Abstract
Human immunodeficiency virus (HIV) protease inhibitors (PIs) have been used successfully in extending the life span of people infected with HIV. The use of PIs has also been associated with dyslipidemia and an increased risk of cardiovascular disease, but the underlying mechanisms remain elusive. Several PIs have been implicated in activating the nuclear receptor pregnane X receptor (PXR), which acts as a xenobiotic sensor to regulate xenobiotic metabolism in the liver and intestine. Recent studies indicate that PXR may also play an important role in the regulation of lipid homeostasis. In the present study, we identified amprenavir, a widely used HIV PI, as a potent PXR-selective agonist. Computational docking studies combined with site-direct mutagenesis identified several key residues within the ligand-binding pocket of PXR that constitute points of interaction with amprenavir. Amprenavir efficiently activated PXR and induced PXR target gene expression in vitro and in vivo. Short-term exposure to amprenavir significantly increased plasma total cholesterol and atherogenic low-density lipoprotein cholesterol levels in wild-type mice, but not in PXR-deficient mice. Amprenavir-mediated PXR activation stimulated the expression of several key intestinal genes involved in lipid homeostasis. These findings provide critical mechanistic insight for understanding the impact of PIs on cardiovascular disease and demonstrate a potential role of PXR in mediating the adverse effects of HIV PIs in humans.
Introduction
The introduction of highly active antiretroviral therapy (HAART) has led to a marked increase in survival for people infected with human immunodeficiency virus (HIV) (Bhaskaran et al., 2008). However, HAART has also been associated with dyslipidemia and an increased risk of cardiovascular disease (Carr et al., 1998; Periard et al., 1999; Distler et al., 2001; Stein et al., 2001; Friis-Moller et al., 2003, 2007; Lang et al., 2010). For example, findings from the Data Collection on Adverse Events of Anti-HIV Drugs (DAD) study suggested that the incidence of cardiovascular disease increased with longer exposure to HAART (Friis-Moller et al., 2003). Another large-scale study that included more than 23,000 HIV patients analyzed the combination of drugs used in HAART and concluded that protease inhibitors (PIs) are associated with an increased risk of cardiovascular disease, which is partly explained by dyslipidemia (Friis-Moller et al., 2007). A Swiss HIV Cohort Study found that PIs are the cause of dyslipidemia in patients (Periard et al., 1999), and several other studies have reached the same conclusion (Stein et al., 2001; Lang et al., 2010). Despite the strong evidence linking PIs with dyslipidemia and cardiovascular disease, the underlying mechanisms responsible for the adverse effects of PIs remain elusive.
Several PIs such as ritonavir have been implicated to activate the pregnane X receptor (PXR, also known as steroid and xenobiotic receptor) (Dussault et al., 2001; Zhou et al., 2009b). PXR is a nuclear receptor activated by numerous endogenous hormones, dietary steroids, pharmaceutic agents, and xenobiotic chemicals (Blumberg et al., 1998; Kliewer et al., 1998; Zhou et al., 2009b). PXR functions as a xenobiotic sensor that induces expression of genes required for xenobiotic metabolism in the liver and intestine, including cytochrome P450s, conjugating enzymes (e.g., glutathione transferase), and ABC family transporters (e.g., multidrug resistance 1 [MDR1]) (Kliewer et al., 2002; Zhou et al., 2009b). In the past decade, the role of PXR as a xenobiotic sensor has been well established (Zhou et al., 2009b).
Recent evidence indicates that PXR may also play an important role in the regulation of lipid homeostasis (Zhou et al., 2006b, 2009a; de Haan et al., 2009; Gao and Xie, 2012). It is well known that many clinically relevant PXR ligands can elevate plasma lipid levels in patients and may increase their risk of cardiovascular disease (Khogali et al., 1974; Eiris et al., 1995; Lutjohann et al., 2004; Shafran et al., 2005; Zhou et al., 2009a). A recent meta-analysis of seven genomewide association studies indicated that common genetic variants in PXR can affect plasma low-density lipoprotein (LDL) cholesterol levels in humans (Lu et al., 2010). Modulation of PXR activity has been found to alter lipid homeostasis and affect plasma lipid levels in several animal models. We recently reported that chronic activation of PXR led to increased levels of plasma total cholesterol and atherogenic LDL and very-low-density lipoprotein (VLDL) in wild-type (WT) mice, but not in PXR-deficient (PXR−/−) mice (Zhou et al., 2009a). Activation of PXR also increased plasma total cholesterol and VLDL levels in apolipoprotein E (ApoE)*3-Leiden mice which exhibit a human-like lipoprotein distribution on a cholesterol-rich diet (de Haan et al., 2009). Another report showed that acute activation of PXR increased plasma triglyceride levels in both LDL receptor-deficient and ApoE-deficient (ApoE−/−) mice but decreased LDL cholesterol levels in LDL receptor-deficient mice (Hoekstra et al., 2009). Activation of PXR can regulate many genes involved in lipid homeostasis including CD36, stearoyl-CoA desaturase-1,7-dehydrocholesterol reductase, S14, and lipin-1 in the liver, intestine, or macrophages of several animal models (Zhou et al., 2006b, 2009b; de Haan et al., 2009; Hoekstra et al., 2009; Moreau et al., 2009; Cheng et al., 2012; He et al., 2013). These studies indicate that PXR can mediate cholesterol and lipid homeostasis at multiple levels.
We report that amprenavir, a widely used HIV PI, is a potent PXR-selective agonist. Computational docking studies together with site-directed mutagenesis identified several key residues within PXR’s ligand-binding pocket that constitute points of interaction with amprenavir. Amprenavir efficiently activated PXR and induced PXR target gene expression in vitro and in vivo. Short-term exposure to amprenavir significantly increased plasma total cholesterol and atherogenic lipoprotein LDL cholesterol levels in WT mice, but not in PXR−/− mice. Amprenavir-mediated PXR activation significantly regulated several key intestinal genes involved in lipid homeostasis. These findings provide critical mechanistic insight for understanding the impact of PIs on cardiovascular disease and demonstrate a potential role of PXR in mediating the adverse effects of PIs in humans.
Materials and Methods
Reagents and Plasmids.
Pregnenolone 16α-carbonitrile (PCN) and rifampicin (RIF) were purchased from Sigma-Aldrich (St. Louis, MO). Amprenavir, ritonavir, nelfinavir, saquinavir, and indinavir sulfate were obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. All chemicals were dissolved in dimethyl sulfoxide. Human (h) and mouse (m) PXR expression vectors; GAL4 DNA-binding domain-linked nuclear receptor ligand-binding domain (LBD) vectors (GAL4-hPXR, GAL4-mPXR, GAL4-rPXR, GAL4- retinoid acid receptor α, GAL4-retinoid X receptor, GAL4-farnesoid X receptor, GAL4-liver X receptor, GAL4- peroxisome proliferator-activated receptor (PPAR)α, GAL4-PPARγ, and GAL4-vitamin D receptor); and cytomegalovirus X-β-gal expression vectors have been described before elsewhere (Zhou et al., 2006a, 2007; Sui et al., 2012). VP16-PXR, GAL4- nuclear receptor corepressor (NCoR), GAL4-silencing mediator of retinoid and thyroid hormone (SMRT), GAL4-steroid receptor coactivator-1 (SRC1), GAL4- PPAR-binding protein (PBP), and PXR-dependent CYP3A4 promoter reporter (CYP3A4XREM-luciferase); CYP3A2 promoter reporter [(CYP3A2)3-luciferase]; and GAL4 reporter (MH100-luciferase) have also been described previously elsewhere (Zhou et al., 2004, 2006a, 2007; Sui et al., 2012).
Cell Culture and Transfections.
The human hepatic cell line HepG2 and intestine epithelial cell line LS180 were obtained from the American Type Culture Collection (Manassas, VA). The human hepatoma HepaRG cells were purchased from Life Technologies (Carlsbad, CA). Transfection assays were performed as described previously elsewhere (Zhou et al., 2007; Sui et al., 2012). The cells were transfected with various expression plasmids or hPXR mutants as well as the corresponding luciferase reporter plasmids, together with cytomegalovirus X-β-galactosidase control plasmids using FuGENE 6 (Roche Diagnostics, Indianapolis, IN). The cells were then incubated with the corresponding ligands as indicated in the figure legends for 24 hours, and β-gal and luciferase assays were performed as described elsewhere (Zhou et al., 2007; Sui et al., 2012). Fold activation was calculated relative to the solvent controls.
Each data point represents the average of triplicate experiments ± S.D. and was replicated in three to five independent experiments. EC50 values were calculated by curve fitting of data, using Prism software (GraphPad Software, San Diego, CA). For the mammalian two-hybrid assays, HepG2 cells were transfected with GAL4 reporter, VP16-hPXR, and GAL-SRC1, GAL-PBP, GAL-NCoR, and GAL-SMRT (Zhou et al., 2007; Sui et al., 2012). The cells were then treated with compounds at the indicated concentrations.
Computational Docking Studies.
Preparation of the protein structure for subsequent ligand-receptor docking studies was conducted as previously described elsewhere (Sui et al., 2012). Briefly, the structural coordinates of the tethered hPXR linker PXR/SRC-1 were retrieved from the RSCB Protein Data Bank entry 3HVL. The larger PXR fragment of chain A, Gly142–Glu458, was extracted for molecular modeling using MOE 2010 software (Chemical Computing Group, Montreal, QC, Canada), and for ligand-receptor docking studies using GOLD software (version 5.0) (Payne and Glen, 1993). Water molecules, salt ions, ligand [SR12813, tetraethyl 2-(3,5-di-tert-butyl-4-hydroxyphenyl)ethenyl-1,1-bisphosphonate], and coreceptor fragments were deleted. After the addition of hydrogen atoms and assigning of the AMBER99 force-field charges to the protein, the hydrogen atomic positions were allowed to relax (Wang et al., 2008). The resulting protein structural coordinates were saved in Tripos mol2 format and used later for GOLD docking.
The ligands were docked to the 3HVL chain A using semiflexible docking whereby the ligand has full conformational flexibility and the hydroxyl groups of designated protein side chains in the binding pocket can rotate to optimize hydrogen bond contacts. Each ligand was docked 50 independent times. The binding pocket was defined as all atoms within an 8-Å radius around the bound ligand SR12813.
Site-Directed Mutagenesis.
The hPXR full-length plasmid was used as a wild-type template to generate a series of mutant plasmids using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer-supplied protocol as described before (Sui et al., 2012). The primers used for mutant generation are listed in Supplemental Table 1.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction Analysis.
Total RNA was isolated from mouse tissues, intestinal LS180, and HepaRG cells using TRIzol Reagent (Life Technologies) per the manufacturer-supplied protocol. Quantitative real-time polymerase chain reaction was performed using gene-specific primers and the SYBR green polymerase chain reaction kit (Life Technologies), as described previously elsewhere (Zhou et al., 2007; Sui et al., 2012). The primer sets used in this study are listed in Supplemental Table 2.
Animals.
C57BL/6 WT mice were purchased from The Jackson Laboratory (Bar Harbor, ME). PXR-deficient mice (PXR−/−) on C57BL/6 background were employed as described previously elsewhere (Zhou et al., 2009a). All the animals were housed in the Division of Laboratory Animal Resources, University of Kentucky, and the protocol was approved by the Institutional Animal Care and Use Committee in the specific pathogen-free environment with a light-dark cycle. We fed 8-week-old male WT or PXR−/− mice a semisynthetic low-fat AIN76 diet containing 0.02% cholesterol (Research Diet, New Brunswick, NJ) (Teupser et al., 2003; Zhou et al., 2009a; Sui et al., 2011) and treated them by oral gavage with vehicle (corn oil) or 10 mg/kg body weight amprenavir daily for 7 days. On the day of they were killed, the mice were anesthetized by intraperitoneal injection with ketamine (Fort Dodge Animal Health, Fort Dodge, IA). The mice were exsanguinated by left ventricular puncture, and the blood was collected into EDTA-containing syringes. Plasma was prepared by spinning at 16,000g for 10 minutes. The circulation was flushed with phosphate-buffered saline, and the intestinal and liver tissues were collected and stored in RNAlater solution (Life Technologies).
Plasma Analysis.
Plasma total cholesterol and triglyceride concentrations were determined enzymatically by colorimetric methods as described previously elsewhere (Roche Diagnostics) (Zhou et al., 2009a; Park et al., 2012). Plasma from multiple mice (n = 4–7) was pooled, and the plasma lipoprotein cholesterol distributions were determined by fast-performance liquid chromatography (FPLC) (Sui et al., 2011). The lipoprotein fractions were isolated by spinning 60 μl of plasma in a TL-100 ultracentrifuge (Beckman Coulter, Brea, CA) at its own density (1.006 g/ml) at 70,000g for 3 hours to harvest the supernatant, and then after adjusting the infranatant with solid KBr to a density of 1.063 g/ml spinning it for 70,000g for 18 hours to harvest the supernatant (Zhou et al., 2011). The cholesterol content of each supernatant and the final infranatant were measured and taken to be VLDL (<1.006 g/ml), LDL (1.006 ≤ d ≤ 1.063 g/ml), and high-density lipoprotein (HDL) (d > 1.063 g/ml) cholesterol, respectively. Cholesterol concentrations in all three fractions were then determined enzymatically by a colorimetric method (Roche Diagnostics). Plasma triglyceride levels were determined enzymatically in the original plasma sample.
Statistical Analysis.
All data are expressed as mean ± S.D. unless otherwise noted. The statistical analysis was performed using a two-sample, two-tailed Student’s t test unless otherwise noted, with P < 0.05 considered statistically significant. One-way analysis of variance was used when multiple comparisons were made, followed by Dunnett’s t test for multiple comparisons with a control.
Results
Amprenavir Is a Potent PXR-Selective Agonist.
We tested several widely used HIV PIs, including amprenavir, ritonavir, nelfinavir, saquinavir, and indinavir sulfate, for PXR activation by use of transfection assays. Because PXR exhibits considerable differences in its pharmacology across species (namely, mouse versus human) (Blumberg et al., 1998; Zhou et al., 2009b), the potent PXR ligands RIF and PCN were used as the positive control for human (h) and mouse (m) PXR, respectively. Consistent with a previous report (Dussault et al., 2001), ritonavir activated PXR and induced PXR-mediated reporter activity (Fig. 1, A and B). Interestingly, amprenavir was a more potent PXR agonist than any of the other tested PIs (Fig. 1, A and B). Amprenavir can activate both human and mouse PXR and induced reporter gene activity in a dose-dependent manner (Fig. 1, A and B). Dose-response analysis indicated that amprenavir was able to activate PXR at concentrations as low as 1 μM, reaching peak activation at 30 μM (Fig. 1C). The EC50 for amprenavir activation of PXR-mediated CYP3A4 promoter activity was 8.6 μM (Fig. 1C). We next tested the ability of amprenavir to activate a panel of other nuclear receptors, including human retinoid acid receptor α, retinoid X receptor, farnesoid X receptor, liver X receptor α, PPARα, PPARγ, vitamin D receptor, estrogen receptor α, and estrogen receptor β (Fig. 1D). Amprenavir was unable to activate any of these other nuclear receptors. Results from this panel emphasize the importance of studying the effects of amprenavir-mediated PXR agonism.
Fig. 1.
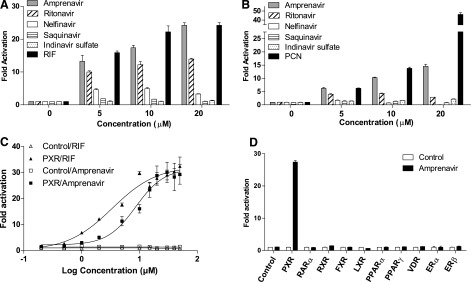
Amprenavir is a potent PXR-selective agonist. (A and B) HepG2 cells were transfected with (A) full-length hPXR together with a hPXR reporter (CYP3A4-luc) or (B) full-length mPXR together with a mPXR reporter [(CYP3A2)3-luc] and cytomegalovirus X (CMX)-β-galactosidase control plasmid. Cells were then treated with dimethyl sulfoxide (DMSO) control, amprenavir, ritonavir, nelfinavir, saquinavir, indinavir sulfate, RIF (a hPXR ligand), or PCN (a mPXR ligand) at the indicated concentrations for 24 hours. (C) HepG2 cells were cotransfected with hPXR together with CYP3A4-luc reporter and CMX-β-galactosidase plasmid. Cells were then treated with amprenavir or RIF at the indicated concentrations for 24 hours. (D) HepG2 cells were cotransfected with a GAL4 reporter and a series of GAL4 constructs in which the GAL4 DNA-binding domain (DBD) is linked to the indicated nuclear receptor LBD. Cells were treated with DMSO control or 10 μM amprenavir for 24 hours. Data are shown as fold induction of normalized luciferase activity compared with DMSO treatment and represent the mean of triplicate experiments. ER, estrogen receptor; FXR, farnesoid X receptor; LXR, liver X receptor; RAR, retinoid acid receptor; RXR, retinoid X receptor.
Amprenavir Promotes PXR Coactivator Recruitment and Corepressor Disassociation.
Nuclear receptor coregulators play critical roles in nuclear receptor signaling (Kliewer et al., 2002). We used a mammalian two-hybrid assay (Zhou et al., 2007; Sui et al., 2012) to evaluate the effects of amprenavir on PXR coregulator interaction. Similar to the potent PXR ligand RIF, amprenavir promoted the specific interactions between PXR and the coactivators SRC-1 and PBP (Fig. 2A). Consistent with our previous reports (Zhou et al., 2007; Sui et al., 2012), unliganded PXR interacted with corepressors, NCoR and SMRT. Amprenavir disrupted this interaction, as did RIF (Fig. 2B). Binding of amprenavir to PXR inhibits PXR/corepressor interaction and promotes PXR/coactivator recruitment, thereby inducing PXR transcriptional activation in a concentration-dependent manner.
Fig. 2.
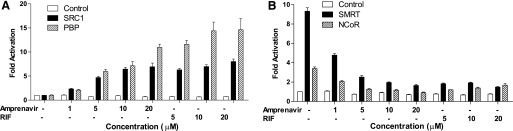
Amprenavir promotes PXR coactivator recruitment and corepressor disassociation. HepG2 cells were transfected with a GAL4 reporter and VP16-hPXR as well as expression vector for GAL4 DNA-binding domain (DBD) or GAL4 DBD linked to the receptor interaction domains of the indicated PXR coactivators (GAL-SRC1 and GAL-PBP) (A) or corepressor (GAL-SMRT and GAL-NCoR) (B). Cells were then treated with dimethyl sulfoxide (DMSO) control, amprenavir, or RIF at the indicated concentrations for 24 hours.
Computational Docking and Modeling Studies.
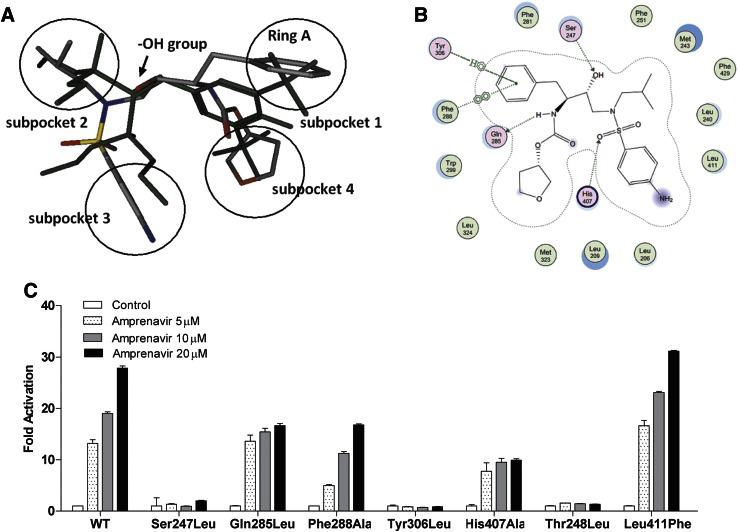
To investigate the potential interaction pattern between amprenavir and PXR, a structure-based approach was employed. Amprenavir was docked into the high-resolution crystal structure of human PXR in complex with SR12813 (Wang et al., 2008). The results from GOLD docking are shown in Fig. 3A. The binding pose of amprenavir in the PXR ligand-binding domain (LBD) is similar to SR12813. The PXR LBD appears to possess four hydrophobic subpockets and a connection region in the middle. Amprenavir occupies all four subpockets, and its hydroxyl group forms a hydrogen bond with Ser247, which is located in the connection region of PXR, to help to position the drug in the optimal orientation inside the receptor. Another two hydrogen bonds were formed between amprenavir and PXR (Gln285 and His407) that provide additional electrostatic interactions.
Fig. 3.
Key residues of PXR LBD are required for amprenavir’s agonistic activity. (A) Overlay of docked amprenavir and SR12813 (green) inside ligand-binding pocket of PXR. Atoms in amprenavir were colored by atom type (oxygen in red, phosphor in yellow, nitrogen in blue, and carbon in gray), and all the atoms in SR12813 were colored in green for clear comparison. Four subpockets were presented by black circles. (B) Interaction map between amprenavir and PXR. Amprenavir was computationally docked in the ligand-binding pocket of the hPXR X-ray crystal structure (PDB ID 3hvl). The amprenavir-PXR complex was stabilized by three hydrogen bonds with residues Ser247, Gln285, and His407, respectively. The noncovalent contacts with the aromatic side chains Phe288, Trp299, and Tyr306 lining in the subpocket 1 contributed significantly to amprenavir binding. Phe288 was well positioned to engage in π-stacking interactions with the phenyl ring of amprenavir. The interaction diagram reveals that amprenavir occupied the major portion of the large, flexible PXR ligand-binding pocket. (C) HepG2 cells were cotransfected with a full-length PXR WT plasmid or mutant PXR plasmids as indicated, together with CYP3A4-luciferase reporter and cytomegalovirus X (CMX)-β-galactosidase plasmid. After transfection, cells were treated with control medium or medium containing amprenavir at indicated concentrations for 24 hours.
Nonpolar contacts also play a key role in stabilizing amprenavir within the ligand-binding pocket of PXR. As displayed in Fig. 3B, the interaction map of amprenavir and PXR revealed an important π-π stacking between ring A of amprenavir and Phe288 in subpocket 1 of PXR. Tyr306 forms a 3.5-Å edge-to-face contact to the same phenol ring of the drug that also engages in hydrophobic interactions with Trp299, the other hydrophobic residue in subpocket 1. In subpocket 2, amprenavir forms direct contacts with one residue on αAF of the PXR activation function-2 (AF-2) surface, Phe429, which may stabilize the active AF-2 conformation of the receptor and contribute to the agonist activity of amprenavir on PXR. Subpockets 3 and 4 of PXR are located on the bottom of the cavity, which corresponds to the most structurally flexible region of the PXR LBD. This region is dominated by hydrophobic residues (e.g., Met323 and Leu209) that form van der Waals contacts with amprenavir.
Key LBD Residues of PXR Are Required for Amprenavir’s Agonistic Activity.
To validate the results of the docking analysis, we mutated the key amino acids responsible for amprenavir’s agonist activity including Ser247, Gln285, Phe288, and Tyr306. We also included Thr248, a key amino acid known to be important for PXR/coactivator interaction (Ueda et al., 2005; Sui et al., 2012), and Leu411, an amino acid predicted by the docking analysis not to interact with amprenavir, as the putative positive and negative controls for our mutagenesis study, respectively. As shown in Fig. 3C, Ser247Leu and Tyr306Leu mutations completely blocked amprenavir’s agonist activity at all tested drug concentrations. Gln285Leu and His407Ala exerted little or no effects on the drug at low concentration (5 μM). At higher concentrations (10 and 20 μM), these mutations showed inhibitory effects on amprenavir’s activity. Phe288Ala showed some inhibitory effects on amprenavir’s activity. As expected, the Thr248Leu mutation abolished the activity of amprenavir; Leu411Phe did not inhibit but rather slightly promoted amprenavir as an agonist. In summary, our site-directed mutagenesis analysis confirmed the docking model and revealed the key residues within PXR’s binding pocket responsible for amprenavir’s agonist effects.
Amprenavir Induces PXR Target Gene Expression in Human Cells.
We next used human hepatoma HepaRG cells (Kanebratt and Andersson, 2008) and intestinal LS180 cells (Zhou et al., 2006a, 2007) to test the effects of amprenavir exposure on PXR activity and target gene expression. Similar to the known human PXR ligand RIF, amprenavir induced the expression of bona fide PXR target genes involved in phase I (CYP3A4), phase II (UGT1A1), and phase III (MDR1) metabolism in both HepaRG cells (Fig. 4A) and LS180 cells (Fig. 4B). These results suggest that amprenavir can activate PXR and induce PXR target gene expression in human cells.
Fig. 4.
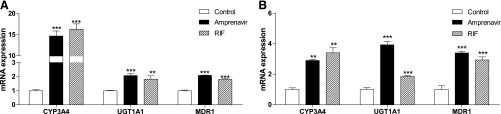
Amprenavir induces PXR target gene expression in human cells. Human HepaRG hepatoma cells (A) and LS180 intestinal cells (B) were treated with control medium or medium containing 10 μM amprenavir or RIF for 24 hours. The total RNA was isolated, and the gene expression levels were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) with primers for human CYP3A4, UGT1A1, or MDR1 (n = 3; **P < 0.01; ***P < 0.001).
Amprenavir Elevates Plasma Lipid Levels in WT Mice, but Not in PXR−/− Mice.
To further investigate the effects of amprenavir on PXR activity in vivo, WT and PXR−/− mice were treated with vehicle (corn oil) or 10 mg/kg body weight of amprenavir daily by oral gavage for 1 week. Amprenavir-treated WT mice had significantly increased total cholesterol levels compared with the control WT mice (Fig. 5A). By contrast, amprenavir did not affect plasma cholesterol levels in the PXR−/− mice (Fig. 5B). The FPLC analysis of the cholesterol distribution pattern revealed that amprenavir increased atherogenic LDL cholesterol fractions in WT mice, but not in PXR−/− mice (Fig. 5, C and D). The lipoprotein fractions (VLDL, LDL, and HDL) were isolated by ultracentrifugation (Zhou et al., 2011), and the cholesterol concentrations in all three fractions were then measured. Consistent with the FPLC results (Fig. 5, C and D), amprenavir treatment did not affect the HDL or VLDL cholesterol levels but significantly increased the LDL cholesterol levels in WT mice (Fig. 6A). By contrast, amprenavir did not affect any of the lipoprotein levels in PXR−/− mice (Fig. 6B). These results suggest that the adverse effects of amprenavir on plasma cholesterol levels are mediated, at least in part, through PXR in mice.
Fig. 5.

Amprenavir elicits hyperlipidemia in WT, but not in PXR−/− mice. (A and B) We treated 8-week-old WT (A) and PXR−/−(B) mice with vehicle (corn oil) or 10 mg/kg of amprenavir by oral gavage for 1 week. Plasma total cholesterol levels were measured by a standard method (n = 4–7; *P < 0.05 compared with control group). (C and D) The plasma cholesterol distribution of WT (C) and PXR−/− mice (D) was analyzed by FPLC.
Fig. 6.
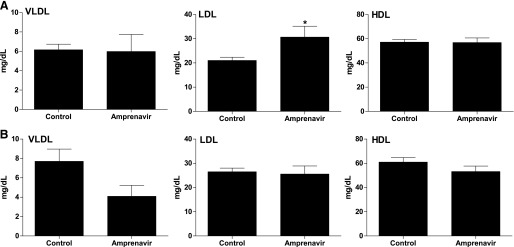
Amprenavir elevates atherogenic LDL cholesterol levels in WT mice. We treated 8-week-old WT (A) and PXR−/− (B) mice with vehicle or 10 mg/kg of amprenavir by oral gavage for 1 week. The pipoprotein fractions (VLDL, LDL, and HDL) were isolated, and the cholesterol levels of each fraction were measured by a standard method (n = 4–7 per group; *P < 0.05 compared with control group).
Amprenavir Affects Genes Involved in Lipid Homeostasis.
To determine whether amprenavir can activate PXR in these mice, the expression levels of known PXR target genes were measured in the intestine and liver. Amprenavir stimulated expression of known PXR target genes, including CYP3A11, glutathione transferase A1, and MDR1a, in the intestine (Fig. 7A) of WT mice but not in PXR−/− mice. Interestingly, amprenavir did not affect the expression of hepatic PXR target genes (Fig. 7B), which might be due to the insufficient accumulation of amprenavir in the liver. To elucidate the possible molecular mechanisms through which amprenavir-mediated intestinal PXR activation might induce dyslipidemia, the expression levels of genes involved in lipid homeostasis were measured (Fig. 7C). Of the genes surveyed, CD36 showed significant change, with its expression levels increased more than 10-fold. CD36, a class B scavenger receptor, plays an important role in intestinal lipid absorption (Abumrad and Davidson, 2012).
Fig. 7.
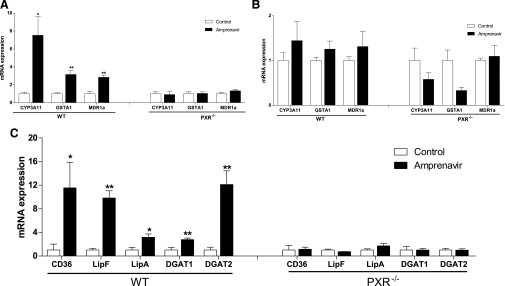
Activation of PXR by amprenavir stimulates the expression of intestinal genes involved in lipid homeostasis. We treated 8-week-old WT and PXR−/− mice with vehicle (corn oil) or 10 mg/kg of amprenavir by oral gavage for 1 week. The total RNA was isolated from the liver and small intestine (duodenum). (A and B) The mRNA levels of known PXR target genes, CYP3A11, glutathione transferase A1, and MDR1a, in the intestine (A) and liver (B) were measured by quantitative real-time polymerase chain reaction (qRT-PCR). (C) The expression levels of intestinal genes involved in lipid homeostasis were measured by qRT-PCR (n = 4–5 per group; *P < 0.05; **P < 0.01).
Consistent with a recent report (Cheng et al., 2012), the expression levels of diacylglycerol acyltransferase 1 and 2 (DGAT1 and 2), two enzymes involved in intestinal lipid transportation and chylomicron secretion (Ables et al., 2012), were also induced by amprenavir-mediated PXR activation in WT mice. It was recently reported that a Drosophila PXR ortholog DHR96 regulates expression of the intestine lipase Magro (CG5932) which mediates cholesterol and triglyceride homeostasis in Drosophila. Magro protein is most similar to mammalian gastric lipase (LipF) (56% similarity) and lysosomal lipase (LipA) (50% similarity) (Sieber and Thummel, 2012). Interestingly, amprenavir-mediated PXR activation stimulated the expression of both LipF and LipA in the intestine of WT mice, but not in PXR−/− mice (Fig. 7C), indicating a possible role of intestinal PXR in mediating dietary lipid breakdown and absorption in mammals. Taken together, upregulation of those genes by PXR activation contribute to amprenavir-elicited hyperlipidemia.
Discussion
PIs have been associated with dyslipidemia and an increased risk of cardiovascular disease (Periard et al., 1999; Friis-Moller et al., 2007; Lang et al., 2010), but the underlying mechanisms are still unknown. Several PIs have been shown to activate PXR which functions as a xenobiotic sensor to regulate genes required for xenobiotic metabolism in the liver and intestine (Kliewer et al., 2002; Zhou et al., 2009b). We have identified a widely used PI amprenavir as a potent and selective PXR agonist. Amprenavir is usually taken by patients with or without ritonavir. Interestingly, a case-control study suggested that the risk of cardiovascular disease was increased by exposure to all PIs and, in particular, to amprenavir with or without ritonavir (Lang et al., 2010). We found that amprenavir activated both human and mouse PXR but did not affect the activity of other nuclear receptors. Short-term exposure to amprenavir significantly increased plasma total cholesterol and LDL cholesterol levels in WT mice, but not in PXR−/− mice. Amprenavir-mediated PXR activation regulated several key intestinal genes involved in lipid homeostasis. These findings provide critical mechanistic insight for understanding the impact of PIs on cardiovascular disease and demonstrate a potential role of PXR in mediating adverse effects of HIV PIs in humans.
Using in silico ligand-PXR docking studies combined with site-directed mutagenesis, we identified Tyr306, Ser247, His407, and Phe288 as participating in amprenavir-PXR interaction (Fig. 3). A critical polar interaction with Ser247 helps to orient amprenavir in the ligand-binding cavity that enables formation of nonpolar interactions with a highly hydrophobic region of PXR’s ligand-binding pocket lined by Phe288, Trp299, and Tyr306. These three residues are highly conserved among different species and interact with structurally diverse PXR agonists, as shown in multiple published PXR X-ray crystal structures. This Ser247-directed orientation of amprenavir also guided the selection of the most plausible binding alignment among the multiple poses generated by our docking simulations. Based on our previous modeling results on diverse ligands with PXR, we conclude that two essential features need to be satisfied for ligand binding to PXR, namely, (1) one key hydrogen bond with Ser247 or His407 and (2) van der Waals contacts with subpocket 1. Besides the interactions formed by these two essential features, numerous hydrophobic contacts between amprenavir and PXR further stabilize the ligand binding and indirectly promote contact with the AF-2 region, thus forming the optimal orientation of αAF helix to interact with coactivators. Our observations regarding the key pharmacophoric features for PXR binding will hopefully provide guidance useful for PXR ligand screening in the future.
The role of PXR in xenobiotic metabolism has been well established; however, recent studies have revealed the role of PXR in dyslipidemia and atherosclerosis. We recently demonstrated the proatherogenic effects of PXR in animals (Zhou et al., 2009a; Sui et al., 2011). We previously reported that chronic activation of PXR elicited by feeding mice the mouse PXR ligand PCN led to increased levels of plasma total cholesterol and VLDL and LDL in WT mice, but not in PXR−/− mice (Zhou et al., 2009a). In our present study, mice were treated with 10 mg/kg per day of amprenavir to investigate its potential effects on lipid levels. Patients usually take a daily dose of 1400 mg or 2800 mg of the amprenavir prodrug fosamprenavir, which is rapidly and extensively converted to amprenavir after oral administration (Wire et al., 2006). Pharmacokinetic studies have demonstrated that the mean peak plasma concentration of amprenavir is approximately 18 μM after a 1200-mg single dose (Sadler et al., 1999; Sadler and Stein, 2002), which is sufficient to activate PXR based on our results (Fig. 1). Therefore, we believe that amprenavir can activate PXR and regulate PXR target genes in patients taking this drug routinely.
The dose of 10 mg/kg per day of amprenavir used in our present study to treat animals is considered low or modest. Nevertheless, after 1 week of treatment, amprenavir significantly increased plasma total cholesterol and LDL cholesterol levels in WT mice, but not in PXR−/− mice, indicating the significant role of PXR in mediating the adverse effects of amprenavir (Figs. 5 and 6). Interestingly, amprenavir regulated the PXR target genes in the intestine but not in the liver (Fig. 7). This could be explained by the relatively low dose of amprenavir and the short term of treatment in this study. In addition, amprenavir is metabolized by the CYP3A4 enzyme in the liver (Barry et al., 1999). Therefore, the relatively low dose of amprenavir used in our present study may be insufficient for amprenavir to reach the high concentrations required to activate PXR in the liver.
Intestinal lipid transportation plays a central role in lipid homeostasis. In addition to prototypic PXR target genes (e.g., CYP3A11, MDR1a), amprenavir significantly stimulated the expression of several key genes involved in intestinal lipid homeostasis in WT mice, including CD36, DGAT1, and DGAT2. We and others have previously reported that activation of PXR induces CD36 expression and increases lipid accumulation in the liver (Zhou et al., 2006b), intestine (Cheng et al., 2012), and macrophages (Zhou et al., 2009a). The role of CD36 in mediating intestinal fatty acid uptake has been well established (Nauli et al., 2006; Nassir et al., 2007; Abumrad and Davidson, 2012); several studies also have indicated that CD36 mediates cholesterol uptake in the intestine (Nauli et al., 2006; Nassir et al., 2007). For example, cholesterol uptake was significantly decreased in the enterocytes isolated from CD36-deficient (CD36−/−) mice (Nassir et al., 2007). In a lipid-infusion study, CD36−/− mice exhibited accumulation of dietary cholesterol in the intestinal lumen and reduction of cholesterol transport into the lymph (Nauli et al., 2006). Therefore, PXR-mediated CD36 upregulation may contribute to amprenavir-stimulated elevation of cholesterol levels.
In addition, Liang et al. (Liang et al., 2001) previously reported that PIs such as ritonavir inhibit degradation and secretion of apolipoprotein B (ApoB), the primary apolipoprotein of chylomicrons and LDL. Interestingly, the inhibitory effects of PI on ApoB secretion were reversed by oleic acid, a fatty acid that stimulates neutral-lipid biosynthesis (Liang et al., 2001). In the presence of oleic acid, PI treatment even increased ApoB secretion above controls (Liang et al., 2001). Because CD36 plays an important role in the absorption of oleic acid (Nassir et al., 2007), it is plausible that PXR-mediated CD36 expression increases oleic acid absorption and contributes to PI-induced ApoB secretion and hyperlipidemia.
The precise mechanisms through which PXR modulates lipid homeostasis and cholesterol levels in various animal models as well as in humans remain to be determined. By contrast, the Drosophila PXR ortholog DHR96 has been established to be a key regulator in mediating cholesterol and triglyceride homeostasis (Horner et al., 2009; Sieber and Thummel, 2009, 2012). Recent studies have shown that DHR96 regulates expression of the intestine lipase Magro, which is most similar to mammalian LipA and LipF (Sieber and Thummel, 2012). Both LipA and LipF were upregulated by amprenavir-mediated PXR activation in the intestine of WT mice but not in PXR−/− mice (Fig. 7C).
LipA is an enzyme responsible for hydrolysis of cholesterol esters and triglycerides within lipoprotein particles internalized by receptor-mediated endocytosis (Anderson and Sando, 1991; Sheriff et al., 1995). Previous studies have demonstrated an important role of LipA in maintaining cholesterol levels in mice (Du et al., 2001), and mutations in LipA result in cholesterol ester storage disease, which is associated with hyperlipidemia in humans (Pisciotta et al., 2009). LipF also contributes to lipid catabolism by hydrolysis of dietary triglycerides in the stomach and intestine, sequentially producing free fatty acids and diacylglycerol (Gargouri et al., 1986; Carriere et al., 1993). It would be interesting to further investigate the detailed mechanisms by which PXR transcriptionally regulates LipA and LipF expression and mediates intestinal lipid homeostasis. Upregulation of those key genes by PXR activation contribute to amprenavir-elicited hyperlipidemia.
In summary, we demonstrated that amprenavir is a selectively potent PXR agonist. Amprenavir efficiently activated PXR and stimulated PXR target gene expression in vitro and in vivo. Computational docking studies and site-directed mutagenesis analysis identified key residues within PXR’s ligand binding pocket that interact with amprenavir. Our study provides critical insight for understanding the mechanism by which clinically relevant PXR ligands interact and activate PXR. We also showed, for the first time, that amprenavir-mediated activation of PXR elevated lipid levels in an animal model, which has direct clinical consequences for patients under long-term treatment with amprenavir and other PIs. Activation of PXR may present a key mechanism for adverse effects of PIs in humans.
Supplementary Material
Acknowledgments
The authors thank B. Blumberg (UC, Irvine) and B. M. Forman (City of Hope National Medical Center) for plasmids, R. M. Evans (Salk Institute) and B. Blumberg for the PXR−/− mice, A. Daugherty and D. Howatt for the FPLC analysis, and R. Greenberg for valuable discussions.
Abbreviations
- AF-2
activation function-2
- ApoB
apolipoprotein B
- ApoE
apolipoprotein E
- DGAT
diacylglycerol acyltransferase
- FPLC
fast-performance liquid chromatography
- HAART
highly active antiretroviral therapy
- HDL
high-density lipoprotein
- HIV
human immunodeficiency virus
- LBD
ligand-binding domain
- LDL
low-density lipoprotein
- LipA
lysosomal lipase
- LipF
gastric lipase
- MDR1
multidrug resistance 1
- NCoR
nuclear receptor corepressor
- PBP
PPAR-binding protein
- PCN
pregnenolone 16α-carbonitrile
- PI
protease inhibitor
- PPAR
peroxisome proliferator-activated receptor
- PXR
pregnane X receptor
- RIF
rifampicin
- SMRT
silencing mediator of retinoid and thyroid hormone
- SR12813
tetraethyl 2-(3,5-di-tert-butyl-4-hydroxyphenyl)ethenyl-1,1-bisphosphonate
- SRC-1
steroid receptor coactivator-1
- VLDL
very-low-density lipoprotein
- WT
wild-type
Authorship Contributions
Participated in research design: Helsley, Sui, Welsh, Zhou.
Conducted experiments: Helsley, Sui, Ai, Park, Zhou.
Contributed new reagents or analytic tools: Helsley, Sui, Ai, Zhou.
Performed data analysis: Helsley, Sui, Ai, Park, Zhou.
Wrote or contributed to the writing of the manuscript: Helsley, Ai, Welsh, Zhou.
Footnotes
This work was supported in part by the National Institutes of Health [Grants P20GM103527 and P30HL101300]; a training grant from the National Institutes of Health [Grant T32HL072743] (to R.N.H.); and the American Heart Association [Grant 09SDG2150176] (to C.Z.).
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Ables GP, Yang KJ, Vogel S, Hernandez-Ono A, Yu S, Yuen JJ, Birtles S, Buckett LK, Turnbull AV, Goldberg IJ, et al. (2012) Intestinal DGAT1 deficiency reduces postprandial triglyceride and retinyl ester excursions by inhibiting chylomicron secretion and delaying gastric emptying. J Lipid Res 53:2364–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abumrad NA, Davidson NO. (2012) Role of the gut in lipid homeostasis. Physiol Rev 92:1061–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RA, Sando GN. (1991) Cloning and expression of cDNA encoding human lysosomal acid lipase/cholesteryl ester hydrolase. Similarities to gastric and lingual lipases. J Biol Chem 266:22479–22484 [PubMed] [Google Scholar]
- Barry M, Mulcahy F, Merry C, Gibbons S, Back D. (1999) Pharmacokinetics and potential interactions amongst antiretroviral agents used to treat patients with HIV infection. Clin Pharmacokinet 36:289–304 [DOI] [PubMed] [Google Scholar]
- Bhaskaran K, Hamouda O, Sannes M, Boufassa F, Johnson AM, Lambert PC, Porter K, CASCADE Collaboration (2008) Changes in the risk of death after HIV seroconversion compared with mortality in the general population. JAMA 300:51–59 [DOI] [PubMed] [Google Scholar]
- Blumberg B, Sabbagh W, Jr, Juguilon H, Bolado J, Jr, van Meter CM, Ong ES, Evans RM. (1998) SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev 12:3195–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A, Samaras K, Burton S, Law M, Freund J, Chisholm DJ, Cooper DA. (1998) A syndrome of peripheral lipodystrophy, hyperlipidaemia and insulin resistance in patients receiving HIV protease inhibitors. AIDS 12:F51–F58 [DOI] [PubMed] [Google Scholar]
- Carriere F, Barrowman JA, Verger R, Laugier R. (1993) Secretion and contribution to lipolysis of gastric and pancreatic lipases during a test meal in humans. Gastroenterology 105:876–888 [DOI] [PubMed] [Google Scholar]
- Cheng J, Krausz KW, Tanaka N, Gonzalez FJ. (2012) Chronic exposure to rifaximin causes hepatic steatosis in pregnane X receptor-humanized mice. Toxicol Sci 129:456–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan W, de Vries-van der Weij J, Mol IM, Hoekstra M, Romijn JA, Jukema JW, Havekes LM, Princen HM, Rensen PC. (2009) PXR agonism decreases plasma HDL levels in ApoE3-Leiden.CETP mice. Biochim Biophys Acta 1791:191–197 [DOI] [PubMed] [Google Scholar]
- Distler O, Cooper DA, Deckelbaum RJ, Sturley SL. (2001) Hyperlipidemia and inhibitors of HIV protease. Curr Opin Clin Nutr Metab Care 4:99–103 [DOI] [PubMed] [Google Scholar]
- Du H, Heur M, Duanmu M, Grabowski GA, Hui DY, Witte DP, Mishra J. (2001) Lysosomal acid lipase-deficient mice: depletion of white and brown fat, severe hepatosplenomegaly, and shortened life span. J Lipid Res 42:489–500 [PubMed] [Google Scholar]
- Dussault I, Lin M, Hollister K, Wang EH, Synold TW, Forman BM. (2001) Peptide mimetic HIV protease inhibitors are ligands for the orphan receptor SXR. J Biol Chem 276:33309–33312 [DOI] [PubMed] [Google Scholar]
- Eirís JM, Lojo S, Del Río MC, Novo I, Bravo M, Pavón P, Castro-Gago M. (1995) Effects of long-term treatment with antiepileptic drugs on serum lipid levels in children with epilepsy. Neurology 45:1155–1157 [DOI] [PubMed] [Google Scholar]
- Friis-Møller N, Reiss P, Sabin CA, Weber R, Monforte Ad, El-Sadr W, Thiébaut R, De Wit S, Kirk O, Fontas E, et al. DAD Study Group (2007) Class of antiretroviral drugs and the risk of myocardial infarction. N Engl J Med 356:1723–1735 [DOI] [PubMed] [Google Scholar]
- Friis-Møller N, Sabin CA, Weber R, d’Arminio Monforte A, El-Sadr WM, Reiss P, Thiébaut R, Morfeldt L, De Wit S, Pradier C, et al. Data Collection on Adverse Events of Anti-HIV Drugs (DAD) Study Group (2003) Combination antiretroviral therapy and the risk of myocardial infarction. N Engl J Med 349:1993–2003 [DOI] [PubMed] [Google Scholar]
- Gao J, Xie W. (2012) Targeting xenobiotic receptors PXR and CAR for metabolic diseases. Trends Pharmacol Sci 33:552–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargouri Y, Pieroni G, Rivière C, Lowe PA, Saunière JF, Sarda L, Verger R. (1986) Importance of human gastric lipase for intestinal lipolysis: an in vitro study. Biochim Biophys Acta 879:419–423 [DOI] [PubMed] [Google Scholar]
- He J, Gao J, Xu M, Ren S, Stefanovic-Racic M, O’Doherty RM, Xie W. (2013) PXR ablation alleviates diet-induced and genetic obesity and insulin resistance in mice. Diabetes DOI: [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra M, Lammers B, Out R, Li Z, Van Eck M, Van Berkel TJ. (2009) Activation of the nuclear receptor PXR decreases plasma LDL-cholesterol levels and induces hepatic steatosis in LDL receptor knockout mice. Mol Pharm 6:182–189 [DOI] [PubMed] [Google Scholar]
- Horner MA, Pardee K, Liu S, King-Jones K, Lajoie G, Edwards A, Krause HM, Thummel CS. (2009) The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes Dev 23:2711–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanebratt KP, Andersson TB. (2008) Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab Dispos 36:1444–1452 [DOI] [PubMed] [Google Scholar]
- Khogali AM, Chazan BI, Metcalf VJ, Ramsay JHR. (1974) Hyperlipidaemia as a complication of rifampicin treatment. Tubercle 55:231–233 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Goodwin B, Willson TM. (2002) The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr Rev 23:687–702 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82 [DOI] [PubMed] [Google Scholar]
- Lang S, Mary-Krause M, Cotte L, Gilquin J, Partisani M, Simon A, Boccara F, Costagliola D, Clinical Epidemiology Group of the French Hospital Database on HIV (2010) Impact of individual antiretroviral drugs on the risk of myocardial infarction in human immunodeficiency virus-infected patients: a case-control study nested within the French Hospital Database on HIV ANRS cohort CO4. Arch Intern Med 170:1228–1238 [DOI] [PubMed] [Google Scholar]
- Liang JS, Distler O, Cooper DA, Jamil H, Deckelbaum RJ, Ginsberg HN, Sturley SL. (2001) HIV protease inhibitors protect apolipoprotein B from degradation by the proteasome: a potential mechanism for protease inhibitor-induced hyperlipidemia. Nat Med 7:1327–1331 [DOI] [PubMed] [Google Scholar]
- Lu Y, Feskens EJ, Boer JM, Müller M. (2010) The potential influence of genetic variants in genes along bile acid and bile metabolic pathway on blood cholesterol levels in the population. Atherosclerosis 210:14–27 [DOI] [PubMed] [Google Scholar]
- Lütjohann D, Hahn C, Prange W, Sudhop T, Axelson M, Sauerbruch T, von Bergmann K, Reichel C. (2004) Influence of rifampin on serum markers of cholesterol and bile acid synthesis in men. Int J Clin Pharmacol Ther 42:307–313 [DOI] [PubMed] [Google Scholar]
- Moreau A, Téruel C, Beylot M, Albalea V, Tamasi V, Umbdenstock T, Parmentier Y, Sa-Cunha A, Suc B, Fabre JM, et al. (2009) A novel pregnane X receptor and S14-mediated lipogenic pathway in human hepatocyte. Hepatology 49:2068–2079 [DOI] [PubMed] [Google Scholar]
- Nassir F, Wilson B, Han X, Gross RW, Abumrad NA. (2007) CD36 is important for fatty acid and cholesterol uptake by the proximal but not distal intestine. J Biol Chem 282:19493–19501 [DOI] [PubMed] [Google Scholar]
- Nauli AM, Nassir F, Zheng S, Yang Q, Lo CM, Vonlehmden SB, Lee D, Jandacek RJ, Abumrad NA, Tso P. (2006) CD36 is important for chylomicron formation and secretion and may mediate cholesterol uptake in the proximal intestine. Gastroenterology 131:1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Sui Y, Gizard F, Xu J, Rios-Pilier J, Helsley RN, Han SS, Zhou C. (2012) Myeloid-specific IκB kinase β deficiency decreases atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol 32:2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne AW, Glen RC. (1993) Molecular recognition using a binary genetic search algorithm. J Mol Graph 11:74–91, 121–123 [DOI] [PubMed] [Google Scholar]
- Périard D, Telenti A, Sudre P, Cheseaux JJ, Halfon P, Reymond MJ, Marcovina SM, Glauser MP, Nicod P, Darioli R, et al. (1999) Atherogenic dyslipidemia in HIV-infected individuals treated with protease inhibitors. The Swiss HIV Cohort Study. Circulation 100:700–705 [DOI] [PubMed] [Google Scholar]
- Pisciotta L, Fresa R, Bellocchio A, Pino E, Guido V, Cantafora A, Di Rocco M, Calandra S, Bertolini S. (2009) Cholesteryl ester storage disease (CESD) due to novel mutations in the LIPA gene. Mol Genet Metab 97:143–148 [DOI] [PubMed] [Google Scholar]
- Sadler BM, Hanson CD, Chittick GE, Symonds WT, Roskell NS. (1999) Safety and pharmacokinetics of amprenavir (141W94), a human immunodeficiency virus (HIV) type 1 protease inhibitor, following oral administration of single doses to HIV-infected adults. Antimicrob Agents Chemother 43:1686–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler BM, Stein DS. (2002) Clinical pharmacology and pharmacokinetics of amprenavir. Ann Pharmacother 36:102–118 [DOI] [PubMed] [Google Scholar]
- Shafran SD, Mashinter LD, Roberts SE. (2005) The effect of low-dose ritonavir monotherapy on fasting serum lipid concentrations. HIV Med 6:421–425 [DOI] [PubMed] [Google Scholar]
- Sheriff S, Du H, Grabowski GA. (1995) Characterization of lysosomal acid lipase by site-directed mutagenesis and heterologous expression. J Biol Chem 270:27766–27772 [DOI] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. (2009) The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab 10:481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. (2012) Coordination of triacylglycerol and cholesterol homeostasis by DHR96 and the Drosophila LipA homolog magro. Cell Metab 15:122–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JH, Klein MA, Bellehumeur JL, McBride PE, Wiebe DA, Otvos JD, Sosman JM. (2001) Use of human immunodeficiency virus-1 protease inhibitors is associated with atherogenic lipoprotein changes and endothelial dysfunction. Circulation 104:257–262 [DOI] [PubMed] [Google Scholar]
- Sui Y, Ai N, Park SH, Rios-Pilier J, Perkins JT, Welsh WJ, Zhou C. (2012) Bisphenol A and its analogues activate human pregnane X receptor. Environ Health Perspect 120:399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui Y, Xu J, Rios-Pilier J, Zhou C. (2011) Deficiency of PXR decreases atherosclerosis in apoE-deficient mice. J Lipid Res 52:1652–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teupser D, Persky AD, Breslow JL. (2003) Induction of atherosclerosis by low-fat, semisynthetic diets in LDL receptor-deficient C57BL/6J and FVB/NJ mice: comparison of lesions of the aortic root, brachiocephalic artery, and whole aorta (en face measurement). Arterioscler Thromb Vasc Biol 23:1907–1913 [DOI] [PubMed] [Google Scholar]
- Ueda A, Matsui K, Yamamoto Y, Pedersen LC, Sueyoshi T, Negishi M. (2005) Thr176 regulates the activity of the mouse nuclear receptor CAR and is conserved in the NR1I subfamily members PXR and VDR. Biochem J 388:623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Prosise WW, Chen J, Taremi SS, Le HV, Madison V, Cui X, Thomas A, Cheng KC, Lesburg CA. (2008) Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability. Protein Eng Des Sel 21:425–433 [DOI] [PubMed] [Google Scholar]
- Wire MB, Shelton MJ, Studenberg S. (2006) Fosamprenavir : clinical pharmacokinetics and drug interactions of the amprenavir prodrug. Clin Pharmacokinet 45:137–168 [DOI] [PubMed] [Google Scholar]
- Zhou C, King N, Chen KY, Breslow JL. (2009a) Activation of PXR induces hypercholesterolemia in wild-type and accelerates atherosclerosis in apoE deficient mice. J Lipid Res 50:2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Poulton EJ, Grün F, Bammler TK, Blumberg B, Thummel KE, Eaton DL. (2007) The dietary isothiocyanate sulforaphane is an antagonist of the human steroid and xenobiotic nuclear receptor. Mol Pharmacol 71:220–229 [DOI] [PubMed] [Google Scholar]
- Zhou C, Pridgen B, King N, Xu J, Breslow JL. (2011) Hyperglycemic Ins2AkitaLdlr−/− mice show severely elevated lipid levels and increased atherosclerosis: a model of type 1 diabetic macrovascular disease. J Lipid Res 52:1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Tabb MM, Nelson EL, Grün F, Verma S, Sadatrafiei A, Lin M, Mallick S, Forman BM, Thummel KE, et al. (2006a) Mutual repression between steroid and xenobiotic receptor and NF-κB signaling pathways links xenobiotic metabolism and inflammation. J Clin Invest 116:2280–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Tabb MM, Sadatrafiei A, Grün F, Blumberg B. (2004) Tocotrienols activate the steroid and xenobiotic receptor, SXR, and selectively regulate expression of its target genes. Drug Metab Dispos 32:1075–1082 [DOI] [PubMed] [Google Scholar]
- Zhou C, Verma S, Blumberg B. (2009b) The steroid and xenobiotic receptor (SXR), beyond xenobiotic metabolism. Nucl Recept Signal 7:e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, Ren S, Evans RM, Xie W. (2006b) A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem 281:15013–15020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.