Abstract
Plant innate immunity relies on successful detection of trespassing pathogens through recognizing their microbe-associated molecular patterns (MAMPs) by pattern recognition receptors (PRRs) at the cell surface. We recently reported two rice lysin motif (LysM)-containing proteins, OsLYP4 and OsLYP6, as dual functional PRRs sensing bacterial peptidoglycan (PGN) and fungal chitin. Here we further demonstrated the important roles of OsLYP4 and OsLYP6 in rice defense signaling, as silencing of either LYP impaired the defense marker gene activation induced by either bacterial pathogen Xanthomonas oryzaecola or fungal pathogen Magnaporthe oryzae. Moreover, we found that OsLYP4 and OsLYP6 could form homo- and hetero-dimers, and could interact with CEBiP, suggesting an unexpected complexity of chitin perception in rice.
Keywords: lysin motif-containing proteins, rice, innate immunity, defense-related gene, pattern recognition receptors
OsLYP4 and OsLYP6 Affected Pathogen-Induced Defense-Related Gene Activation in Rice
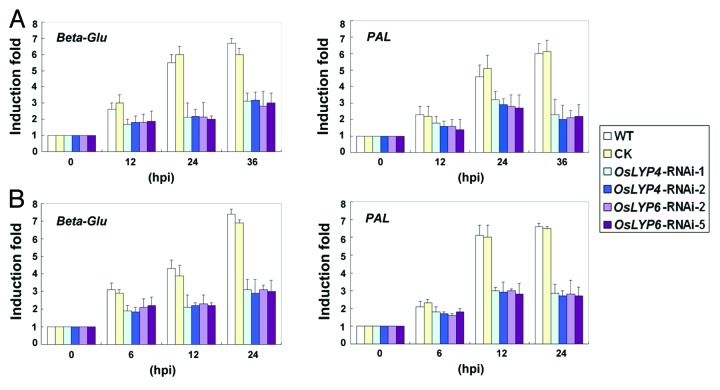
LysM is a ubiquitous protein motif found in virtually every living organism except for Archaea.1 The characterized LysM-containing proteins in high plants were mostly involved in rhizobial symbiosis or perception of chitin signals,2 and could be roughly categorized into LysM-containing receptor-like kinases (LysM-RLKs or LYKs) and non-receptor kinase LysM proteins (LYPs).3,4 The LysM-RLKs such as NFR1 and NFR5 are genetically defined as receptors for the chitin-derived rhizobial nodulation factor and are coupled to the symbiosis progress.5,6 On the other hand, rice chitin receptor complex components CEBiP and co-receptor OsCERK1 both contain LysMs, and intact LysM domain is necessary for chitin binding in Arabidopsis CERK1.7,8 Interestingly, Arabidopsis LYM1/LYM3 was found to contribute to PGN sensing but not chitin.9 Recently, we identified two rice LysM proteins, OsLYP4 and OsLYP6, play dual function in chitin and PGN perception.10 In this research, to investigate the influence of OsLYP4 and OsLYP6 in pathogen-induced defense-related gene activation in rice, we tested the selected marker genes, such as Beta-Glu (Os05 g0495900) and PAL (Os02 g0627100) in OsLYP4 and OsLYP6 RNAi rice. The induction of the selected marker genes was monitored by qPCR at different time points after the fungal blast pathogen Magnaporthe oryzae, or the bacterial streak pathogen X. oryzaecola treatment. As expected, the upregulation of selected defense marker genes were suppressed to different extents in OsLYP4-RNAi and OsLYP6-RNAi transgenic lines compared with that in control (CK) rice after M. oryzae inoculation (Fig. 1A) or X. oryzaecola inoculation (Fig. 1B). These results are consistent with our former data, in which knockdown of either OsLYP genes could increase rice susceptibility to both fungal and bacterial pathogens.10 Considering that these microbial pathogens contain a cocktail of MAMPs,11 these data suggested that PGN or chitin signaling mediated by OsLYP4 and OsLYP6 contributes significantly to rice defense responses against bacteria and fungi.
Figure 1. Suppression of pathogen-induced gene expression in OsLYP4- and OsLYP6-RNAi rice plants. (A) The fourth leafs at the five-leaf stage from each rice line were inoculated with M. oryzae (106 colony-forming units/mL) and the induction fold of Beta-Glu or PAL gene was examined by qPCR at 0, 12, 24 and 36 h after inoculation. (B) The induction fold of Beta-Glu or PAL gene was examined by qPCR at 0, 6, 12 and 24 h after X. oryzaecola (106 colony-forming units/mL) inoculation. The induction fold of each gene was calculated by the gene expression level in pathogen–treated seedlings relative to that in mock-treated seedlings at the same time point. hpi, hour post inoculation. Three biological repeats were conducted, and the data represent means ± SD (10 leaves in each transgenic line, three independent experiments). WT: wild type, CK: control transgenic rice transformed with empty vector, OsLYP4-RNAi-line -1, OsLYP4-RNAi -2, OsLYP6-RNAi line -2 and OsLYP6-RNAi line -5 were compared.
OsLYP4 and OsLYP6 May Cooperate With Other LysM-Containing Receptors to Transduce PGN or Chitin Signal
Our previous study has suggested that OsLYP4 and OsLYP6 are not functionally redundant in chitin and PGN signaling pathways.10 Moreover, another lysin motif-containing protein, CEBiP has been reported to play important role in chitin sensing and signal transduction.12,13 Therefore, we investigated the protein-protein interactions between these LYP proteins with yeast two-hybrid (Y2H).
In Y2H assay, we indeed detected positive interactions between OsLYP4 and OsLYP6 (Fig. 2A), and between CEBiP and OsLYP4 (Fig. 2B). The false positives were excluded by the negative control (data not shown). Furthermore, the CEBiP and OsLYP4 interaction was further verified in rice protoplasts by bimolecular fluorescence complementation (BiFC) (Fig. 2B). Interestingly, we also found that OsLYP4 and OsLYP6 could form homodimers in Y2H (Fig. 2A).
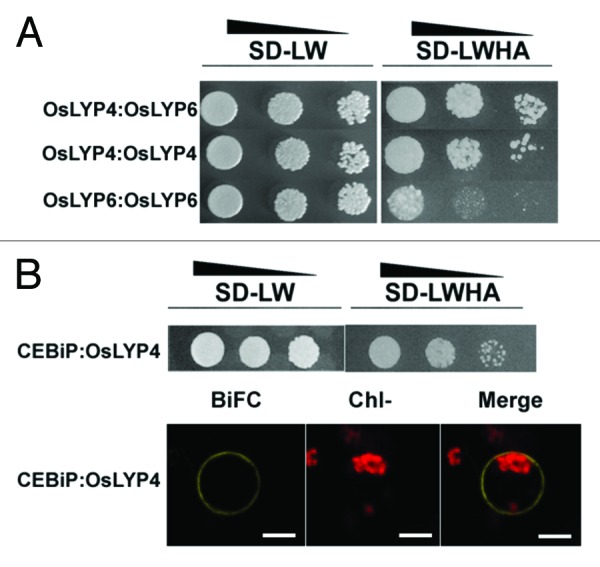
Figure 2. OsLYP4 and OsLYP6 Can Interact with other LYPs. (A) Yeast two-hybrid analysis of the protein-protein interaction between OsLYP4, and OsLYP6. Yeast cells harboring the indicated bait (cloned into the pGADT7) and prey plasmids (cloned into the pGBKT7) were diluted by 1:1, 1:10 or 1:100 and spotted on the selective medium SD-LW (medium without leucine and tryptophan) and SD-LWHA (medium without leucine, tryptophan, histidine and adenine). Growth on the SD-LWHA medium indicates a positive interaction. (B) Interactions between OsLYP4 and CEBiP proteins in yeast and rice protoplasts. BiFC (bimolecular fluorescence complementation) indicated the complementary yellow fluorescence, Chl- indicated the chloroplast autofluorescence in the protoplast. Bar = 10 μm.
These data consistently support a cooperative relationship between OsLYP4 and OsLYP6 as our previous genetic analyze. Therefore, we propose that OsLYP4 and OsLYP6 work in the same complex for PGN or chitin perception and subsequent signal transduction. The identification of OsLYP4 and OsLYP6 as additional chitin receptors in rice raises the question regarding the relationship between the OsLYP4-OsLYP6 chitin receptor complex and the reported CEBiP-OsCERK1 chitin receptor complex. Shimizu et al. has found that a major portion of CEBiP proteins were visualized as homodimers by the blue native PAGE analysis.13 However, a small fraction of CEBiP proteins did exist as larger-size oligo-mers,13 which may suggest that some CEBiP proteins can be in the same complexes with OsLYP4 and OsLYP6. Our data that CEBiP interacted with OsLYP4 in yeast and in plant cells (Fig. 2B) supported such a possibility. As CEBiP, OsLYP4, and OsLYP6 could all form homodimers (Fig. 2A, and reviewed in ref. 13), and these OsLYPs apparently need a signaling partner (e.g., OsCERK1)13 with an intracellular kinase domain for defense signal transduction, the composition and the stoichiometry of different chitin or PGN receptor complexes in rice await further investigation, which will shed new light on the rice innate immunity.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No.30571069 and No.31100874), the Natural Science Foundation of Guangdong province, PR China (No.S2011040001868), the project of Science and Technology new star in Zhujiang city (2012J2200024) and the Fundamental Research Funds for the Central Universities (12lgpy33).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/22980
References
- 1.Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–47. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 2.Knogge W, Scheel D. LysM receptors recognize friend and foe. Proc Natl Acad Sci U S A. 2006;103:10829–30. doi: 10.1073/pnas.0604601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang XC, Wu X, Findley S, Wan J, Libault M, Nguyen HT, et al. Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol. 2007;144:623–36. doi: 10.1104/pp.107.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang XC, Cannon SB, Stacey G. Evolutionary genomics of LysM genes in land plants. BMC Evol Biol. 2009;9:183. doi: 10.1186/1471-2148-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–40. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 6.Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–92. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 7.Iizasa E, Mitsutomi M, Nagano Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J Biol Chem. 2010;285:2996–3004. doi: 10.1074/jbc.M109.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem. 2010;285:28902–11. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D, Tsuda K, et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci U S A. 2011;108:19824–9. doi: 10.1073/pnas.1112862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B, Li JF, Ao Y, Qu J, Li Z, Su J, et al. Lysin Motif-Containing Proteins LYP4 and LYP6 Play Dual Roles in Peptidoglycan and Chitin Perception in Rice Innate Immunity. Plant Cell. 2012;24:3406–19. doi: 10.1105/tpc.112.102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 12.Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci U S A. 2006;103:11086–91. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64:204–14. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]