Abstract
Initiation of DNA replication in eukaryotes, archea, and eubacteria requires interaction of structurally conserved ATP-binding initiator proteins and origin DNA to mediate assembly of replisomes. However, the specific requirement for ATP in the early steps of initiation remains unclear. This is true even for the well studied Escherichia coli replication origin, oriC, where the ATP form of initiator DnaA is necessary and sufficient for initial DNA strand separation, but the five DnaA-binding sites (R boxes) with consensus sequence 5′TGTGNAT/AAA bind both active ATP-DnaA and inactive ADP-DnaA with equal affinity. By using dimethyl sulfate footprinting, we recently identified two initiator-binding sites, I2 and I3, with sequence 5′TG/TGGATCAG/A. We now show that sites I2 and I3 preferentially bind DnaA-ATP and are required for origin unwinding. Guanine at position 3 determines DnaA-ATP preference, and changing this base to thymine at both I sites allows DnaA-ADP to bind and open oriC, although DNA strand separation is not precisely localized in the AT-rich region. These observations indicate that specific initiator binding sites within a replication origin can be important determinants of an ATP-dependent molecular switch regulating DNA strand separation.
One of the earliest steps in triggering new rounds of DNA synthesis requires binding of initiator proteins to replication origin DNA (1). Several origin-binding proteins in eukaryotes, archea, and eubacteria are members of the AAA+ family of ATPases, which are active in the ATP-bound form and are inactivated by hydrolysis of ATP to ADP (1–4). These proteins form complexes that mediate subsequent steps of initiation, including DNA strand separation and recruitment of replisome components (1). The recent discovery of structural conservation in AAA+ initiation proteins in bacteria, archea, and yeast (3–5) suggests that aspects of the initiation mechanism may be similar across all domains of life. However, additional information on the specific function of ATP in the early steps of initiation is required before common initiation mechanisms can be inferred.
For a variety of bacterial chromosomal and plasmid origins, as well as some eukaryotic viral origins, the first step of initiation requires binding of multiple copies of an initiator protein that serves to melt the DNA duplex in an AT-rich region (1, 6). This process has been extensively studied in Escherichia coli, where initial strand opening requires multiple copies of the AAA+ initiator DnaA (7, 8). Only ATP-DnaA is active in formation of the open complex (8, 9). Within oriC are five 9-mer DnaA-binding sites (R boxes; see Fig. 1A) with consensus sequence 5′TGTGNAT/AAA (10, 11). Although R boxes have different affinities for DnaA based on slight differences in the nucleotide sequence, each R box binds inactive DnaA-ADP and active DnaA-ATP with equal affinity (8, 12). Based on this finding, two scenarios seem likely with regard to the requirement for DnaA-ATP in oriC unwinding. One possibility is that DnaA-ATP and DnaA-ADP form structurally different complexes when bound to R boxes, and only the ATP form of the complex can make the conformation needed to open the DNA duplex. The second possibility is that oriC contains binding sites specific for DnaA-ATP. DnaA-ATP binding to such sites would be analogous to the interaction of Saccharomyces cerevisiae origins with Orc1, which requires ATP (2).
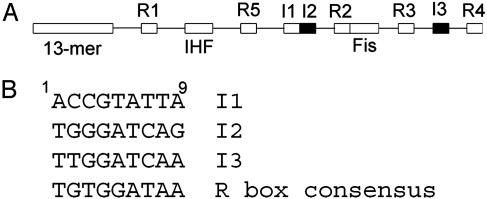
Fig. 1.
Location of R boxes and I sites in oriC. (A) Relative position of DnaA-binding sites are marked above the line representing oriC DNA. Additional landmarks, including Factor for inversion stimulation (Fis)- and integration host factor (IHF)-binding sites and the13-mer AT-rich region, are marked below the line. Dark boxes represent I2 and I3. (B) The 9-mer sequences for R boxes and I sites.
If binding sites for DnaA-ATP exist within oriC, where might they be found? We previously identified several 9-mer DnaA-binding sites flanking the center of oriC (I sites, Fig. 1 A and refs. 13 and 14) that may be candidates for preferential binding of DnaA-ATP. I sites differ from the R box consensus at 3–4 bases (Fig. 1B and ref. 14), but both R boxes and I sites are highly conserved among the oriC regions of the Enterobacteriaceae (15).
In the present study, we analyzed DnaA–ATP and –ADP interactions with I sites by dimethyl sulfate (DMS) footprint analysis and measured the effect of single-base changes on DnaA-binding specificity and oriC function. We found that I2 and I3 strongly preferred DnaA-ATP. Single-base changes in I2 and I3 produced two different classes of phenotypes. One class of mutations blocked oriC unwinding in vitro due to reduction of DnaA binding in the affected I site. These plasmids were unable to replicate from oriC in vivo. The second class reduced the ATP specificity of I sites, allowing binding by DnaA-ADP. Mutant origins with one nondiscriminatory I box retained a requirement for DnaA-ATP to unwind in vitro. Dual mutations in I2 and I3 produced origins that were unwound by DnaA-ADP, although these origins did not localize strand separation in the AT-rich region as precisely as did wild-type in vitro. Mutations that allowed DnaA-ADP to bind to I sites were functional in vivo, although the plasmid harboring mutations in both I2 and I3 perturbed host growth and integrated into the chromosome with high frequency. Our findings are consistent with a crucial role for I sites as cis-acting ATP-dependent regulators of oriC strand separation. Similar cis-acting sites may be found on replication origins in other cell types.
Materials and Methods
Strains and Plasmids. pOC170 was used as the starting plasmid for all mutant oriC constructions. It is 3,852 base pairs long and carries replication origins from both pBR322 and oriC (16). Supercoiled pOC170 or mutant plasmid used for the in vitro analysis of DnaA binding and open complex formation was isolated by using the QIAPrep Spin plasmid preparation kit (Qiagen). Mutagenesis of oriC was performed by using the QuikChange site-directed mutagenesis kit (Stratagene) with oligonucleotide primers (BioSource International) of 20–28 base pairs carrying the mutation in the center. After DpnI restriction endonuclease digestion to remove parental template, mutant plasmids were transformed into XL1-Blue endA1 gyrA46 hsdR17 lac recA1 relA1 supE44 thi; F′lac: lacIq Δ(lacZ) M15 Tn10 proA+ proB+ (17). DNA sequence analysis of mutant oriC was performed by using an Applied Biosystems 373 DNA sequencer with XL upgrade and Bio-Rad sequence analysis software, version 3.4
For evaluation of replication of oriC plasmids in vivo, pOC170 or mutant plasmids were transformed into either P3478 polA1, thyA36 deoC2 IN(rrnD-rrnE)1, λ-, or its isogenic parent, W3110 (18).
Chemicals, Proteins, and Enzymes. Reagent grade chemicals were purchased from either Fisher Scientific or Sigma. Media components were from Difco. All enzymes, except P1 endonuclease, were from New England Biolabs.
Amino-terminal His10-tagged DnaA was purified as described by Li and Crooke (19).
DNA Modification and Primer Extension. DMS modification of DNA (0.75 μg) in vitro was performed as described (13). Reactions contained either 5 mM ATP or ADP, as noted in Figs. 2, 3, 4, 5. DnaA was preincubated in reaction buffer with 5 mM ATP or ADP before addition to reactions at the concentrations indicated in Figs. 2, 3, 4, 5. All experiments were repeated at least three times.
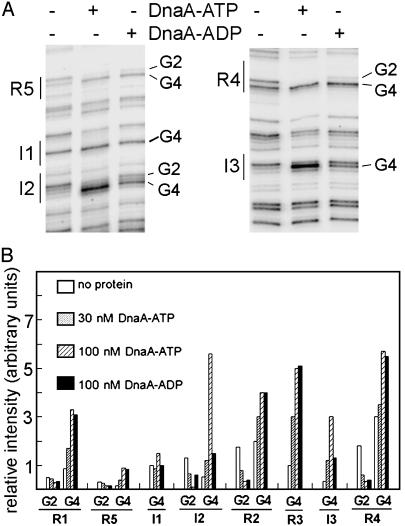
Fig. 2.
DnaA-ATP binds preferentially to I sites but not to R boxes. (A) DMS modification patterns after incubating 0 nM DnaA, 100 nM DnaA-ATP, or 100 nM DnaA-ADP with pOC170, measured by primer extension. Sites R5, I1, I2, R4, and I3, and guanines at position 2 or 4 in the recognition site (Fig. 1) are marked. (B) Relative intensities of DMS modification at G2 and G4 within all eight DnaA-binding sites on plasmid incubated with 0 nM DnaA, 30 nM Dna-ATP, 100 nM DnaA-ATP, or 100 nM DnaA-ADP. Sites I1, I3, and R3 lack a guanine in position 2.
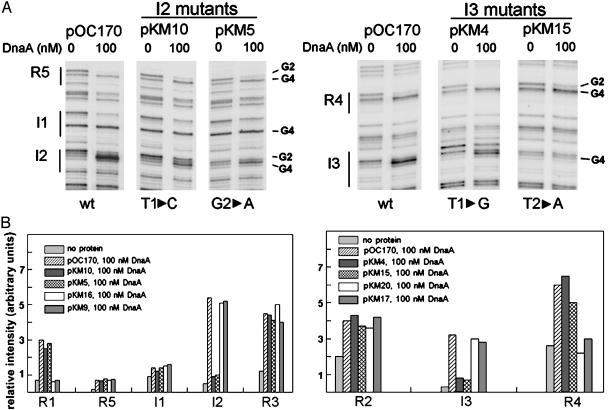
Fig. 3.
Single-base changes in positions 1 or 2 in I sites and R boxes reduce DnaA binding. (A) DMS modification patterns after incubating 0 or 100 nM DnaA-ATP with pOC170 or derivative plasmids mutated in position 1 of I2 (pKM10), and I3 (pKM4), or position 2 of I2 (pKM5) and I3 (pKM15). The base changes in the mutants are indicated at the bottom. Sites R5, I1, I2, R4, and I3, and guanines at position 2 and 4 in the recognition site are marked. (B) Relative intensities of DMS modification (G4) for DnaA-binding sites with 0 nM DnaA or 100 nM DnaA-ATP incubated with pOC170 or 100 nM DnaA incubated with mutant plasmids. R1 mutants are pKM16 (T1 to C), and pKM9 (G2 to A). R4 mutants are pKM20 (T1 to G) and pKM17 (G2 to A).
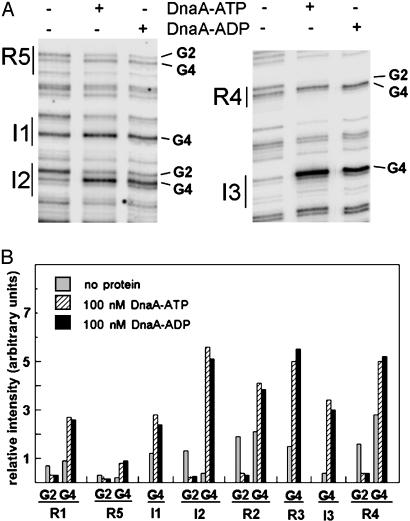
Fig. 4.
Changing G to T at position 3 of the binding site removes discrimination for DnaA-ATP at I2 and I3. (A) DMS modification patterns after incubating 0 nM DnaA, 100 nM DnaA-ATP, or 100 nM DnaA-ADP with plasmids in which G at position 3 of I2 (Left)orI3(Right) is changed to a T. Sites R5, I1, I2, R4, and I3, and guanines at position 2 or 4 in the recognition site (see Fig. 1) are marked. (B) Relative intensities of DMS modified G2 and G4 within all eight DnaA-binding sites.
Fig. 5.
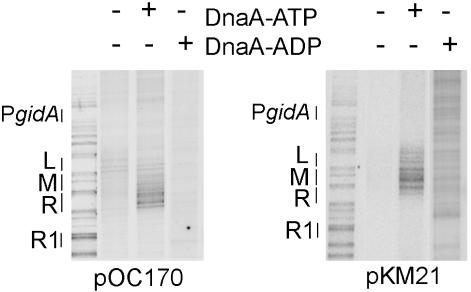
An oriC plasmid mutated in position 3 of I2 and I3 can be unwound with DnaA-ADP, but the region of DNA strand separation is not localized to the 13-mer region. pOC170, or the I2/I3 mutant pKM21 (changing G3 to T) incubated with 0 nM DnaA, 200 nM DnaA-ATP, or 200 nM DnaA-ADP was treated with P1 endonuclease, and cleavage sites were mapped by primer extension. The positions of the right (R), middle (M), and left (L) 13-mer regions, as well as R1 and the promoter for gidA are marked. No cutting induced by DnaA-ATP or DnaA-ADP on either plasmid was detected in oriC to the right of the regions shown.
P1 endonuclease (Roche Diagnostics) digestions were performed as described (13). After digestion, supercoiled, relaxed circular, or linear forms of the plasmid were resolved by electrophoresis through 1% agarose gels in Tris borate buffer (20). Ethidium bromide-stained gels were photographed and the photographs were scanned by using a Bio-Rad GS700 densitometer. Relative amounts of DNA in each form were determined by using Bio-Rad quantity one software.
DMS- and P1-treated samples were extended with 32P-labeled primer as described (13). Two primers were used, a left primer hybridizing at bases 272–290 to analyze top strand modifications of plasmid template, and a right primer hybridizing at bases 124–142 to analyze bottom-strand modifications. Extension products were resolved on 6% polyacrylamide sequencing gels as described (13), and dried gels were scanned on a Bio-Rad Molecular Imager FX PhosphorImager. Representative scans are shown in the Figures. Images were analyzed by using Bio-Rad quantity one software. Ratios of intensities of bands in binding sites to internal standard bands were calculated to yield relative intensity of modified guanines. Deviations in band intensities among experiments were <10%. Nucleotide positions of reference bands used for determining relative intensities were at the following oriC bases: 92, 93, 98, 99 (R1); 182, 180 (I1, I2, and R5); 196, 198, 204, 209 (R2); 217 (R3); and 275, 277, 279 (I3 and R4). The complete nucleotide sequence of oriC is shown in ref. 13.
Results
Sites I2 and I3 Preferentially Bind DnaA-ATP. To determine whether I sites interact with both DnaA-ADP and DnaA-ATP, we performed DMS footprint analysis by using purified DnaA and supercoiled pOC170, an oriC plasmid (16). At levels of DnaA that support open complex formation, DnaA binding to R boxes and I sites causes specific and reproducible changes in DNA cleavage or modification patterns produced by several reagents, including Dnase I (10, 11, 21), 1–10-phenanthroline-copper (22), and DMS (13, 14, 23). In DMS-treated oriC DNA, the DnaA footprint is sharply defined, with both R boxes and I sites showing a distinctive DnaA-induced DMS hypersensitivity at position 4 (Fig. 1B) of their respective consensus sequences. DnaA binding also suppresses DMS reactivity at the G in position 2, if it is present (Fig. 2 and refs. 13, 14, 23, and 24). This characteristic DMS “footprint,” also seen when DnaA is bound to templates other than oriC (25), is equivalent on supercoiled and linear templates (14). The G4 enhancement and G2 suppression become more pronounced as DnaA levels increase (13, 14), so that quantification of the intensity changes can be used as a measure of the extent of site filling.
Footprints obtained by using 100 nM DnaA incubated with 0.75 μg of pOC170 DNA reflect conditions where DNA strand separation in the AT-rich region is detected by single-strand scission reagents (13). At this DnaA level, DMS footprints for DnaA-ATP were identical to those generated with DnaA-ADP at all R boxes (Fig. 2), which is consistent with measurements made by using other techniques (8, 12). In contrast, DnaA-ATP and DnaA-ADP DMS footprints at I2 and I3 were not identical. The DnaA-ADP footprints at I2 and I3 were equivalent in intensity to those seen with 3- to 4-fold less DnaA-ATP (scan, Fig. 2 A), indicating that I2 and I3 preferentially bind DnaA-ATP.
The DMS footprint of one additional site, previously identified in our laboratory (13), and termed I1 (see Figs. 1 A and 2), may also show a subtle preference for DnaA-ATP, but the change in the intensity of footprints generated by the two nucleotide forms at higher (100 nM) DnaA levels was <1.5-fold, and binding could not be detected at lower (30 nM) DnaA levels. Although the last five nucleotides of the 9-bp I1 site are similar to those in R boxes (Fig. 1B), the first three nucleotides of I1, thought to be important for DnaA contact (26, 27), differ from both R boxes and other I sites, and this difference may account for the lower amount of observed binding. Because the <2-fold change in footprint intensity generated by DnaA-ATP and DnaA-ADP in I1 make it difficult to assess changes in binding caused by our subsequent mutagenesis analyses, we focused the studies in this paper on I2 and I3.
We could not identify any DnaA-specific DMS footprints within the AT-rich region of oriC (data not shown). It is likely that the levels of DnaA-ATP used in our study were not high enough to detect such binding, because 4-fold higher concentrations of DnaA-ATP have previously been reported to protect the AT-rich region from Dnase I (28).
A Single-Base Substitution at Positions 1 and 2 of I2 or I3 Reduces DnaA-ATP Binding, but a Substitution at Position 3 Enhances DnaA-ADP Binding. DnaA contacts its 9 bp recognition site at the beginning and end of the sequence, but not in the middle, with T1, T3, A6, A8 (see Fig. 1B), playing an important role for contact with R boxes (26, 27). Furthermore, in previous studies, single-base substitutions (G to A) at position 2 of R box consensus sequences inactivated oriC (29). To examine the DnaA contacts in the I site sequence more closely, we attempted to reduce binding of DnaA-ATP at I2 and I3 by substituting bases at position 1 and position 2 (mutations described in the Fig. 3 legend and Table 2). We found that I sites mutated in position 1 or 2 bound less DnaA-ATP than to wild-type I sites (Fig. 3), similar to results seen with R box position 1 and 2 mutants (shown in scans in Fig. 3). Thus, bases at positions 1 and 2 of both I sites and R boxes play an important role in contacting DnaA. The mutations in I2 or I3 did not affect the binding of DnaA to the remaining recognition sites, suggesting that loss of one individual site does not change interaction of DnaA at other oriC sites. However, the data do not rule out potential cooperative interactions among DnaA molecules, because our footprints detect only DnaA-DNA contacts, not DnaA-DnaA interaction. In fact, we have shown previously that binding to weak R boxes and I sites can be stimulated by integration host factor binding to oriC, presumably by causing the DnaA oligomers at R1 and R4 to be redistributed or shared with the weaker sites (13). Recent studies have also shown that DnaA oligomerization appears to be necessary for oriC function in vivo (30).
Table 2. In vivo replication activity of oriC plasmids mutated in DnaA-binding sites.
| Site | Mutation | Mnemonic | Binding | polA† | oriC activity§ |
|---|---|---|---|---|---|
| Wild-type | pOC170 | Normal | 1.0 | +++ | |
| R1 | T1 to A | pKM16 | Reduced | 0.01 | +/– |
| R1 | G2 to A | pKM9 | Reduced | <0.005 | – |
| R4 | T1 to G | pKM20 | Reduced | 0.06 | + |
| R4 | G2 to A | pKM17 | Reduced | 0.07 | + |
| I2 | T1 to C | pKM10 | Reduced | 0.005 | – |
| I2 | G2 to A | pKM5 | Reduced | <0.005 | – |
| I2 | G3 to T | pKM13 | Normal* | 0.3 | ++ |
| I3 | T1 to G | pKM4 | Reduced | <0.005 | – |
| I3 | T2 to A | pKM15 | Reduced | 0.005 | – |
| I3 | G3 to T | pKM3 | Normal* | 0.4 | ++ |
| 12/13 | G3 to T | pKM21 | Normal* | 0.6‡ | +++ |
ATP-DnaA binding was normal, but sites also bound ADP-DnaA
Transformants of P3478 polA/W3110 by test plasmid, normalized for transformation by pOC170
Colonies of transformed W3110 were very small, and <10% of cells in the primary colonies harbored plasmid
Based on relative transformation of the polA strain: –, 0.5% or lower; +/–, 0.6–5%; +, 6–10%; ++, 11–50%; +++, 50–100%
Guanine is found at position 3 of I sites, but thymine is found at this position in R boxes. We anticipated that substituting G to T would cause I sites to become similar to R boxes and bind DnaA-ATP and DnaA-ADP with equal affinity. In fact, the G3 to T substitution did increase the affinity of I2 and I3 for DnaA-ADP, although the affinity for DnaA-ATP was essentially unchanged (see Figs. 2 and 4).
Reduced Binding of DnaA-ATP at an I Site Decreases DNA Strand Opening and in Vivo Function. Once bound to an origin, the role of the initiator protein in several systems is to separate DNA strands in an adjacent AT-rich region (1). The resulting single-stranded DNA is susceptible to cleavage by P1 endonuclease (7, 13, 14, 21). On supercoiled templates, P1 endonuclease makes opposing cuts on both strands of the open complex, resulting in linearization of the molecule. In oriC, the AT-rich region contains three direct 13-mer repeats to the left of R1 (Fig. 1). After incubation with sufficient DnaA-ATP, strand opening begins within the right 13-mer and usually remains focused within the middle and right 13-mers (7, 13, 14). To test the effect of binding-defective I sites on oriC function, DnaA-ATP-catalyzed strand opening was detected by using P1 endonuclease. The data are summarized in Table 1. In the presence of ≈30 copies of DnaA-ATP per oriC, 25–30% of supercoiled wild-type oriC plasmids (pOC170) are converted to linear form by P1, similar to levels previously reported in the absence of integration host factor or histone-like protein (7, 13). In contrast, this level of DnaA-ATP did not separate DNA strands in supercoiled mutant oriC plasmids with reduced binding at I2 and I3 (Fig. 4), because P1 cutting yielded only background levels (8%) of linearization. (Table 1). Furthermore, a lack of P1-induced linearization indicates that DnaA-ADP did not catalyze open complex formation on wild-type oriC plasmid (Table 1). These results suggest that both I2 and I3 must interact with DnaA-ATP to melt the DNA duplex in the AT-rich region.
Table 1. Open complex formation caused by DnaA-ATP or DnaA-ADP binding to oriC plasmids mutated in 12 or 13.
| Site | Mutation | Mnemonic | Incubated with | Percent linear |
|---|---|---|---|---|
| Wild-type | pOC170 | No DnaA | 8 | |
| 13 | T1 to G | pKM4 | No DnaA | 6 |
| 12 | T1 to C | pKM10 | No DnaA | 5 |
| 13 | G3 to T | pKM3 | No DnaA | 7 |
| 12/13 | G3 to T | pKM21 | No DnaA | 10 |
| Wild-type | pOC170 | 200 nM ATP-DnaA | 25 | |
| 13 | T1 to G | pKM4 | 200 nM ATP-DnaA | 6 |
| 12 | T1 to C | pKM10 | 200 nM ATP-DnaA | 7 |
| 13 | G3 to T | pKM3 | 200 nM ATP-DnaA | 25 |
| 12/13 | G3 to T | pKM21 | 200 nM ATP-DnaA | 30 |
| Wild-type | pOC170 | 200 nM ADP-DnaA | 9 | |
| 13 | G3 to T | pKM3 | 200 nM ADP-DnaA | 5 |
| 12/13 | G3 to T | pKM21 | 200 nM ADP-DnaA | 28 |
Plasmids were incubated with protein and digested with P1 endonuclease, and the formation of linear molecules, indicating cutting in single-stranded regions, was analyzed.
Mutations in individual R boxes on oriC plasmids that decreased DnaA binding are reported to decrease or eliminate in vivo replication (31). To determine whether decreased binding mutations in I2 and I3 also perturbed oriC function, we transformed a polA-defective strain, with pOC170-derived plasmids containing the mutant oriC. Mutant polA strains cannot initiate replication from pBR322 origins of replication; thus plasmid replication depends on functional oriC. Plasmids carrying mutations in I2 or I3, previously shown to reduce DnaA-ATP binding, were defective for replication in polA backgrounds (Table 2). Similar single-base pair mutations resulting in decreased DnaA binding to R1 and R4 also decreased oriC function. These results would be expected if oriC was not able to form an unwound complex in vivo unless all R boxes and I sites were bound to DnaA.
The DNA Duplex in the AT-Rich Region of I2/I3 Double Mutants Is Opened with DnaA-ADP, but Strand Separation Is Less Precisely Localized than in Wild-Type Origins. Substituting G for T at position 3 produced I sites with greater affinity for DnaA-ADP, thus reducing discrimination. Mutant origins containing only a single G3 substitution at either I2 (pKM10) or I3 (pKM3) were able to replicate in polA strains, due to the availability of DnaA-ATP to fill all of the DnaA-binding sites in vivo (Table 2). However, these origins could not be unwound by DnaA-ADP (Table 1), presumably because only the mutated I site could interact with DnaA-ADP. Combining both nondiscriminatory I site mutations onto a single copy of oriC produced a replication origin that bound DnaA-ADP at I2 and I3 in addition to R boxes. If complete loading of DnaA at all sites is a requirement for initial strand opening in the AT-rich region, the oriC I2/I3 double mutant (pKM21) should become unwound with DnaA-ADP in vitro. Interestingly, incubations with DnaA-ADP caused P1 endonuclease to convert 28% of supercoiled pKM21 to linear form, which is similar to the amount of unwinding detected after incubating DnaA-ATP with wild-type oriC (Table 1). Linearization of pKM21 was equivalent to wild-type oriC with DnaA-ATP, and no significant linearization was observed in the absence of DnaA (Table 1), suggesting that the two single-base pair mutations did not simply destabilize the DNA double-helix. Rather, these results provide evidence that strand opening in the AT-rich region can be achieved with either the ATP or the ADP form of DnaA, as long as DnaA fills I2 and I3 as well as the R boxes.
Measurement of plasmid linearization by P1 endonuclease reveals the presence of single-stranded regions, but does not identify the location of the unwound regions on the plasmid. On wild-type oriC plasmids, unwinding is localized to the AT-rich 13-mer regions (7, 13). To map the single-stranded region in the I2/I3 double mutant pKM21 and to compare unwinding by DnaA-ATP and DnaA-ADP, the positions of P1 endonuclease cutting were determined by primer extension (13, 14). When either pOC170 or pKM21 oriC is incubated with DnaA-ATP, P1 endonuclease cutting is highly localized within the 13-mers (Fig. 5 and refs. 7 and 13), but not elsewhere in oriC. No localized P1 cutting was observed when DnaA-ADP was incubated with pOC170 (Fig. 5), which was consistent with the low level of linearization by P1 (Table 2). In contrast, even though DnaA-ADP incubated with pKM21 produced sites of P1 endonuclease sensitivity that could be mapped within the AT-rich region (Fig. 5), these cut sites were not tightly localized. Rather, the P1 cleavage sites were more dispersed than those observed after incubation with DnaA-ATP, with cut sites extending outside the AT-rich region to the left into an adjacent promoter (gidA), and to the right, ending near R1 (Fig. 5). Thus, DnaA-ATP appears to play two roles: to assemble the appropriate complex needed to melt the AT-rich DNA duplex, and to restrict the region of unwinding to only the AT-rich region. Assembling the complex needed for DNA strand separation can be accomplished by DnaA binding to all I sites and R boxes, but restricting strand separation to a localized region within the AT-rich sequence may require additional DnaA–oriC interactions (28) or a conformation attainable with only DnaA-ATP.
The plasmid containing the I2/I3 double mutant oriC, pKM21, successfully transformed a mutant polA strain, which is consistent with the expectation that this mutant origin, which could bind DnaA at all R boxes and I sites, would function in vivo (Table 2). However, wild-type cells harboring this plasmid formed very small colonies, and only 10% of cells in the primary transformed colonies could form secondary colonies when passed onto selective media. Transformed cells grown in non-selective media had increased and heterogeneous cell sizes relative to wild type, and often the cells failed to divide or had abnormal septum placement. Further, PCR analysis of chromosomal DNA in transformed cells indicated that the I2/I3 double mutant integrates into the chromosome with high frequency (data not shown). These defects were not seen in cells transformed with the wild-type plasmid, which is stably maintained in the autonomous state, and does not appreciably affect host cell growth. It appears that the presence of plasmid oriC that can be opened by DnaA-ADP causes competition with the wild-type chromosomal copy. The nature of this competition is currently being investigated; however, the frequency with which the I2/I3 double mutant integrates into the chromosome is consistent with the results seen with oriC plasmids that replicate more than once per cell cycle (32).
Discussion
The first step of initiation of DNA replication in all systems is the binding of initiator protein to replication origin DNA (1). In a number of bacterial and viral systems, this binding melts the origin DNA duplex in a nearby AT-rich region (1, 6), although the biochemical mechanism that achieves unwinding is not yet known. Replication of all known bacterial chromosomes, as well as a variety of plasmids, requires the AAA+ initiator protein DnaA (6). DnaA has structural homology to archaeal and eukaryotic initiator proteins (3, 4). The strong conservation of DnaA-like proteins among all domains of life suggest that studies elucidating how DnaA mediates strand opening should reveal common mechanisms of initiation.
In E. coli, only the ATP bound form of DnaA is active for DNA strand opening in the AT-rich region (8, 9), although the role of ATP in the process was unclear. We report here that two DnaA-binding sites in oriC (I2 and I3) interact preferentially with DnaA-ATP. Preferential binding of the ATP form of an initiator protein has also been observed for budding yeast, Drosophila, and human origins (2). In E. coli, filling of I sites plays a critical role in determining the requirement for the DnaA-ATP-bound form for duplex unwinding. By constructing single-base substitutions in sites of interest, we show that there are at least two classes of I site mutant phenotypes: one that prevents binding of DnaA-ATP and inactivates oriC, and one that allows DnaA-ADP to bind to I2, I3, and oriC.
Single-base substitutions in the first two nucleotides of the recognition site for I2 and I3 reduced DnaA-ATP binding and also inactivated oriC unwinding in vitro and oriC function in vivo. Similar mutations in R1 and R4 also reduced binding and inactivated oriC. Previously, Langer et al. (31) reported that mutations in R boxes that reduced binding inactivated oriC plasmids. Our results indicate the importance of filling I sites as well as R boxes in forming unwound complexes at oriC.
A critical difference between I sites and R boxes is their binding affinity for DnaA-ADP. We find that position 3 is key to the ability of I2 and I3 to discriminate between the nucleotide forms of DnaA. Replacing G with T at position 3 of either I2 or I3 eliminated the discrimination and allowed DnaA-ADP to bind. DnaA-ADP and DnaA-ATP very likely have different conformations (4, 6, 9), but details of the differences have not yet been revealed. Future studies on the structure of different nucleotide forms of initiator proteins should help explain the ATP requirement for binding to I sites, as well as for binding sites in other systems.
The mutant oriC that bound DnaA-ADP at both I2 and I3 (pKM21) formed an open complex with DnaA-ADP, but DNA strand separation was no longer localized. It is difficult to understand how the switch to DnaA-ADP binding would change the position of strand separation unless the position was determined by some mechanism that requires DnaA-ATP. There are at least two possible explanations for the increased region of unwinding in the mutant origin. The first is that the altered conformation of DnaA-ADP produces a complex that destabilizes the DNA duplex in a different position in oriC. A second possibility is that DnaA-ATP must be available to bind to other sites in addition to I2 and I3. Speck and Messer (28) recently reported sites in the AT-rich region with consensus 5′-AGATCT that preferentially bind DnaA-ATP and stabilize single-stranded regions. These sites may also restrict the region of unwinding.
Levels of DnaA-ATP remain low during cell growth, but rise abruptly around the time DNA synthesis is initiated (33). This sudden increase is compatible with a model in which filling I2 and I3 with DnaA-ATP triggers a molecular switch needed to melt the DNA duplex and raises questions about the nucleotide form of DnaA bound to oriC during the cell cycle. Previous reports (9, 34) indicate that oriC will unwind with a mixture of ATP- and ADP-DnaA. For wild-type oriC, R1, R2, and R4 are bound to DnaA throughout the cell cycle (23, 24), and can bind DnaA-ADP, as well as DnaA-ATP. Thus, a stable platform of mixed DnaA-ATP and DnaA-ADP at strong R boxes, perhaps inherited from a previous generation, would rapidly convert to an initiation complex produced by newly synthesized DnaA-ATP filling I sites. This scenario is reminiscent of eukaryotic DNA replication, where a stable origin–recognition complex is retained throughout the cell cycle, and transitions to an active complex at the correct time in the cell cycle by a mechanism that remains to be identified (35, 36). Closer analysis of assembly of replication complexes in eukaryotes may reveal important protein–DNA interactions, analogous to those seen here for bacterial origins, which are required for initiation.
Acknowledgments
We thank Walter Messer for providing pOC170 and Elliott Crooke for providing the plasmid pZL411 used to express His10-tagged DnaA. This work was supported by National Institutes of Health Grant GM54042.
Abbreviation: DMS, dimethyl sulfate.
References
- 1.Baker, T. A. & Bell, S. P. (1998) Cell 92, 295-305. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. P. & Dutta, A. (2002) Annu. Rev. Biochem. 71, 333-374. [DOI] [PubMed] [Google Scholar]
- 3.Liu, J., Smith, C. L., DeRyckere, D., DeAngelis, K., Martin, G. S. & Berger, J. M. (2000) Mol. Cell 6, 637-648. [DOI] [PubMed] [Google Scholar]
- 4.Erzberger, J. P., Pirruccello, M. M. & Berger, J. M. (2002) EMBO J. 21, 4763-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giraldo, R. (2003) FEMS Microbiol. Rev. 26, 533-554. [DOI] [PubMed] [Google Scholar]
- 6.Messer, W. (2002) FEMS Microbiol. Rev. 26, 355-374. [DOI] [PubMed] [Google Scholar]
- 7.Bramhill, D. & Kornberg, A. (1988) Cell 52, 743-755. [DOI] [PubMed] [Google Scholar]
- 8.Sekimizu, K., Bramhill, D. & Kornberg, A. (1987) Cell 50, 259-265. [DOI] [PubMed] [Google Scholar]
- 9.Yung, B. Y. M., Crooke, E. & Kornberg, A. (1990) J. Biol. Chem. 265, 1282-1285. [PubMed] [Google Scholar]
- 10.Fuller, R. S., Funnell, B. E. & Kornberg, A. (1984) Cell 38, 889-900. [DOI] [PubMed] [Google Scholar]
- 11.Matsui, M., Oka, A., Takanami, M., Yasuda, S. & Hirota, Y. (1985) J. Mol. Biol. 184, 529-533. [DOI] [PubMed] [Google Scholar]
- 12.Schaper, S. & Messer, W. (1995) J. Biol. Chem. 270, 17622-17626. [DOI] [PubMed] [Google Scholar]
- 13.Grimwade, J. E., Ryan, V. T. & Leonard, A. C. (2000) Mol. Microbiol. 35, 835-844. [DOI] [PubMed] [Google Scholar]
- 14.Ryan, V. T., Grimwade, J. E., Nievera, C. J. & Leonard, A. C. (2002) Mol. Microbiol. 46, 113-124. [DOI] [PubMed] [Google Scholar]
- 15.Zyskind, J. W., Cleary, J. M., Brusilow, W. S., Harding, N. E. & Smith, D. W. (1983) Proc. Natl. Acad. Sci. USA 80, 1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weigel, C., Schmidt, A., Ruckery, B., Lurz, R. & Messer, W. (1997) EMBO J. 16, 6574-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullock, W. O., Fernandez, J. M. & Short, J. M. (1987) BioTechniques 5, 376-379. [Google Scholar]
- 18.DeLucia, P. & Cairns, J. (1969) Nature 224, 1164-1166. [DOI] [PubMed] [Google Scholar]
- 19.Li, Z. & Crooke, E. (1999) Protein Expression Purif. 17, 41-48. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd. Ed.
- 21.Hwang, D. S. & Kornberg, A. (1992) J. Biol. Chem. 267, 23083-23086. [PubMed] [Google Scholar]
- 22.Margulies, C. & Kaguni, J. M. (1996) J. Biol. Chem. 271, 17035-17040. [DOI] [PubMed] [Google Scholar]
- 23.Samitt, C. E., Hansen, F. G., Miller, J. F. & Schaechter, M. (1989) EMBO J. 8, 989-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cassler, M. R., Grimwade, J. E. & Leonard, A. C. (1995) EMBO J. 14, 5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay, G., Carr, K. M., Kaguni, J. M. & Chattoraj, D. K. (1993) EMBO J. 12, 4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speck, C., Weigel, C. & Messer, W. (1997) Nucleic Acids Res. 25, 3242-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujikawa, N., Kurumizaka, H., Nureki, O., Terada, T., Shirouzu, M., Katayama, T. & Yokoyama, S. (2003) Nucleic Acids Res. 31, 2077-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speck, C. & Messer, W. (2001) EMBO J. 20, 1469-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka, A., Sasaki, H., Sugimoto, K. & Takanami, M. (1984) J. Mol. Biol. 176, 443-458. [DOI] [PubMed] [Google Scholar]
- 30.Simmons, L. A., Felczak, M. & Kaguni, J. M. (2003) Mol. Microbiol. 49, 849-858. [DOI] [PubMed] [Google Scholar]
- 31.Langer, U., Richter, S., Roth, A., Weigel, C. & Messer, W. (1996) Mol. Microbiol. 21, 301-311. [DOI] [PubMed] [Google Scholar]
- 32.Skarstad, K. & Lobner-Olesen, A. (2003) EMBO J. 22, 140-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurokawa, K., Nishida, S., Emoto, A., Sekimizu, K. & Katayama, T. (1999) EMBO J. 18, 6642-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crooke, E., Castuma, C. E. & Kornberg, A. (1992) J. Biol. Chem. 267, 16779-16782. [PubMed] [Google Scholar]
- 35.Diffley, J. F., Cocker, J. H., Dowell, S. J., Harwood, J. & Rowley, A. (1995) J. Cell Sci. Suppl. 19, 67-72. [DOI] [PubMed] [Google Scholar]
- 36.Ritzi, M. & Knippers, R. (2000) Gene 245, 13-20. [DOI] [PubMed] [Google Scholar]