Abstract
Although protein folding domains are generally conserved for function across distant homologous sequences, one crucial structural feature is not conserved: the wrapping of backbone hydrogen bonds, that is, the extent to which they are intramolecularly desolvated and thereby protected from water attack. Extensive data on protein complex interfaces led us to postulate that insufficiently wrapped backbone hydrogen bonds in monomeric domains must be adhesive, and therefore determinants of interactivity, a result that has been experimentally confirmed. Here, we show that the wrapping of certain conserved folds becomes progressively poorer as species diverge in some lineages. This trend is thus concurrent with a progressive enhancement of the interactivity of individual domains sharing the conserved fold. Such increase in interactivity is predicted to impose an “evolutionary brake” on the overall speed of sequence divergence. This phenomenon follows when more and more residues become engaged in protein associations and thus become functionally indispensable. For complete proteomes for which statistically significant structural data are available, scale-free network statistics based solely on the distribution of folding domains, catalogued by their number of wrapping defects, best describe the proteomic connectivity. Thus, the intermolecular connectivity may be effectively used as a measure of species complexity. Our results might contribute to explaining how interactome complexity may be achieved without a dramatic increase in genome size.
A paradigmatic discovery links protein fold and biological function: the conservation of the fold across distant homologous sequences (1, 2). However, not all structural features are preserved, and in particular, a crucial factor defining protein interactivity is not conserved. This article describes this phenomenon and its consequences for the evolution of proteomic connectivity. Specifically, we address the question: Are there gene-encoded signals of structure that determine lower or higher interactivity as the same fold is examined across species?
We recently introduced an indicator of protein interactivity, the underdehydrated or underwrapped hydrogen bond (3–9), recently termed dehydron (7), which is encoded in the 3D structure of protein folded domains. Dehydrons are sites of structural vulnerabilities resulting from incomplete wrapping or intramolecular desolvation of backbone hydrogen bonds (7–9); they have been previously shown statistically (5) and experimentally (6, 8) to signal adhesive sites, potentially determinant of protein–protein associations. A systematic mining of such signals is used here to identify patterns of proteomic connectivity.
By examining folds conserved for function across species we found significant differences in the number and distribution of dehydrons. Within a conserved fold, the number of dehydrons in higher eukaryotes is consistently greater than in species of lower complexity. This finding indicates a higher interactivity because, as shown here, domain connectivity is proportional to the average number of dehydrons in the family. Furthermore, the dehydron patterns associated with structural domains for a given species consistently define a scale-free interaction network for the species proteomes.
The biological complexity of higher eukaryotes does not result from their genome or even the estimated transcriptome sizes (10–12). Furthermore, the combinatorial multiplicity arising from domain shuffling cannot properly account for qualitative differences in power-law descriptions of proteomic connectivity (10). This statement refers to an underlying picture in which interactions between protein domains are mapped as connections on a network, where each domain is represented as a node, and highly connected nodes are scarce, whereas nodes with few connections are common, as fitted by a power-law distribution.
Methods
The extent of intramolecular hydrogen-bond desolvation in a monomeric structure may be quantified by determining the number of hydrophobic carbonaceous groups (CHn, n = 1, 2, or 3) within a desolvation domain typically defined as two inter-secting balls of fixed radius centered at the α-carbons of the hydrogen-bonded residues (3–9). The details of these calculations have been extensively discussed (3–9) and thus only need to be sketched here. The precise statistics of hydrogen-bond wrapping vary according to the desolvation radius adopted, but the tails of the distribution single out the same dehydrons in a given structure over a 6.0- to 7.4-Å range in the adopted desolvation radius. In this work the value 6.4 Å was adopted.
In most [≈92% of Protein Data Bank (PDB) entries] stable protein folds, the backbone hydrogen bonds are on average wrapped by ρ = 18.7 ± 5.9 hydrophobic groups (or 15.0 ± 3.7 if we count only side-chain groups and exclude those from the hydrogen-bonded residue pair, as indicated in ref. 3). The sole exception to these statistics are certain cellular prion proteins (9), with average wrapping as low as ρ = 11.70 (Table 1), and some neurotoxins held together by a profusion of disulfide bridges (5, 9). Dehydrons are then defined as hydrogen bonds in the tails of the distribution, i.e., with <12 hydrophobic groups in their desolvation domains. Because there is a considerable thermodynamic advantage associated with the removal of water surrounding dehydrons, such bonds have been identified, together with the overexposed hydrophobic groups that attach to the dehydrons, as determinants of protein binding sites, conferring specificity to protein interactions. They are virtually ubiquitous factors, occurring as points of contact in 38% of the PDB complexes. For those complexes, the density of dehydrons at the protein–protein interface is >1.5 times the average density for individual monomeric partners. Furthermore, dehydrons constitute a significant factor (interface dehydron density larger than average) in 92.9% of all PDB complexes (5).
Table 1. Examples of the most dramatic variations in the extent of desolvation of backbone hydrogen bonds for the same folding domains examined across different species.
| Protein or domain | Species/PDB code | G, Mb | N | NHB | ρ | Y | rd/HB (×100) |
|---|---|---|---|---|---|---|---|
| DHFR | Haloferax volcanii (archaea)/1vdr | ≈1.8 | 157 | 82 | 21.84 | 4 | 4.8 |
| DHFR | Thermotoga maritima (bacteria)/1dlg | 1.8 | 164 | 87 | 21.78 | 5 | 5.7 |
| DHFR | E. coli/1dra | 4.6 | 159 | 84 | 21.11 | 5 | 5.9 |
| DHFR | Lactobacillus casei/3dfr | 162 | 90 | 19.62 | 11 | 12.2 | |
| DHFR | H. sapiens/1hfp | ≈3,000 | 186 | 95 | 18.21 | 16 | 16.8 |
| Ankyrin repeat | M. musculus (mouse)/lap7 | ≈3,000 | 168 | 77 | 17.08 | 11 | 14.3 |
| Ankyrin repeat | H. sapiens/1bd8 | ≈3,000 | 156 | 88 | 16.72 | 16 | 18.2 |
| Cytochrome c | Chlamydomonas reinhardtii (algae)/1cyi | ≈100 | 89 | 52 | 19.74 | 6 | 11.5 |
| Cytochrome c | Rhodopila globiformis (bacteria)/1hro | 105 | 50 | 17.52 | 7 | 14.0 | |
| Cytochrome c | Oryza sativa (rice)/1ccr | 430 | 111 | 55 | 14.94 | 11 | 20.0 |
| Cytochrome c | Thunnus alalunga (tuna)/5cyt | 103 | 53 | 14.25 | 13 | 24.5 | |
| Cytochrome c | Katsuo (bonito)/1cyc | 103 | 41 | 14.03 | 12 | 29.2 | |
| Cytochrome c | Equus caballus/1giw | ≈3,000 | 104 | 44 | 14.01 | 14 | 31.8 |
| Hemoglobin | Vitreoscilla stercoraria (bacteria)/2vhb | ≈4 | 136 | 102 | 23.50 | 0 | 0 |
| Hemoglobin | Lupinus luteus (pea)/1gdj | 153 | 109 | 23.43 | 0 | 0 | |
| Hemoglobin | Paramecium caudatum/1dlw | 116 | 77 | 22.02 | 0 | 0 | |
| Hemoglobin | (Nonsymbiotic) Oryza sativa (rice)/1d8u | 430 | 165 | 106 | 23.58 | 2 | 1.8 |
| Hemoglobin | Equus caballus/1gob | ≈3,000 | 146 | 101 | 21.45 | 2 | 2.0 |
| Hemoglobin | H. sapiens/1bz0 | ≈3,000 | 146 | 103 | 21.45 | 3 | 2.9 |
| Hsp90 chaperone | Saccharomyces cerevisiae (yeast)/1amw | 12.1 | 213 | 147 | 20.07 | 20 | 13.6 |
| Hsp90 chaperone | H. sapiens/1byq | ≈3,000 | 213 | 139 | 19.37 | 26 | 18.7 |
| Lysozyme | Coliphage T4 (phage)/109L | 4–5 | 160 | 120 | 18.68 | 18 | 15.0 |
| Lysozyme | Gallus gallus (hen egg white)/132L | 129 | 85 | 17.42 | 19 | 22.3 | |
| Lysozyme | Canis familiaris/1ell | ≈3,000 | 130 | 90 | 17.34 | 20 | 22.2 |
| Lysozyme | H. sapiens/133L | ≈3,000 | 130 | 86 | 16.38 | 29 | 33.7 |
| Myoglobin | Aplysia limacina (mollusc)/1mba | 146 | 106 | 23.42 | 0 | 0 | |
| Myoglobin | Chironomus thummi thummi (insect)/1eca | ≈200 | 136 | 101 | 21.31 | 3 | 2.9 |
| Myoglobin | Thunnus albacares (tuna)/1myt | 146 | 110 | 21.15 | 8 | 7.2 | |
| Myoglobin | Caretta caretta (sea turtle)/1lht | 153 | 110 | 21.09 | 11 | 10.0 | |
| Myoglobin | Physeter catodon (whale)/1bz6 | 153 | 113 | 20.98 | 11 | 9.7 | |
| Myoglobin | Sus scrofa (pig)/1mwc | ≈2,700 | 153 | 113 | 19.95 | 12 | 10.6 |
| Myoglobin | Equus caballus/1dwr | ≈3,000 | 152 | 112 | 18.90 | 14 | 12.5 |
| Myoglobin | Elephas maximus (Asian elephant)/1emy | 153 | 115 | 18.90 | 15 | 13.0 | |
| Myoglobin | Phoca vitulina (seal)/1mbs | 153 | 109 | 18.84 | 16 | 14.7 | |
| Myoglobin | H. sapiens/2hbc | ≈3,000 | 146 | 102 | 18.80 | 16 | 15.7 |
| PDZ | Drosophila melanogaster/1ihj | 137 | 94 | 47 | 17.88 | 3 | 6.4 |
| PDZ | Rattus norvegicus/1qlc | ≈3,000 | 95 | 41 | 17.80 | 8 | 19.5 |
| PDZ | H. sapiens/1g90 | ≈3,000 | 91 | 44 | 16.29 | 9 | 20.4 |
| PrPc | S. cerevisiae/1koa | 12.1 | 233 | 148 | 22.33 | 13 | 8.7 |
| PrPc | S. cerevisiae/1kod | 12.1 | 220 | 137 | 22.95 | 10 | 7.3 |
| PrPc | M. musculus/1ag2 | ≈3,000 | 103 | 53 | 12.42 | 29 | 54.7 |
| PrPc | Mesocricetus auratus (Syrian hamster)/1b10 | 104 | 59 | 11.79 | 35 | 59.3 | |
| PrPc | Bos taurus/1dwy | ≈3,000 | 104 | 59 | 11.76 | 35 | 59.3 |
| PrPc | H. sapiens/1qm0 | ≈3,000 | 104 | 59 | 11.71 | 35 | 59.3 |
| Reverse transcriptase | Moloney murine leukemia virus/1mml | ≈10-2 | 251 | 158 | 19.71 | 12 | 7.6 |
| Reverse transcriptase | HIV-1 (RT domains 1,2)/1rth | ≈10-2 | 209 | 120 | 16.68 | 21 | 17.5 |
| SH3 | C. elegans (worm)/3sem | 97 | 57 | 31 | 18.48 | 1 | 3.2 |
| SH3 | Gallus gallus/1hd3 | 58 | 29 | 18.40 | 2 | 6.9 | |
| SH3 | H. sapiens/5hck | ≈3,000 | 61 | 24 | 16.74 | 4 | 16.6 |
| Ubiquitin | E. coli/1foz | 4.6 | 66 | 39 | 18.69 | 4 | 10.2 |
| Ubiquitin | M. musculus/1u9b | ≈3,000 | 158 | 104 | 18.54 | 19 | 18.2 |
| Ubiquitin | H. sapiens/1ubi | ≈3,000 | 76 | 48 | 16.56 | 9 | 18.7 |
The notations are as follows: G, estimated genome size when known; N, polypeptide chain length; NHB, total no. of backbone hydrogen bonds; ρ, no. of desolvating carbonaceous groups per backbone hydrogen bond averaged over all hydrogen bonds in protein structure; Y, total no. of dehydrons in the structure; and rd/HB(×100), percentage ratio of dehydrons (no. of dehydrons every hundred hydrogen bonds).
The desolvation spheres could alternatively be defined as centered at N and O, the hydrogen-bond heavy atoms; this is not the criterion adopted here. Defined in terms of N and O, their 78% overlap and the geometric hindrance associated with bringing a third residue side chain to proximity with the backbone makes the statistics less revealing: most hydrogen bonds appear nearly equally protected, by 8–11 nonpolar groups.
Results
Dehydrons as Determinants of Proteomic Connectivity. Now we are in a position to answer the question posed at the outset: there is at least one kind of structural characteristic that does seem to change in a recognizable, somewhat systematic way in interspecies comparisons. Although folds are generally preserved for function, we find vast differences in the way conserved folds pack their backbone hydrogen bonds. Within a conserved domain fold, the number of dehydrons in higher eukaryotes is consistently greater than in, say, bacteria, and appears to be a consistent signature of the species complexity. Because dehydrons have been found to be adhesive and to determine binding sites (3–6) (further validation of this result will be given in this work) they will be used here to assess proteomic interactivity. Furthermore, the defects in the packing of structural domains within a given species will be shown to consistently determine a characteristic exponent that describes the node distribution within a scale-free interaction network (13–15).
The extent of intramolecular hydrogen-bond protection within a monomeric structure is quantified as described in Methods. With the desolvation-sphere radius fixed at 6.4 Å, the parameter ρ unambiguously determines the extent of underwrapping (ρ < 12) that makes a hydrogen bond a dehydron.
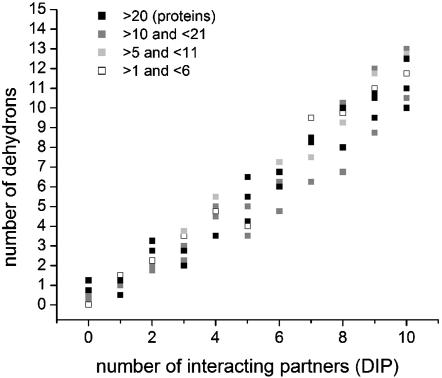
From a proteomics perspective, the role of dehydrons as determinants of interactivity is clearly validated. Fig. 1 shows a correlation between the number of dehydrons in all monomeric PDB domains from the yeast proteome and the number of interacting partners of such domains, inferred from large-scale two-hybrid experiments, deposited in the Database of Interacting Proteins (DIP) (16), a unique database derived from the yeast proteome. The correlation is statistically significant, with correlation coefficient 0.88 and dispersion 0.29. The DIP is unlikely to report binding partnerships exhaustively. Despite these shortcomings, the correlation clearly argues for dehydrons as markers for interactivity.
Fig. 1.
Correlation between the number of dehydrons of a given domain fold averaged over all proteins in the domain and its number of Database of Interacting Proteins (DIP)-reported interactive partners. The domains are binned according to their number of interactive partners and, given a fixed number of partners, they are subsequently grouped according to the abundance of PDB proteins in each domain. Each domain has an associated (averaged) number of dehydrons. Thus, black squares correspond to domains with >20 proteins; dark gray squares correspond to domains containing between 10 and 21 proteins; light gray squares correspond to domains between 5 and 11 proteins; and empty squares indicate domains with <6 representatives. The correlation is significant, with a correlation coefficient of 0.88 and dispersion 0.29.
Dehydrons are not in vitro artifacts (7) and, on the other hand, it is unlikely that underwrapped structure would prevail in vivo: the preservation of the structural integrity of functional proteins requires intermolecular contacts to prevent water attack on the hydrogen bonds. Unfortunately, no database of in vivo protein associations seems to exist at present.
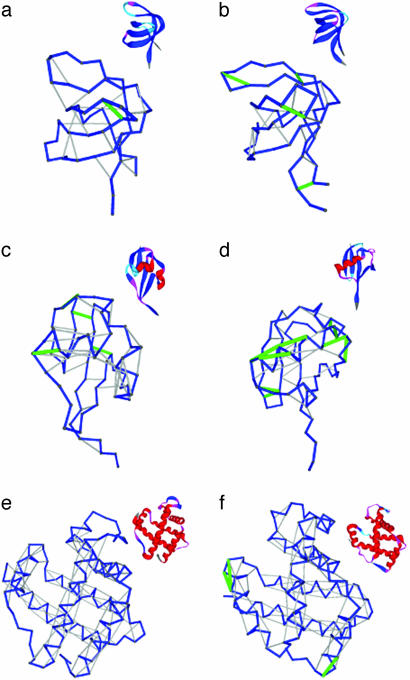
Dehydron Patterns Across Species. We noticed (Table 1) that when the same folding domain within a protein family is examined across different species, there are marked nontrivial differences in the percentage ratio r = rd/HB of dehydrons to backbone hydrogen bonds. Thus, for instance, the Src homology 3 (SH3) domain in the nematode Caenorhabditis elegans (pdb.3sem) has an r of 3.2% dehydrons in contrast with 16.6% in the human SH3 domain (pdb.5hck) (Fig. 2 a and b). Because dehydrons determine protein interactivity, this difference suggests a far more complex signal-transduction network in the latter species. Likewise, on the same basis, the human ubiquitin (pdb.1ubi, r = 18.7) is more interactive than its Escherichia coli counterpart (pdb.1foz, r = 10.2) (Fig. 2 c and d).
Fig. 2.
Illustrative comparative analysis of the packing of backbone hydrogen bonds for the same domains in different species. The dehydrons are indicated as green segments joining α-carbons, properly desolvated backbone hydrogen bonds are shown as gray segments, and the backbone conformation is displayed as a blue virtual-bond polygonal joining α-carbons. SH3 domains are from nematode C. elegans (pdb.3sem) (a) and H. sapiens (pdb.5hck) (b); ubiquitin is from E. coli (pdb.1foz) (c) and H. sapiens (pdb.1ubi) (d); and hemoglobin is from Paramecium (pdb.1dlw) (e) and H. sapiens β-subunit (pdb.1bz0, chain B) (f).
Even within the relatively noninteractive hemoglobin domain there are significant differences: the paramecium “hemoglobin” (pdb.1 dlw) is a perfect desolvator of its hydrogen bonds (r = 0), and is monomeric in vivo. The analogous-fold hemoglobin β-subunit (pdb.1bz0, r = 2.9) in humans possesses three dehydrons (Fig. 2 e and f) and occurs as a tetramer. Between these is the dimeric hemoglobin of mollusks. Within the natural interactive context of the human Hb subunit, the dehydrons signal crucial binding sites: dehydrons (90,94), (90,95) are associated with the β-FG corner involved in the quaternary α1β2 interface, whereas dehydron (5,9) is adjacent to Glu-6, which in sickle cell anemia mutates to Val-6, and is located at the protein–protein Glu-6-(Phe-85, Leu-88) interface in the deoxy-HbS fiber (17).
Fig. 2 and Table 1 suggest that as more complex species diverge, the conserved fold associated with a given function becomes more interactive. We thus predict that a “brake” must apply to the sequence evolutionary speed caused by the increasing number of binding-related residues (cf. refs. 1 and 2).
As dehydrons occur in diverging species, they turn the hydrogen-bonded residues into potential interactors and thus are likely to be conserved.
Dehydrons and Proteomic Networks. The broad separation in the extent of hydrogen-bond protection for different structural domains shown in Table 1 suggests the need to correlate the interactivity of structural families [or Structural Classification of Proteins (scop) superfamilies] (18, 19) with their average r value determined over all representative domains in the family reported in the PDB. Interactivity is here defined based on a structural criterion, i.e., by the concurrence of different domains in PDB protein complexes.
Although we are constrained to adopt a structural database to compute the average r, the number of connections ν of the family may be inferred independently from a broader database such as pfam, which covers at least 65% of the present swissprot release (20). The parameter ν of a family can be inferred from a structural database by enumerating all of the partner families that contain domains found in PDB entries to be engaged in complexation or intramolecular interaction, in the case of multidomain proteins, with a domain in the given family (21–25).
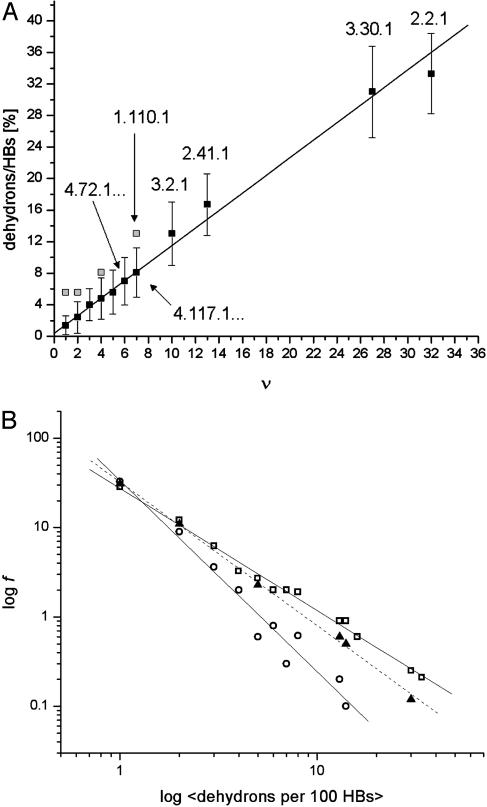
The strong correlation between the average ratio of dehydrons to well wrapped hydrogen bonds in a family and its parameter ν is displayed in Fig. 3A for all of the families with domains represented in the PDB. Although a family might be found in the PDB, its interactions might not be exhaustively reported in this database (such families are indicated as gray squares) and connectivities need to be obtained from the broader nonstructural pfam database. The families whose interactions were exhaustively explored in the PDB correspond to the dark squares in Fig. 3A.
Fig. 3.
(A) Correlation between the average dehydron ratio and the connectivity ν of scop families represented in the PDB. Of the 831 families interrogated, many share identical (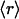 , ν) points, albeit with different r dispersions. The error bars on the ordinates represent the dispersion in the r ratios across all members of each family subsequently averaged over all families sharing the same (
, ν) points, albeit with different r dispersions. The error bars on the ordinates represent the dispersion in the r ratios across all members of each family subsequently averaged over all families sharing the same (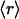 , ν) point. The connectivities of families marked by solid squares were determined by examining PDB complexes where at least one domain belongs to the family, and independently from a nonstructural database (pfam). The gray squares denote families with a well determined r ratio but whose connectivity is underreported in the PDB. They are located on the correlation line once their connectivity is independently determined from a nonstructural database (pfam). The most interactive such family, with 8% dehydrons, is 1.115.1 whose PDB ν value is 4, but an independent nonstructural assessment using pfam gives ν = 7. (B) scop families distributed according to their average ratio
, ν) point. The connectivities of families marked by solid squares were determined by examining PDB complexes where at least one domain belongs to the family, and independently from a nonstructural database (pfam). The gray squares denote families with a well determined r ratio but whose connectivity is underreported in the PDB. They are located on the correlation line once their connectivity is independently determined from a nonstructural database (pfam). The most interactive such family, with 8% dehydrons, is 1.115.1 whose PDB ν value is 4, but an independent nonstructural assessment using pfam gives ν = 7. (B) scop families distributed according to their average ratio 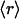 of dehydrons per 100 hydrogen bonds (HBs). The quantity f = f(
of dehydrons per 100 hydrogen bonds (HBs). The quantity f = f(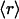 ), with f = fraction of total number of families, gives the distribution here plotted in log–log scale. □, H. sapiens; ▴, M. musculus; ○, E. coli.
), with f = fraction of total number of families, gives the distribution here plotted in log–log scale. □, H. sapiens; ▴, M. musculus; ○, E. coli.
Of the 831 scop families interrogated, many share identical (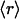 , ν) points in the plot displayed in Fig. 3A, albeit with different r dispersions. The error bars on the ordinates represent the dispersion in the r ratios across all members of each family subsequently averaged over all families partaking the same (
, ν) points in the plot displayed in Fig. 3A, albeit with different r dispersions. The error bars on the ordinates represent the dispersion in the r ratios across all members of each family subsequently averaged over all families partaking the same (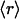 , ν) point. The actual statistics of the abundance of families grouped according to
, ν) point. The actual statistics of the abundance of families grouped according to 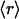 values is reported below. Although r values are computed on individual proteins and then averaged over a given family to obtain the
values is reported below. Although r values are computed on individual proteins and then averaged over a given family to obtain the 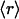 parameter, the interactivity ν is only an attribute of the family as a whole, as it arises by interrogating individual PDB complexes and determining the concurrence of protein domains from different families. A strong proportionality (correlation parameter: 0.92; dispersion: 0.10%) exists between the average ratio r and the connectivity parameter ν for those families whose interactivity is properly represented in the PDB (in such cases, the ν value has been found to coincide with that obtained from the pfam database). The linear correlation encompasses highly interactive (ν > 20) Ig and P-loop NTP hydrolase families (scop ID nos. 2.2.1 and 3.30.1, respectively), middle-interactive families (6 < ν ≤ 20), such as the serine proteases, Rossman domains, kinases, and signal-transduction SH2 domain (scop 1.48 ID nos. 2.41.1, 3.2.1, 4.117.1, and 4.72.1, respectively) and even all of the sparsely interactive 743 families with ν in the range 1 ≤ ν ≤ 6.
parameter, the interactivity ν is only an attribute of the family as a whole, as it arises by interrogating individual PDB complexes and determining the concurrence of protein domains from different families. A strong proportionality (correlation parameter: 0.92; dispersion: 0.10%) exists between the average ratio r and the connectivity parameter ν for those families whose interactivity is properly represented in the PDB (in such cases, the ν value has been found to coincide with that obtained from the pfam database). The linear correlation encompasses highly interactive (ν > 20) Ig and P-loop NTP hydrolase families (scop ID nos. 2.2.1 and 3.30.1, respectively), middle-interactive families (6 < ν ≤ 20), such as the serine proteases, Rossman domains, kinases, and signal-transduction SH2 domain (scop 1.48 ID nos. 2.41.1, 3.2.1, 4.117.1, and 4.72.1, respectively) and even all of the sparsely interactive 743 families with ν in the range 1 ≤ ν ≤ 6.
This correlation is meaningful only in a statistical sense and does not hold at the individual protein level for a number of reasons: (i) the interactivity at the molecular level is in general underreported (see above); (ii) the cytosolic or cytoplasmic environment is a masking factor that affects the number of dehydrons without bearing significantly on proteomic interactivity (8); (iii) the spatial proximity of several dehydrons within a single structure might tie them up to a single binding mode (5), and (iv) even though an adhesive dehydron might arise on the protein surface, there is no guarantee that a proper geometric match would exist for that region. Only further studies will reveal specific implications for living organisms.
The ν value for the most underrepresented major interactive family, the armadillo (ARM) repeat (scop ID no. 1.110.1), falls along the proportionality line when its connectivity is inferred from the pfam database. The same is true for some ν = 5 and 7 families, like 1.115.1 and 3.3.1, whose connections are underreported in the PDB. The fact that the Ig family has more interactivity than would be expected on the basis of its average number of dehydrons (Fig. 3A) probably implies that it is using binding sites in a relatively unselective way, although this idea needs to be established by identifying complexes with different partners using overlapping binding sites.
Given that, as shown in Fig. 3A, the average number of dehydrons per 100 hydrogen bonds in a given family, 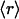 , is a measure of its connectivity, we can determine the interactive complexity of a species by mapping the distribution of protein families according to their average r value. Thus, Fig. 3B shows the fraction f = f(
, is a measure of its connectivity, we can determine the interactive complexity of a species by mapping the distribution of protein families according to their average r value. Thus, Fig. 3B shows the fraction f = f(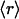 ) of the total number of families having on average
) of the total number of families having on average 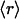 dehydrons per 100 backbone hydrogen bonds in their structural domains. The data are shown for three species chosen based on the number of family representatives in the PDB. The species selection criterion adopted is that at least two-thirds of the scop families for the species must have enough representative domains in the PDB so that the quantity
dehydrons per 100 backbone hydrogen bonds in their structural domains. The data are shown for three species chosen based on the number of family representatives in the PDB. The species selection criterion adopted is that at least two-thirds of the scop families for the species must have enough representative domains in the PDB so that the quantity 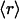 becomes statistically significant (its dispersion over the family is smaller than
becomes statistically significant (its dispersion over the family is smaller than 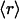 /2). Thus, the family distribution according to their dehydron abundances for three species selected are shown in Fig. 3B: Homo sapiens, Mus musculus, and E. coli.
/2). Thus, the family distribution according to their dehydron abundances for three species selected are shown in Fig. 3B: Homo sapiens, Mus musculus, and E. coli.
The results displayed in a log–log plot reveal a scale-free distribution (14, 24, 26) best approximated as f(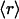 ) = 0.36
) = 0.36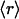 -γ, with γ = 1.44 for H. sapiens (broadest distribution), γ = 1.49 for M. musculus, and γ = 2.1 for E. coli. These results provide a structurally based assessment of the modulation of proteomic interactivity across different species resulting from the dramatic differences in the packing of conserved folds.
-γ, with γ = 1.44 for H. sapiens (broadest distribution), γ = 1.49 for M. musculus, and γ = 2.1 for E. coli. These results provide a structurally based assessment of the modulation of proteomic interactivity across different species resulting from the dramatic differences in the packing of conserved folds.
Discussion
Although the protein fold is mostly conserved for function across species, genotypic variation brings about a variability in the extent of molecular association needed to sustain the protein structure. This variability arises from differences in the extent to which intramolecular hydrogen bonds are shielded from water attack. Thus, as hydrogen bonds become less well wrapped intramolecularly, the conserved fold becomes more interactive, a trend observed as one examines species of increasing complexity. This result hinges on the fact that underwrapped hydrogen bonds (dehydrons) are inherently adhesive and their number on a given domain increases with species complexity.
Whether such greater levels of complexation are adventitious to certain functions or invariably inherent to their regulation remains to be determined, but the increment in interactivity must surely impose a slowdown in sequence variability that deserves further study.
A bewildering feature of higher organisms remains how complex physiologies can be achieved without a dramatic increase in genome size (the number of genes in the human genome proved to be deceptively low). Rice, for instance, has a relatively large genome, a mere order of magnitude smaller than higher mammals while probably possessing a far less complex physiology. On the other hand, most of the rice folds for specific functional domains are considerably better wrapped than their animal counterparts, as illustrated in Table 1. This trend cannot be statistically quantified at this point because of the dearth of high-resolution domains from the rice proteome in the PDB. Nevertheless our results imply that the interactome determined by the wrapping deficiencies in protein folds might represent a measure of complexity, helping explain how complex physiologies may be achieved without a significant increase in genome size.
Abbreviations: PDB, Protein Data Bank; SH3, Src homology 3.
References
- 1.Wilson, A. C. (1985) Sci. Am. 253 (4), 164-173. [DOI] [PubMed] [Google Scholar]
- 2.Moore, G. R. & Pettigrew, G. W. (1990) Cytochromes c: Evolutionary, Structural, and Physico-Chemical Aspects (Springer, Berlin).
- 3.Fernández, A., Colubri, A. & Berry, R. S. (2002) Physica A 307, 235-259. [Google Scholar]
- 4.Fernández, A. & Berry, R. S. (2002) Biophys. J. 83, 2475-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández, A. & Scheraga, H. A. (2003) Proc. Natl. Acad. Sci. USA 100, 113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández, A. & Scott, L. R. (2003) Phys. Rev. Lett. 91, 018102-018105. [DOI] [PubMed] [Google Scholar]
- 7.Fernández, A. & Scott, L. R. (2003) Biophys. J. 85, 1914-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández, A. & Berry, R. S. (2003) Proc. Natl. Acad. Sci. USA 100, 2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernández, A., Kardos, J., Scott, L. R., Goto, Y. & Berry, R. S. (2003) Proc. Natl. Acad. Sci. USA 100, 6446-6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koonin, E. V., Wolf, Y. I. & Karev, G. P. (2002) Nature 420, 218-223. [DOI] [PubMed] [Google Scholar]
- 11.Wuchty, S. (2001) Mol. Biol. Evol. 18, 1694-1702. [DOI] [PubMed] [Google Scholar]
- 12.Vitkup, D., Melamud, E., Moult, J. & Sander, C. (2001) Nat. Struct. Biol. 8, 559-566. [DOI] [PubMed] [Google Scholar]
- 13.Barabasi, A. & Albert, R. (1999) Science 286, 509-512. [DOI] [PubMed] [Google Scholar]
- 14.Barabasi, A. L. (2002) Linked: The New Science of Networks (Perseus, New York).
- 15.Apic, G., Gough, J. & Teichmann, S. A. (2001) Bioinformatics 17, Suppl. 1, S83-S89. [DOI] [PubMed] [Google Scholar]
- 16.Xenarios, I., Salwinski, L., Duan, X. J., Higney, P., Kim, S. M. & Eisenberg, D. (2002) Nucleic Acids Res. 30, 303-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voet, D. & Voet, J. G. (1995) Biochemistry (Wiley, New York).
- 18.Murzin, A., Brenner S. E., Hubbard, T. J. P. & Chothia, C. (1995) J. Mol. Biol. 247, 536-540. [DOI] [PubMed] [Google Scholar]
- 19.LoConte, L., Brenner, S. E., Hubbard, T. J., Chothia, C. & Murzin, A. G. (2002) Nucleic Acids Res. 30, 264-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bateman, A., Birney, E., Durbin, R., Eddy, S., Howe, K. & Sonnhammer, E. (2000) Nucleic Acids Res. 28, 263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park, J., Lappe, M. & Teichmann, S. A. (2001) J. Mol. Biol. 307, 929-938. [DOI] [PubMed] [Google Scholar]
- 22.Janin, J., Miller, S. & Chothia, C. (1998) J. Mol. Biol. 204, 155-164. [DOI] [PubMed] [Google Scholar]
- 23.Jones, S., Marin, A. & Thornton, J. M. (2000) Protein Eng. 13, 77-82. [DOI] [PubMed] [Google Scholar]
- 24.Aloy, P. & Russell, R. B. (2002) Proc. Natl. Acad. Sci. USA 99, 5896-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan, X. J., Xenarios, I. & Eisenberg, D. (2002) Mol. Cell. Proteomics 1, 104-116. [DOI] [PubMed] [Google Scholar]
- 26.Wolf, Y. I., Brenner, S. E., Bash, P. A. & Koonin, E. V. (1999) Genome Res. 9, 17-26. [PubMed] [Google Scholar]