ref. 16). However, when Alzheimer's disease brains were compared with those of controls, BACE1 mRNA levels were not significantly different from normal (17–19), suggesting that β-secretase may be overexpressed in this disease because of an increase in the rate of translation of the BACE1 mRNA.
The possibility that the translation efficiency of BACE1 mRNA is increased in Alzheimer's disease prompted the present investigation of the 5′ leader sequence. The 5′ leader sequences of the mouse, rat, and human BACE1 mRNAs are strikingly similar, with three upstream ORFs (uORFs), the first two of which contain AUGs that reside in good contexts to function as initiation codons (20). β-Secretase translation initiates, however, at a codon located downstream of these uORFs. In eukaryotes, translation is hypothesized to initiate predominantly by a cap-binding/scanning mechanism in which ribosomes are recruited at the 5′ cap-structure, scan in a 3′ direction, and initiate translation at the first AUG (21). Although this hypothesis is consistent with the translation of most mRNAs, many others contain one or more AUGs upstream of the initiation codon (reviewed in ref. 22). In some mRNAs, the translation of an uORF blocks the translation of a downstream cistron by diverting the initiation complex from that cistron (22, 23). For other mRNAs, the presence of an uORF does not affect the translation of the downstream cistron as much as might be expected, and thus other mechanisms have been invoked, such as reinitiation, leaky scanning, internal initiation, or shunting (21, 24–31).
BACE1 mRNA provides an opportunity to assess the role of upstream AUGs in the translation of a clinically important cellular mRNA. We investigated the mechanism used to initiate the translation of this mRNA in cell-free lysates, transfected B104 neuroblastoma, and PC12 pheochromocytoma cells. We found that the translation mediated by the BACE1 5′ leader occurs by a cap-dependent mechanism in which ribosomes are recruited to a site located upstream of the uORFs. We also identified sequence elements contained within the BACE1 5′ leader that inhibit translation efficiency. The data indicate that in B104 cells 40S ribosomal subunits completely bypass the three uORFs, possibly by a shunting mechanism, whereas in PC12 cells translation of the second uORF occurs. These results suggest that the translation of the BACE1 mRNA is affected by factors or conditions that alter the efficiency with which 40S ribosomal subunits recognize the upstream AUGs as initiation codons.
Materials and Methods
Isolation and Cloning of BACE1 5′ Leader Sequence. The BACE1 5′ leader was obtained by PCR amplification from rat brain cDNA by using primers based on the rat sequence (accession no. NM_019204): sense primer, 5′-GACTGAATTC CCCCAGCCTG CCTAGGTGC-3′, and antisense primer, 5′-GACTCCATGG TGAGCCCGGG CCTTGTG-3′. An EcoRI restriction site was introduced into the sense primer. The antisense primer contains a naturally occurring NcoI site that overlaps the initiation codon. A reporter construct containing this 5′ leader [pcDNA 3.1(+)-BACE1] was generated by digesting the PCR product with EcoRI and NcoI and cloning it and the Photinus luciferase cistron (the NcoI–XbaI fragment from pGL3-Control; Promega) into pcDNA 3.1(+) (Invitrogen) by using the EcoRI and XbaI restriction sites. A short linker sequence containing the HindIII restriction site (5′-GCTAGCTCAAGCTTCGAATTC-3′) was inserted upstream of the BACE1 5′ leader by using the NheI and EcoRI restriction sites. A control vector containing the 52-nt mouse β-globin 5′ UTR (accession no. J00413) was constructed in a similar fashion. Hairpin-containing constructs were generated by inserting the following sequence into the HindIII restriction site: 5′-AAGCTTGGCA TTCCGGTACT GAATTGATTA GATCTGGTAC CGAGCTCCCC GGGCTGCAGC CCGGGGAGCT CGGTACCCAG ATCTAATCAA GCTCTAGAAG CTT-3′ (complementary sequences are underlined). Deletions were generated by PCR amplification with primers containing EcoRI and NcoI restriction sites and cloned into the pcDNA 3.1(+)-BACE1 vector. Individual and multiple point mutations were introduced into oligonucleotides, amplified by PCR, restriction-digested, and ligated into the pcDNA 3.1(+)-BACE1 vector, replacing the wild-type sequence. Some mutations were generated by using the QuikChange XL II kit (Stratagene). The β-globin/BACE1 upstream AUG vectors were constructed by replacing nucleotides 34–43 of the mouse β-globin 5′ UTR with each of the BACE1 upstream AUGs (uAUG1–uAUG4) and their flanking nucleotides (six nucleotides upstream and one nucleotide downstream), the BACE1 initiation codon, or an optimal consensus sequence. For uAUG1, the flanking nucleotides included two downstream nucleotides. The resulting uORF overlapped the luciferase cistron in a different reading frame for six amino acids. For the dicistronic mRNA analyses, the rat BACE1 5′ leader and β-globin 5′ UTR were cloned into the RP and RPh vectors as described (32).
Analysis of BACE1 5′ Leader in Reporter Gene Constructs by Transient Transfection. The cell lines used in this study were rat B104 neuroblastoma cells, which were derived from nitrosoethylurea-induced tumors in newborn rats (33) and PC12 cells, which were derived from a rat pheochromocytoma and can be induced to express a neuronal phenotype by nerve growth factor (34). Reporter constructs (0.2 μg) were transfected into 1–3 × 105 cells by using FuGENE 6 (Roche Diagnostics) according to manufacturer's instructions. Cells were cotransfected with the pCMVβ vector (Clontech), which expresses the lacZ gene, to normalize for transfection efficiency. Cells were harvested 24 h after transfection, and luciferase and β-galactosidase activities were determined as described (32).
Northern Blot Analyses. Total RNA from transiently transfected cells was prepared with TRIzol (GIBCO/BRL), and Northern blots containing 10–15 μg of total RNA per lane were prepared with Brightstar-Plus nylon membrane (Ambion, Austin, TX) and hybridized with Ultrahyb Northern Blot solution (Ambion) with RNA probes complementary to Photinus luciferase and lacZ-coding regions. Signals on Northern blots were quantified by using a PhosphorImager (Molecular Dynamics).
Cell-Free Translation. Capped or uncapped mRNA templates were transcribed in vitro by using mMessage mMachine and Megascript kits (Ambion), respectively, and quantitated by UV absorption at 260 nm. The size and integrity of transcripts were assessed by electrophoresis of 1 μg on a formaldehyde/Mops denaturing gel with known size standards. In vitro translation reactions with nuclease-treated rabbit reticulocyte lysate (Promega) were performed with 50% (vol/vol) lysate/50 μM amino acids (Promega)/125 mM KCl/2 mM DTT/0.25 unit/μl Superase·In (Ambion)/0.025 μg/μl mRNA. The endogenous Mg2+ concentration of the lysate was 1.93 mM (determined by Promega). m7GpppG cap analogue (Ambion) was added to 200 μM (final concentration) where indicated. Reactions were assembled on ice, lysate was the final component added, and reactions were incubated at 30°C for 60 min.
Results
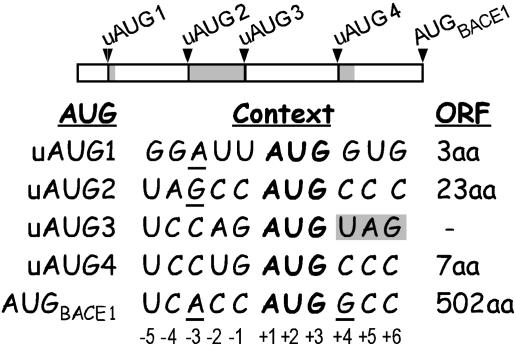
The BACE1 5′ Leader Contains Four Upstream AUGs. The 427-nt rat BACE1 5′ leader was cloned by using primers based on the rat BACE1 mRNA sequence (see Materials and Methods). This 5′ leader contains four upstream AUGs (uAUG1–uAUG4; Fig. 1). In a 5′ to 3′ orientation, the first, second, and fourth AUGs give rise to uORFs containing 3, 23, and 7 codons, respectively, all of which terminate upstream of the BACE1 initiation codon. The third upstream AUG immediately precedes the termination codon of the second uORF. Comparison of the rat, mouse, and human BACE1 5′ leaders revealed that they contain uORFs of the same size and at similar positions (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
Nucleotide context of AUGs contained within the rat BACE1 5′ leader. (Upper) A schematic representation of the BACE1 5′ leader. Arrowheads and black bars are used to designate the positions of the BACE1 initiation codon (AUGBACE1) and four upstream AUGs (uAUG1–uAUG4). The uORFs are indicated as gray bars. (Lower) Table showing the nucleotide sequences flanking the first five AUGs (indicated in bold) and the lengths of the resulting ORFs. Nucleotides that might augment utilization of the upstream AUGs are underlined. The termination codon that follows immediately after uAUG3 is shaded. Nucleotide numbering is indicated below the sequence.
The nucleotide contexts (-3 to + 4) of the upstream AUGs for the three uORFs in rat, mouse, and human BACE1 mRNAs are identical. The first AUG contains a purine at position -3 and a G at position +4, which would ordinarily confer an optimal context for translation initiation; however, it is followed immediately by a U at position +5, which can reduce the effect of a G at position +4 (20). The second AUG also contains a purine at position -3 and therefore resides in good context to function as an initiation codon. In contrast, the third and fourth AUGs contain pyrimidines at both positions -3 and +4, and therefore are in poor contexts to function as initiation codons. The BACE1 initiation codon itself (AUGbace1) resides in an excellent context to function as an initiation codon, with an A at position -3 and a G at position +4.
The rat, mouse, and human BACE1 5′ leaders have GC contents of 66%, 67%, and 76%, respectively, and are predicted to fold into various highly stable structures [up to -125 kcal/mol (1 kcal = 4.18 kJ); ref. 35]. The presence of three uORFs and the potential to form stable RNA secondary structures suggests that the BACE1 mRNA might be translated inefficiently, a possibility we analyzed and which subsequent studies excluded.
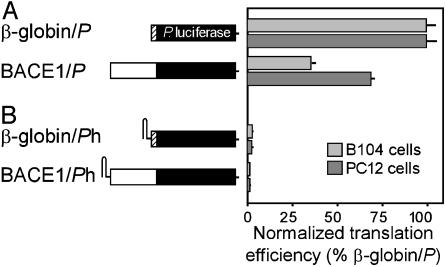
The BACE1 5′ Leader Mediates Translation Relatively Efficiently by a Cap-Dependent Mechanism. The translation efficiency of the BACE1 5′ leader was tested by using reporter constructs containing the Photinus luciferase cistron (BACE1/P; Fig. 2A) in transiently transfected B104 and PC12 cells, two rat cell lines in which we detected expression of BACE1 mRNA. In this experiment, luciferase activities were measured and normalized to the mRNA levels of the reporter construct to obtain translation efficiencies. These translation efficiencies were then compared with those obtained with reporter constructs containing the 5′ leader of the efficiently translated β-globin mRNA (β-globin/P). The results showed that the translation mediated by the BACE1 5′ leader was ≈36% and 69% as efficient as that mediated by the β-globin 5′ UTR in B104 and PC12 cells, respectively.
Fig. 2.
Translation properties of a reporter construct containing the BACE1 5′ leader in transfected cells. A schematic representation of the constructs is indicated to the left. The luciferase cistron is indicated as a black bar, and the β-globin and BACE1 5′ leaders are indicated as hatched and white bars, respectively. (A) Reporter constructs were transiently transfected into B104 or PC12 cells, and translation efficiencies were determined by normalizing luciferase activities to reporter mRNA levels. The translation efficiencies are indicated relative to those obtained with the β-globin/P construct, which is represented as a normalized translation efficiency of 100. (B) Relative translation efficiencies of constructs containing a hairpin structure (h) at the 5′ end of the β-globin and BACE1 5′ leaders.
To determine whether the translation mediated by the BACE1 5′ leader was cap-dependent, we first investigated whether ribosomes were recruited at the 5′ end of this leader. For these experiments a stable stem–loop structure was introduced at the 5′ end of the mRNA to inhibit cap-dependent translation, either by masking the cap structure or blocking scanning (36, 37). In these experiments, the hairpin structure blocked translation by >98% in both cell lines (Fig. 2B). Similar results were obtained with a construct containing the 5′ UTR of the cap-dependent β-globin mRNA, which served as a positive control.
When these mRNAs were transcribed in vitro and translated in a nuclease-treated rabbit reticulocyte lysate, comparable results were obtained, i.e., the hairpin structures blocked the translation of both mRNAs (Table 1). The hairpin structure had a less pronounced effect on the translation of the BACE1/P mRNA in the cell-free lysate than in transfected cells. To test directly whether the BACE1 5′ leader recruits the translation machinery by a cap-dependent mechanism, capped reporter mRNAs were translated in the presence or absence of m7GpppG, which inhibits cap-dependent translation (37–39) by competing for binding to initiation factor eIF4E. The results showed that m7GpppG inhibited the translation of capped mRNAs containing either the BACE1 or β-globin 5′ leader to a similar extent, ≈97% and 94%, respectively (Table 1), but it did not affect the translation of uncapped transcripts (data not shown). These results further indicate that the BACE1 5′ leader initiates translation by a cap-dependent mechanism.
Table 1. Effects of a 5′ stem–loop structure and m7GpppG cap analogue on translation mediated by the BACE1 5′ leader.
| mRNA construct | m7G | Normalized translation efficiencies (SD) |
|---|---|---|
| β-Globin/P | – | 100.0 |
| β-Globin/Ph | – | 0.7 (0.2) |
| β-Globin/P | + | 3.1 (0.4) |
| BACE1/P | – | 100.0 |
| BACE1/Ph | – | 11.5 (6.5) |
| BACE1/P | + | 5.8 (0.7) |
mRNA constructs were in vitro transcribed and translated in rabbit reticulocyte lysate. As indicated, 200 μM m7GpppG (m7G) was included (+) or omitted (–) from reactions. Translation efficiencies were determined by normalizing luciferase activities for β-globin/P and BACE1/P mRNAs to 100. h, hairpin structure.
Although the results above are inconsistent with an internal translation initiation mechanism by an internal ribosome entry site (IRES), this possibility was independently evaluated by testing this 5′ leader in the intercistronic region of various dicistronic mRNAs (as in ref. 39). Using these constructs, we did not detect IRES activity in either transfected cells or cell-free lysates (data not shown).
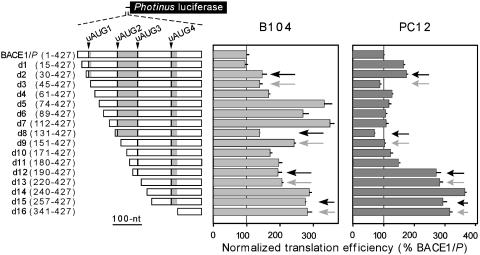
Deletions and Point Mutations of the BACE1 5′ Leader Indicate That Translation of the uORFs Depends on the Cell Type. Sequential deletions from the 5′ end of the BACE1 5′ leader were tested in transiently transfected B104 and PC12 cells to evaluate the effects of the four upstream AUGs on translation efficiency and to identify the general locations of potential cis-acting sequences that may affect the translation efficiency (Fig. 3). Ribosomes initiating translation at the upstream AUGs might be expected to divert ribosomes from the main cistron and decrease its translation. However, comparing pairs of mutations in which an upstream AUG is present or absent, e.g., d2 versus d3, showed no differences or small (<2-fold) differences, suggesting that these upstream AUGs were not recognized efficiently as initiation codons in either cell line. However, the translation efficiencies increased 3-fold with the deletion of nucleotides 61–74 in B104 cells, whereas in PC12 cells, deletion of nucleotides 180–190 increased translation ≈3.5-fold, which was more efficient than the level of translation obtained with the construct containing the β-globin 5′ UTR. The results suggest that these two segments of the BACE1 5′ leader inhibit translation in a cell-type-specific manner.
Fig. 3.
Deletion analysis of the BACE1 5′ leader in transfected cells. The constructs are all based on BACE1/P, which contains the full-length 5′ leader (nucleotides 1–427). Constructs d1–d16 contain sequential deletions of the BACE1 5′ leader. The nucleotides contained within these constructs are indicated in parentheses. The black and gray arrows point to deletions that contain or lack an upstream AUG, respectively. In each cell line, the translation efficiencies are indicated relative to those obtained with the BACE1/P construct, which is represented as a normalized translation efficiency of 100.
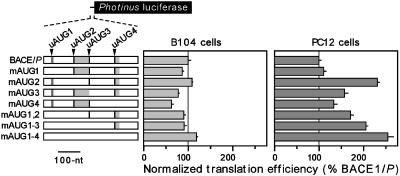
Although it is striking that deletion of the upstream AUGs had little or no effect on translation efficiency, the deletions themselves may have affected translation efficiencies by altering the length of the 5′ leader, the locations of the upstream AUGs relative the 5′ end of the mRNA, or RNA secondary or tertiary structures. We therefore evaluated upstream AUG utilization in the context of the full-length 5′ leader by mutating the upstream AUGs to AUU, either individually or in combination. The effects of these mutations on translation efficiency were determined in transiently transfected cells (Fig. 4). As in the deletion analysis, we expected that translation of the reporter gene would be blocked by translation initiating at an upstream AUG and enhanced by the disruption of actively translated uORFs. However, in B104 cells, only minor differences in translation efficiencies were observed when any or all of the upstream AUGs were mutated, suggesting that the corresponding unmutated AUGs were not used efficiently as initiation codons in these cells. In PC12 cells, mutation of the first upstream AUG had no effect on translation efficiency; however, mutation of the second upstream AUG increased efficiency ≈2-fold. Mutations of the third and fourth upstream AUGs had smaller effects, whereas mutation of all four upstream AUGs increased translation 2.5-fold. These results suggest that the unmutated second, third, and fourth AUGs may be used as initiation codons to some extent, inhibiting translation from the BACE1 initiation codon.
Fig. 4.
Disruption of BACE1 upstream AUGs in transfected cells. The upstream AUGs were mutated to AUU, either individually or in combinations, as indicated by the disappearance of the black and gray bars representing the upstream AUGs and uORFs, respectively. In each cell line, the translation efficiencies are indicated relative to those obtained with the BACE1/P construct, which is represented as a normalized translation efficiency of 100.
The BACE1 Upstream AUGs Function as Initiation Codons in the Context of the β-Globin 5′ UTR. Our results showed that the BACE1 upstream AUGs were not used in B104 cells and that the first upstream AUG was not used in PC12 cells. One possibility is that the upstream AUGs were not present in the mRNAs; for example, shorter mRNAs lacking the upstream AUGs might be generated by a promoter element located 3′ of the upstream AUGs or by nuclease cleavage. However, these possibilities are not consistent with our findings showing that translation could be blocked by a hairpin structure at the 5′ end of the BACE1 5′ leader or by a cap analogue (Fig. 2 and Table 1). The results indicate that the transcripts were full-length, a conclusion supported by performing Northern blot, RT-PCR, and primer extension analyses, which showed that the mRNA transcripts produced from these constructs were full-length and not fragmented (data not shown).
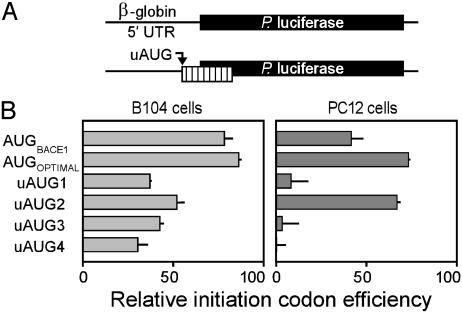
To determine whether the upstream AUGs were bypassed because the translation machinery did not efficiently recognize them as initiation codons, we individually tested (Fig. 5A) each of these initiation codons and their flanking nucleotides in the context of the β-globin 5′ UTR (see Materials and Methods), a UTR consisting of a short, relatively unstructured sequence with an initiation codon that resides in an optimal context. Ribosomes initiating translation at the upstream AUG should be precluded from translating the luciferase cistron, because the upstream AUG yields an ORF that overlaps the luciferase cistron but in a different reading frame. The relative efficiencies of the various initiation codons were determined by their ability to reduce luciferase activity. In addition, we tested the authentic BACE1 initiation codon (AUGBACE1) and an AUG in an optimal context (AUGOPTIMAL; ref. 20).
Fig. 5.
Ability of BACE1 upstream AUGs to function as initiation codons in a different context. (A) The constructs in this study were based on the β-globin/P reporter construct containing the β-globin 5′ UTR, which is indicated as a black line linked to the luciferase reporter gene, which is indicated as a black bar. The AUG codons and flanking regions of the four BACE1 upstream AUGs (uAUG1–uAUG4), the BACE1 initiation codon (AUGBACE1), and an AUG in an optimal context to function as an initiation codon (AUGOPTIMAL) were individually inserted into the β-globin 5′ leader. The uORF derived from these upstream AUGs is indicated as a vertically striped bar. (B) In each cell line, the relative initiation codon efficiencies are indicated relative to those obtained with the β-globin/P construct, which is represented as a normalized translation efficiency of 100.
The results showed that all the upstream AUGs had the ability to be used as initiation codons to various extents (Fig. 5B). The relative efficiency of the authentic BACE1 initiation codon was ≈80% in B104 cells and 40% in PC12 cells. Although this initiation codon lies in an excellent context, i.e., an A at position -3 and a G at position + 4, it was not as efficient as the optimal sequence (AUGOPTIMAL) which contains a C at position -5. In B104 cells, all four upstream AUGs were used as initiation codons, although differences due to nucleotide context appeared to be minor. In PC12 cells, however, only the second upstream AUG was used efficiently; indeed, it was more efficient than the BACE1 initiation codon. Overall, these findings suggest that, in addition to context, other factors lead to differential utilization of these initiation codons in different cell types.
Discussion
In Alzheimer's disease, levels of the β-secretase enzyme increase without corresponding changes in the amount of the BACE1 mRNA. These observations indicate that β-secretase protein levels are modulated by one or more posttranscriptional mechanisms that determine the stability of the protein or the translation efficiency of the BACE1 mRNA. In this article, we investigated factors affecting the mechanism used by the BACE1 mRNA to initiate translation.
We provide evidence suggesting that translation occurs by an unusual mechanism that is not consistent with processive scanning of the 5′ leader or with internal initiation of translation, two mechanisms that are thought to explain the translation of most eukaryotic mRNAs. Rather, the translation mediated by the BACE1 5′ leader appears to occur by a cap-dependent mechanism in which ribosomes are first recruited at the 5′ end and then shunt or skip over segments of the 5′ leader as they move to the initiation codon. Translation mediated by this 5′ leader differed in different cell lines, suggesting that cell-specific factors might affect the efficiency of shunting. These results raise the possibility that comparable mechanistic alterations underlie the increased translation efficiency of the BACE1 mRNA in Alzheimer's disease.
We suggest that in B104 cells the 5′ leader of the BACE1 mRNA mediates translation initiation by shunting, because other mechanisms based on current models (31) are not consistent with the data. The 5′ leader contains four upstream AUGs that give rise to three uORFs (see Fig. 1). Translation of an uORF generally inhibits the translation efficiency of a downstream cistron (40). Indeed, our own data (Fig. 5) showed that the four upstream AUG sequences could actually function as initiation codons when tested in the β-globin 5′ UTR; thus, all four AUG sequences might be expected to be recognized by most of the ribosomes recruited on the mRNA. However, disruption of the upstream AUGs did not affect translation efficiency, indicating that the uORFs were not translated in B104 cells.
Several mechanisms might account for the data. The results might be explained if the translation machinery was recruited at an IRES located 3′ of the upstream AUGs or if the upstream AUGs were bypassed by other mechanisms such as leaky scanning, reinitiation, or shunting. In the following discussion, we consider each of these mechanisms in turn.
The possibility that the upstream AUGs were bypassed by an IRES was not supported by the data; the BACE1 5′ leader did not function as an IRES when tested in dicistronic mRNAs. Moreover, we provide compelling evidence that reporter mRNAs containing the BACE1 5′ leader were translated in a cap-dependent manner (Fig. 2 and Table 1).
The notion that the upstream AUGs were bypassed by a scanning mechanism was also not supported by the data. Indeed, translation mediated by the BACE1 5′ leader in B104 cells appears to be completely independent of the upstream AUGs as shown by the results of our deletion and mutation experiments (Figs. 3 and 4).
Leaky scanning is an extension of the scanning model (reviewed in ref. 21) and suggests that an AUG might be bypassed by scanning ribosomes if it resides in a suboptimal context. If leaky scanning is presumed to account for the results obtained in B104 cells, all four of the BACE1 upstream AUGs would have to be completely bypassed. This possibility is considered unlikely because the nucleotide contexts of all four upstream AUGs were shown to be sufficient to initiate translation in a synthetic reporter construct (Fig. 5).
Reinitiation occurs when some of the ribosomes that terminate translation of an uORF remain associated with the mRNA, scan downstream, and reinitiate translation at another initiation codon. Although inefficient, reinitiation can occur after the translation of a short uORF, and reinitiation rates of up to ≈50% have been reported for the yeast GCN4 mRNA (24) and for synthetic constructs (41). The first uORF in the BACE1 5′ leader is short, and reinitiation was considered a possibility; however, this mechanism appears unlikely inasmuch as disruption of uORF1 by deletion or mutation did not affect translation efficiency, a result which would require that, in the presence of uORF1, the reinitiation efficiency was 100%. Moreover, similarly efficient reinitiation rates from the other uORFs would have had to occur to explain the results obtained with the deletion and mutation constructs (Fig. 3).
Although translation mediated by the BACE1 5′ leader is not consistent with either internal initiation or scanning mechanisms, it remains possible that the upstream AUGs are bypassed by a shunting mechanism. Shunting has been studied extensively in the cauliflower mosaic virus RNA, where ribosomes bypass a stable RNA secondary structure and several upstream AUGs before initiating translation (42). On closer inspection, it was shown that this shunting involved the translation of a short uORF at the base of the stem–loop and thus occurred by an unusual type of reinitiation. Shunting was also postulated to explain the translation of several other mRNAs (reviewed in ref. 31), including the adenovirus and hsp70 mRNAs, for which shunting was thought to be mediated by cis-acting sequences with complementarity to 18S rRNA (43).
Shunting might occur by interactions of the translation machinery with discrete sites in the 5′ leader that involve scanning of segments of the 5′ leader. Alternatively, shunting ribosomes might completely bypass the 5′ leader by base pairing between the initiator Met-tRNA and an accessible initiation codon. We have recently proposed (44) that one of the factors influencing initiation codon recognition is its relative accessibility to the translation machinery. Accessibility of initiation codons might be affected by RNA secondary structures or RNA-binding proteins that act either to increase the accessibility of the initiation codon or to mask upstream AUGs.
The evidence for a shunting mechanism is most clear-cut in B104 cells because none of the uORFs appeared to be translated. In PC12 cells, the data indicated that the second upstream AUG was translated (Fig. 4), a result that is consistent with the relative efficiencies of the four upstream AUGs in these cells (Fig. 5). However, the results of these experiments do not distinguish between leaky scanning and shunting as possible mechanisms used by PC12 ribosomes to initiate translation at the second upstream AUG.
The results obtained in cell lines raise the possibility that the translation properties of the BACE1 mRNA may be altered in Alzheimer's disease. For example, the translation of an uORF might to some extent inhibit BACE1 translation in the normal brain, whereas, during Alzheimer's disease, translation might increase because of a shunting mechanism that enables ribosomes to bypass the upstream AUGs. Testing this notion will require experiments to be performed in neurons obtained from the brains of patients with the disease, a prospect that may be possible given that neurons have been isolated and cultured from elderly postmortem brains (e.g., refs. 45 and 46).
In Alzheimer's disease, it may be that the BACE1 initiation codon becomes more accessible, increasing shunting and translation efficiencies. The accessibility of upstream AUGs and the BACE1 initiation codon are likely to be determined by RNA secondary structures and tertiary interactions that are stabilized by RNA-binding proteins. Another possibility is related to the inhibitory sequences identified in the deletion analysis of the present study. In normal brains, such sequences might adopt particular RNA secondary structures or bind to trans-acting factors that inhibit translation, perhaps by masking the BACE1 initiation codon. In Alzheimer's disease, these sequences might no longer inhibit translation because the RNA secondary structure is modified or because the expression or activity of a binding protein is altered.
Supplementary Material
Acknowledgments
We thank Dr. Jason Pinkstaff for data regarding the expression of the BACE1 mRNA in B104 and PC12 cells, Luke Burman and Holly Collette for excellent technical assistance, and Drs. Frederick Jones and Joseph Gally for critical reading of the manuscript. This work was supported by funding from the G. Harold and Leila Y. Mathers Foundation and by National Institutes of Health Grant GM61725 (to V.P.M.), by U.S. Public Health Service Grant HD09635 (to G.M.E.), and by a Skaggs post-doctoral fellowship (to G.W.R.).
Abbreviations: BACE1, β-site amyloid precursor protein-cleaving enzyme 1; uORF, upstream ORF; IRES, internal ribosome entry site.
References
- 1.Bayer, T. A., Wirths, O., Majtenyi, K., Hartmann, T., Multhaup, G., Beyreuther, K. & Czech, C. (2001) Brain Pathol. 11, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenblum, W. I. (1999) J. Neuropathol. Exp. Neurol. 58, 575-581. [DOI] [PubMed] [Google Scholar]
- 3.Vassar, R., Bennett, B. D., Babu-Khan, S., Kahn, S., Mendiaz, E. A., Denis, P., Teplow, D. B., Ross, S., Amarante, P., Loeloff, R., et al. (1999) Science 286, 735-741. [DOI] [PubMed] [Google Scholar]
- 4.Sinha, S., Anderson, J. P., Barbour, R., Basi, G. S., Caccavello, R., Davis, D., Doan, M., Dovey, H. F., Frigon, N., Hong, J., et al. (1999) Nature 402, 537-540. [DOI] [PubMed] [Google Scholar]
- 5.Yan, R., Bienkowski, M. J., Shuck, M. E., Miao, H., Tory, M. C., Pauley, A. M., Brashier, J. R., Stratman, N. C., Mathews, W. R., Buhl, A. E., et al. (1999) Nature 402, 533-537. [DOI] [PubMed] [Google Scholar]
- 6.Hussain, I., Powell, D., Howlett, D. R., Tew, D. G., Meek, T. D., Chapman, C., Gloger, I. S., Murphy, K. E., Southan, C. D., Ryan, D. M., et al. (1999) Mol. Cell. Neurosci. 14, 419-427. [DOI] [PubMed] [Google Scholar]
- 7.Lin, X., Koelsch, G., Wu, S., Downs, D., Dashti, A. & Tang, J. (2000) Proc. Natl. Acad. Sci. USA 97, 1456-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vassar, R. (2002) Adv. Drug Delivery Rev. 54, 1589-1602. [DOI] [PubMed] [Google Scholar]
- 9.Cai, H., Wang, Y., McCarthy, D., Wen, H., Borchelt, D. R., Price, D. L. & Wong, P. C. (2001) Nat. Neurosci. 4, 233-234. [DOI] [PubMed] [Google Scholar]
- 10.Roberds, S. L., Anderson, J., Basi, G., Bienkowski, M. J., Branstetter, D. G., Chen, K. S., Freedman, S. B., Frigon, N. L., Games, D., Hu, K., et al. (2001) Hum. Mol. Genet. 10, 1317-1324. [DOI] [PubMed] [Google Scholar]
- 11.Luo, Y., Bolon, B., Kahn, S., Bennett, B. D., Babu-Khan, S., Denis, P., Fan, W., Kha, H., Zhang, J., Gong, Y., et al. (2001) Nat. Neurosci. 4, 231-232. [DOI] [PubMed] [Google Scholar]
- 12.Bodendorf, U., Danner, S., Fischer, F., Stefani, M., Sturchler-Pierrat, C., Wiederhold, K. H., Staufenbiel, M. & Paganetti, P. (2002) J. Neurochem. 80, 799-806. [DOI] [PubMed] [Google Scholar]
- 13.Holsinger, R. M., McLean, C. A., Beyreuther, K., Masters, C. L. & Evin, G. (2002) Ann. Neurol. 51, 783-786. [DOI] [PubMed] [Google Scholar]
- 14.Fukumoto, H., Cheung, B. S., Hyman, B. T. & Irizarry, M. C. (2002) Arch. Neurol. 59, 1381-1389. [DOI] [PubMed] [Google Scholar]
- 15.Yang, L. B., Lindholm, K., Yan, R., Citron, M., Xia, W., Yang, X. L., Beach, T., Sue, L., Wong, P., Price, D., et al. (2003) Nat. Med. 9, 3-4. [DOI] [PubMed] [Google Scholar]
- 16.Zohar, O., Cavallaro, S., D'Agata, V. & Alkon, D. L. (2003) Mol. Brain Res. 115, 63-68. [DOI] [PubMed] [Google Scholar]
- 17.Yasojima, K., McGeer, E. G. & McGeer, P. L. (2001) Brain Res. 919, 115-121. [DOI] [PubMed] [Google Scholar]
- 18.Gatta, L. B., Albertini, A., Ravid, R. & Finazzi, D. (2002) NeuroReport 13, 2031-2033. [DOI] [PubMed] [Google Scholar]
- 19.Preece, P., Virley, D. J., Costandi, M., Coombes, R., Moss, S. J., Mudge, A. W., Jazin, E. & Cairns, N. J. (2003) Brain Res. Mol. Brain Res. 116, 155-158. [DOI] [PubMed] [Google Scholar]
- 20.Kozak, M. (1997) EMBO J. 16, 2482-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak, M. (1999) Gene 234, 187-208. [DOI] [PubMed] [Google Scholar]
- 22.Meijer, H. A. & Thomas, A. A. M. (2002) Biochem. J. 367, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geballe, A. P. & Sachs, M. S. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 595-614.
- 24.Hinnebusch, A. G., Wek, R. C., Dever, T. E., Cigan, A. M., Feng, L. & Donahue, T. F. (1993) in Translational Regulation of Gene Expression 2 (Plenum, New York), pp. 87-116.
- 25.Gaba, A., Wang, Z., Krishnamoorthy, T., Hinnebusch, A. G. & Sachs, M. S. (2001) EMBO J. 20, 6453-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herzog, E., Guilley, H. & Fritsch, C. (1995) Virology 208, 215-225. [DOI] [PubMed] [Google Scholar]
- 27.Kos, M., Denger, S., Reid, G. & Gannon, F. (2002) J. Biol. Chem. 277, 37131-37138. [DOI] [PubMed] [Google Scholar]
- 28.Hellen, C. U. & Sarnow, P. (2001) Genes Dev. 15, 1593-1612. [DOI] [PubMed] [Google Scholar]
- 29.Vagner, S., Galy, B. & Pyronnet, S. (2001) EMBO Rep. 2, 893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryabova, L. A., Pooggin, M. M. & Hohn, T. (2002) Prog. Nucleic Acid Res. Mol. Biol. 72, 1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson, R. J. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 127-183.
- 32.Chappell, S. A., Edelman, G. M. & Mauro, V. P. (2000) Proc. Natl. Acad. Sci. USA 97, 1536-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schubert, D., Heinemann, S., Carlisle, W., Tarikas, H., Kimes, B., Patrick, J., Steinbach, J. H., Culp, W. & Brandt, B. L. (1974) Nature 249, 224-227. [DOI] [PubMed] [Google Scholar]
- 34.Greene, L. A. & Tischler, A. S. (1976) Proc. Natl. Acad. Sci. USA 73, 2424-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker, M. (1989) Science 244, 48-52. [DOI] [PubMed] [Google Scholar]
- 36.Kozak, M. (1989) Mol. Cell. Biol. 9, 5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chappell, S. A., Owens, G. C. & Mauro, V. P. (2001) J. Biol. Chem. 276, 36917-36922. [DOI] [PubMed] [Google Scholar]
- 38.Cai, A., Jankowska-Anyszka, M., Centers, A., Chlebicka, L., Stepinski, J., Stolarski, R., Darzynkiewicz, E. & Rhoads, R. E. (1999) Biochemistry 38, 8538-8547. [DOI] [PubMed] [Google Scholar]
- 39.Chappell, S. A. & Mauro, V. P. (2003) J. Biol. Chem. 278, 33793-33800. [DOI] [PubMed] [Google Scholar]
- 40.Morris, D. R. & Geballe, A. P. (2000) Mol. Cell. Biol. 20, 8635-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajkowitsch, L., Vilela, C., Berthelot, K., Ramirez, C. V. & McCarthy, J. E. G. (2004) J. Mol. Biol. 335, 71-85. [DOI] [PubMed] [Google Scholar]
- 42.Hohn, T., Park, H. S., Guerra-Peraza, O., Stavolone, L., Pooggin, M. M., Kobayashi, K. & Ryabova, L. A. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 269-276. [DOI] [PubMed] [Google Scholar]
- 43.Yueh, A. & Schneider, R. J. (2000) Genes Dev. 14, 414-421. [PMC free article] [PubMed] [Google Scholar]
- 44.Mauro, V. P. & Edelman, G. M. (2002) Proc. Natl. Acad. Sci. USA 99, 12031-12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konishi, Y., Lindholm, K., Yang, L. B., Li, R. & Shen, Y. (2002) Am. J. Pathol. 161, 1567-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verwer, R. W., Baker, R. E., Boiten, E. F., Dubelaar, E. J., van Ginkel, C. J., Sluiter, A. A. & Swaab, D. F. (2003) Exp. Gerontol. 38, 167-172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.