Abstract
The human pregnane X receptor (PXR) regulates genes involved in drug metabolism and disposition. PXR associates with multiple corepressors that attenuate and coactivators that enhance its activity. PXR plays a vital role in the drug metabolism pathway, and a comprehensive examination of PXR-associated proteins will provide greater insight into the regulation of the receptor and possible therapeutic implications. We performed a mass spectrometric screen to identify PXR-associated proteins. Here we report that the tumor suppressor protein p53 can associate with PXR and downregulate its activity. A loss-of-function p53 mutant (R175H) interacts with PXR but does not repress its activity. Mutant p53 can relieve the suppressive effect of wild-type p53 by competing with its interaction with PXR, suggesting that protein-protein interaction is required but not sufficient for p53 to repress PXR activity. Interestingly, a PXR variant with a naturally occurring deletion of a conserved, unique sequence in the ligand binding domain (PXR∆174–210) did not interact with p53, indicating that the PXR-p53 interaction is specific. Using a chromatin immunoprecipitation assay, we showed that p53 inhibits the binding of PXR to the CYP3A4 promoter. The loss of p53 function in tumor cells leads to aberrant cell proliferation, apoptosis, carcinogenesis, and altered sensitivity to chemotherapeutic drugs, whereas PXR contributes to chemoresistance in many cancer cells. Our findings show for the first time that wild-type p53 can negatively regulate PXR by physically associating with it. Thus, PXR and p53 appear to play important yet opposing roles in the sensitivity of tumor cells to chemotherapy.
Introduction
Pregnane X receptor (PXR) is a xenobiotic receptor of the nuclear receptor (NR) family. PXR functions mainly in the liver and is activated by a wide range of ligands, which are usually toxic by-products derived from the endogenous metabolism or exogenous chemicals (Blumberg et al., 1998a,b).
The flexible ligand binding domain (LBD) of PXR, from amino acid 142 to 434, can expand to accommodate structurally and pharmacologically diverse ligands, thus contributing to its ability to function as an efficient xenobiotic sensor. Once activated, PXR forms a heterodimer with the retinoid X receptor to engage specific response elements in the promoter region of its target genes to mediate the transcription of a variety of drug-metabolizing enzymes and drug transporters (Wang et al., 2012). A major class of proteins that is regulated by PXR is the 3A subfamily of cytochrome P450 (CYP3A4).
Human PXR has several alternatively spliced isoforms, of which PXR.1 (434 amino acids) is the most abundant and can be transcriptionally activated by ligands (Lamba et al., 2004) (we use “PXR” and “PXR.1” synonymously throughout the text). The second most abundant isoform, PXR.2 (397 amino acids, also referred to as [PXR∆174–210]), constitutes 7% of the mRNA transcript and has a 37 amino acid deletion in the LBD, resulting in attenuated ligand binding capacity and reduced ligand-mediated transcriptional activity, although there is no loss of association with the PXR response elements in the genome (Dotzlaw et al., 1999; Lin et al., 2009). The physiologic role of PXR.2 has not been fully defined.
PXR is regulated by a variety of protein-protein interactions and post-translational modifications. Protein-protein interactions are dynamic processes that, depending on the associated protein, can result in the modulation of activity, subcellular localization, substrate specificity, and stability of the target protein. PXR associates with a variety of protein coregulators that can act as either coactivators or corepressors. Corepressors such as the silencing mediator for retinoid and thyroid receptors (SMRT) and nuclear receptor corepressor (NCoR) attenuate gene transcription through their association with unliganded PXR (Ding and Staudinger, 2005; Johnson et al., 2006). Upon ligand binding, PXR activity is enhanced by the dissociation of corepressors and association instead with coactivators such as forkhead transcription factor and steroid receptor coactivator-1(Watkins et al., 2003; Kodama et al., 2004). To this end, we wanted to identify and expand on the protein partners that associate with PXR and can contribute to its regulation using mass spectrometric analysis. Our study identified tumor suppressor p53, which can bind directly to PXR.
Wild-type p53, once activated by a variety of stressors that threaten the genomic integrity of cells, can induce cell-cycle arrest, DNA repair, senescence, or apoptosis, depending on the severity of the damage; p53 is mutated or inactivated in almost all forms of cancer (Hollstein et al., 1991; Hainaut and Hollstein, 2000). As a transcription factor, p53 regulates the expression or function of a large array of proteins through transcriptional control or protein-protein interactions (Levine et al., 2006).
Also, p53 regulates the activity of several NRs, such as glucocorticoid receptor, androgen receptor, estrogen receptor (ER), liver-X receptor, and hepatocyte nuclear factor-4α1 (Yu et al., 1997; Maeda et al., 2002; Angeloni et al., 2004; Iwano et al., 2006; Alimirah et al., 2007). For androgen receptor, p53 not only modulates its transcriptional activity but negatively regulates its expression (Shenk et al., 2001; Alimirah et al., 2007). p53 interacts directly with glucocorticoid receptor and downregulates its activity, and this downregulation depends on the activity of the wild-type p53 (Yu et al., 1997); p53 is known to enhance the recruitment of ERα to the proximal ER promoter in response to doxorubicin treatment (Angeloni et al., 2004; Shirley et al., 2009). We report here that wild-type p53 can attenuate PXR activity by inhibiting the binding of PXR to its target promoter. Furthermore, we show that the p53-PXR association is specific for PXR.1 and not for PXR.2, which lacks a 37-amino-acid sequence unique for PXR from various species, indicating the specificity of the p53-PXR interaction.
PXR is a master regulator of drug metabolism that contributes to enhanced cellular chemoresistance (Chen et al., 2007; Raynal et al., 2010). On the other hand, cells harboring wild-type p53 are more chemosensitive than are cells where p53 is inactivated (Fan et al., 1994, 1995). Our findings, for the first time, cast p53 in a regulatory role for PXR, a fact that can be exploited for therapeutic purposes given the important roles that both p53 and PXR play in regulating chemosensitivity.
Materials and Methods
Materials.
We obtained p53−/− HCT116 (human colon carcinoma cell line) cells from the Genetic Resources Core Facility at Johns Hopkins University School of Medicine (Baltimore, MD); human osteosarcoma cell line U2OS cells stably expressing green fluorescent protein (GFP)-PXR were obtained from Thermo Scientific (Ashville, NC); human embryonic kidney cells (HEK293T) and liver hepatocellular carcinoma cell line (HepG2) cells were obtained from American Type Culture Collection (Manassas, VA). Anti-p53 antibody was obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); rifampicin, anti-FLAG M2 antibody, EZview Red anti-FLAG M2 affinity gel, and FLAG peptide were obtained from Sigma-Aldrich (St. Louis, MO); GFP-Trap agarose beads were obtained from ChromoTek (Planegg-Martinsried, Germany); charcoal-dextran treated fetal bovine serum (FBS) was obtained from HyClone (Logan, UT); blocking buffer, anti-mouse, and anti-rabbit IRDye secondary antibodies were from LI-COR Biosciences (Lincoln, NE). Mouse IgG used as control in chromatin immunoprecipitation (ChIP) assays was from Cell Signaling Technology, Inc. (Danvers, MA), and protein G-agarose beads were obtained from Sigma-Aldrich.
Cell Culture, Plasmids, and Transfection.
All cells were maintained in a humidified environment at 37°C with 5% CO2. HCT116 cells were maintained in McCoy’s 5A media supplemented with 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. U2OS, HepG2, and HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Additionally, U2OS stable cells were maintained in media containing 0.4 mg/ml G418. pcDNA3.1 containing FLAG-PXR and luciferase reporter gene under the control of the CYP3A4 promoter (CYP3A4-luc) were constructed as previously described (Luo et al., 2002). Wild-type FLAG-p53 and FLAG-p53-RH (R175H) p53 mutants were obtained from Origene (Rockville, MD). p53 mutants containing various mutations at the 22LWKLL26 and 251ILTII255 motifs were generated by Mutagenex Inc. (Hillsborough, NJ). Standard molecular biology methods were used to generate various deletion mutants of PXR. TK-Renilla luciferase plasmid was purchased from Promega (Madison, WI); p53 reporter plasmids PG13-luc and the reporter containing a mutation at the promoter sequence (MG15-luc) were from Addgene (Cambridge, MA) (Kern et al., 1992). All transfections were performed using Lipofectamine 2000 (Life Technologies, Carlsbad, CA) according to the manufacturer’s recommendations.
Immunoprecipitation.
The HEK293T cells were transfected with pcDNA3.1 FLAG-PXR, or pcDNA3.1-FLAG vector using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendation. After 48 hours of incubation, cells were collected in lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, protease inhibitor cocktail (Roche, Indianapolis, IN), and Halt phosphatase inhibitor cocktail (Pierce, Rockford, IL) and incubated for 20 minutes on ice. Lysates were centrifuged at 10,000g for 20 minutes at 4°C to remove cell debris, and the supernatant was incubated with 10 µl of EZview Red anti-FLAG M2 affinity gel (Sigma-Aldrich) for 2 hours at 4°C. The M2 beads with the bound proteins were then washed twice with lysis buffer and twice with TBS and then eluted with 100 ng/µl FLAG peptide (Sigma-Aldrich) in TBS for 1 hour at 4°C. U2OS cells stably expressing GFP-PXR or GFP were collected in lysis buffer and incubated with GFP-Trap agarose beads for 2 hours at 4°C. The beads were washed twice each with lysis buffer and TBS and then boiled in sample loading buffer (Invitrogen) to release the bound proteins.
Western Blot Analysis.
All cell extracts were harvested in lysis buffer, and samples were centrifuged at 10,000g at 4°C for 20 minutes. The samples were then boiled in sample loading buffer (Invitrogen) containing SDS, and equal amounts of samples were resolved on 4–12% SDS-PAGE gradient gel and then transferred onto nitrocellulose membrane. The membrane was blocked and incubated with the indicated antibodies overnight at 4°C. All Western blot analyses and quantitation were performed on the Odyssey Infrared Imaging system (LI-COR Biosciences).
Protein Partner Identification.
After immunoprecipitation using FLAG M2 affinity gel, the eluted protein solution was mixed with ice-cold ethanol and incubated overnight at −20°C to precipitate the proteins. The solution was then centrifuged at 6000g for 30 minutes and dried using a vacuum evaporator. The protein pellet was then submitted for processing and protein identification by the St. Jude Proteomics & Mass Spectrometry Shared Resource using an LTQ-Orbitrap mass spectrometer (ThermoFisher, San Jose, CA); the data were processed using MASCOT searching algorithm (Matrix Science Inc., Boston, MA) and Scaffold (Proteome Software, Portland, OR).
Transactivation Assay.
Cells were transfected with PXR or empty vector control along with CYP3A4-luc and vector expressing TK-Renilla luciferase as transfection control. To check for p53 activity, cells were transfected with plasmids expressing either p53 or empty vector control along with luciferase reporter gene under the control of p53 promoter sequence (PG13) and vector expressing TK-Renilla luciferase. After 48 hours of transfection, the cells were plated in 96-well plates containing either dimethylsulfoxide or the indicated compound in phenol-red free media containing 10% charcoal-dextran–treated FBS. The cells were incubated for 24 hours before processing using the Dual-Glo Luciferase Assay System (Promega). Data are reported in relative luciferase units by normalizing the firefly luciferase activity to Renilla luciferase activity.
ChIP Assay.
HepG2 cells stably expressing FLAG-PXR and CYP3A4-luc, established using the procedure previously described (Lin et al., 2008), were transiently transfected with wild-type p53, p53-R175H mutant, or empty vector. Cells were incubated with 5 µM rifampicin for 24 hours and then treated with 1% formaldehyde to crosslink DNA with the associated proteins. DNA-protein crosslinks were sheared and extracted using sonication at 4°C in lysis buffer containing 10% Triton X-100, 4 M NaCl, 0.5 M EDTA, 1 M Tris, 10% SDS, 100 mM phenylmethylsulfonyl fluooride, and protease inhibitor cocktail. Chromatin from 5 × 107 cells was immunoprecipitated using either IgG as negative control or anti-FLAG M2 beads overnight at 4°C and eluted using FLAG peptide. The eluents were incubated with 200 mM NaCl overnight at 60°C to reverse the crosslinking and then for 2 hours at 45°C with 10 mM EDTA, 40 mM Tris (pH 6.5) and 5 µg/ml Proteinase K. DNA was purified using a QIAquick polymerase chain reaction (PCR) purification kit (Qiagen, Valencia, CA). PCR was performed using primers designed to amplify the DR3 region (the binding site for PXR) of the CYP3A4 promoter (Goodwin et al., 2002) using the following primers: forward 5′-ATC CAC TAG TAA CGG CCG-3′ and reverse 5′-TTC AGC TTG TGA TTC ACC TG-3′. PCR products were resolved on 1% agarose gel and quantified using the ImageJ image processing and analysis tool (NIH, Bethesda, MD).
Statistical Analysis.
In all reporter assays, error bars indicate S.E.M. For ChIP assay, results are expressed as the mean ± standard deviation of at least three independent experiments, and error bars indicate the standard deviation. Statistically significant differences between samples (noted with a line in Fig. 5B: comparisons between empty-vector and wild-type and between empty vector and RH) are indicated by the P value, which was determined using the Student’s t test and considered significant if P < 0.05 (*) or 0.01 (**).
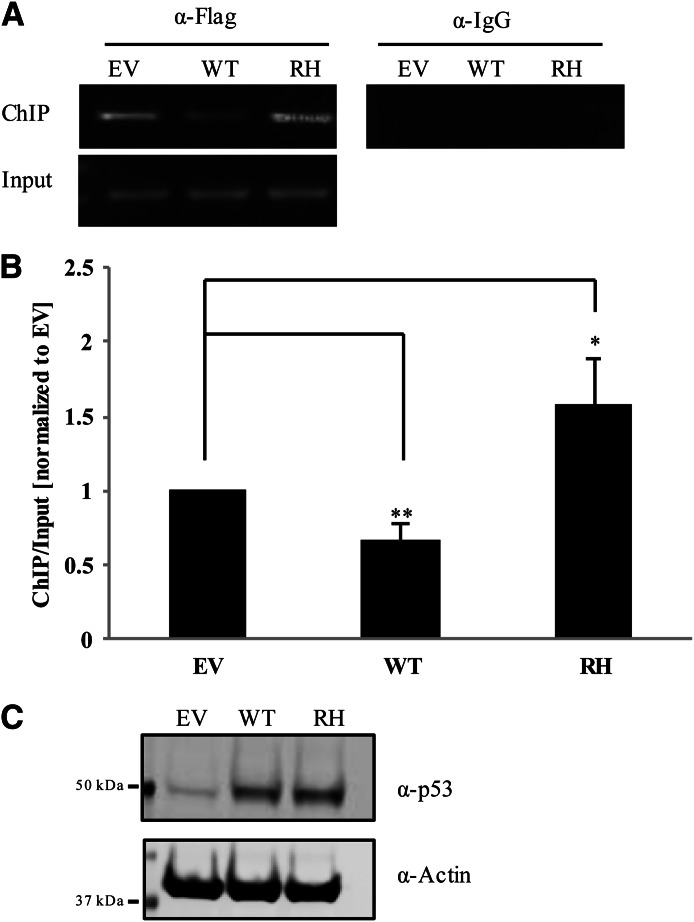
Fig. 5.
Wild-type p53 attenuates the association of PXR with CYP3A4 promoter. (A) Crosslinked FLAG-PXR was immunoprecipitated from HepG2 cells stably expressing FLAG-PXR and CYP3A4 using anti-FLAG M2 agarose beads. Cells were transiently transfected with control vector (EV), wild-type p53 (WT) or p53 R175H mutant (RH). The levels of CYP3A4 promoter associated with PXR were amplified using primers against the DR3 region within the promoter. Mouse IgG was used for control immunoprecipitation (right panel). (B) Band intensity for ChIP was obtained using ImageJ Software and normalized to band intensities generated for the corresponding input sample (lower panel). Error bars indicate ± S.D., and statistical significance was determined by Student’s t test where *P ≤ 0.05; **P ≤ 0.01. (C) Cellular lysates used for ChIP assay were immunoprobed with anti-p53 (α-p53) and anti-actin (α-Actin).
Results
p53 Interacts with PXR.
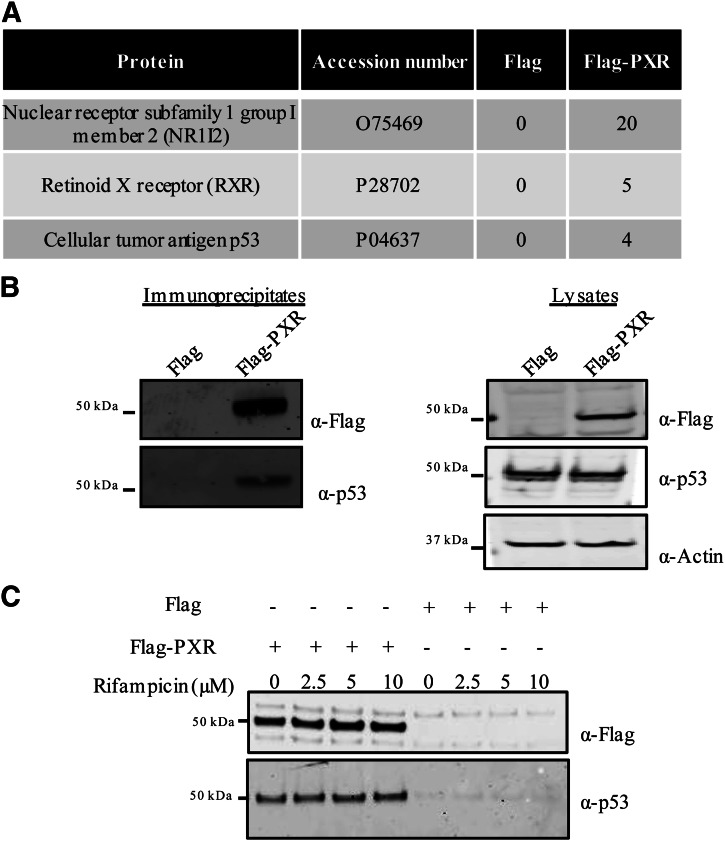
To gain better insight into the regulation of PXR function, we decided to identify protein partners associated with PXR and investigate the effect of such associations on its function. To this end, FLAG-tagged PXR (FLAG-PXR) was transiently overexpressed in HEK293T cells and immunoprecipitated using FLAG-tag-specific antibody covalently attached to agarose beads. The immunoprecipitated samples were then processed, and protein partners associated with FLAG-PXR were identified using mass spectrometry. To identify and eliminate false positives, lysates from cells transiently transfected with plasmid expressing only the FLAG-tag were used as negative controls for immunoprecipitations. A database search using the MASCOT searching algorithm identified 200 proteins associated with PXR. Peptides identified in the negative control samples were eliminated as nonspecific binding partners.
Among the specific binding partners, retinoid X receptor protein, a known PXR binding partner, was consistently detected in only the FLAG-PXR samples. Tumor suppressor protein p53 was one of the associated proteins identified with PXR but not in the negative control sample (Fig. 1A). Using Western blot analysis, we confirmed the association of p53 with FLAG-PXR but not with the FLAG control by using antibody specific for p53 (Fig. 1B). Similar to other NRs, ligand binding to PXR affects its association with coregulators that can regulate PXR signaling. PXR is activated in HEK293T cells in response to rifampicin, a compound known to bind and activate PXR (Supplemental Fig. 1A); however, the association between PXR and p53 was not altered when cells were treated with rifampicin (Fig. 1C).
Fig. 1.
PXR interacts with p53. (A) Proteins and the corresponding number of peptides identified in anti-FLAG immunoprecipitated samples from HEK293T cells transiently overexpressing either FLAG-PXR or FLAG-tag alone using mass spectrometric analysis. (B) Anti-FLAG immunoprecipitated samples were prepared from HEK293T cells transiently overexpressing either FLAG-tag (FLAG) or FLAG-PXR and analyzed using SDS-PAGE. The immunoprecipitates (left panel) and the corresponding lysates (right panel) were immunoprobed using antibodies against p53, FLAG, and β-actin as a loading control. (C) Immunoprecipitates prepared from HEK293T cells overexpressing either FLAG-PXR or FLAG-tag vector and either left untreated or treated with 2.5, 5, or 10 µM rifampicin for 24 hours were probed with antibodies against the FLAG-tag and p53.
Wild-Type p53 Downregulates PXR Activity.
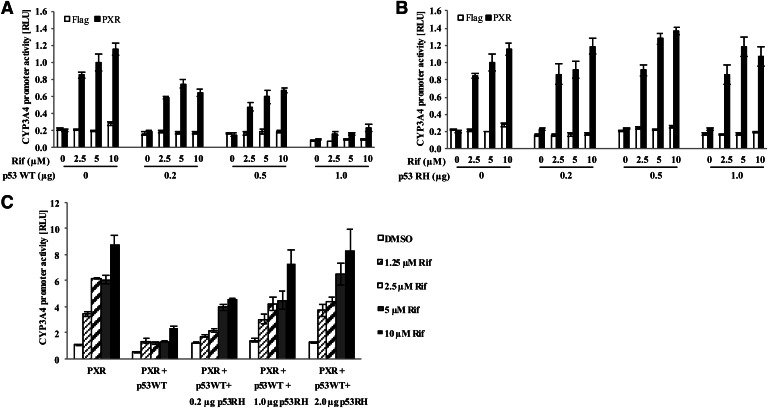
To assess the functional significance of p53-PXR association, the activity of PXR was tested in the presence or absence of wild-type p53. For this purpose, colon cancer cell line HCT116 was used since PXR is functional in this cellular context and isogenic clones with deleted p53 are available (Bunz et al., 1998; Wang et al., 2011). p53−/− HCT116 cells were used to first show that transiently transfected PXR (Supplemental Fig. 1B) and wild-type p53 (Supplemental Fig. 1C) were functional by using the respective reporter gene. Wild-type p53 significantly downregulated PXR reporter activity, and this downregulation occurred in a p53 dose-dependent manner (Fig. 2A). To further test the effect of p53 on PXR activity, a p53 mutant carrying a point mutation at amino acid 175 (R175H) in its DNA binding domain (DBD) was used to determine whether p53 transactivational activity is necessary for its inhibitory effect on PXR. p53 R175H, one of the frequently detected mutants in cancers, is unable to bind to the promoters of p53-regulated genes and is thus considered a nonfunctional protein in regard to transcriptional regulation of p53-dependent genes. As shown in Fig. 2B, the p53 R175H had no effect on PXR reporter activity in p53−/− HCT116 cells, even with increasing amounts of R175H transfected.
Fig. 2.
Wild-type, but not the inactive p53, attenuates PXR activity. HCT116 p53−/− cells were transiently transfected with PXR or an empty vector (FLAG), CYP3A4-luc, TK-Renilla luciferase as transfection control, and either (A) wild-type p53 (p53 WT) or (B) p53 R175H (p53 RH) mutant in the amounts indicated for 48 hours. Cells were then treated with dimethylsulfoxide (DMSO) as control or rifampicin (2.5, 5, or 10 µM) for 24 hours before processing using the Dual-Glo luciferase assay system. (C) HCT116 p53−/− cells were transiently transfected with FLAG-PXR, CYP3A4-luc, TK-Renilla luciferase, 0.2 µg of p53WT, and varying amounts of p53 RH. The firefly luciferase signal was normalized to the corresponding Renilla luciferase signal and reported as relative luciferase units (RLUs).
The effect of p53 R175H was further tested by cotransfecting wild-type p53 along with increasing amounts of p53 R175H. As shown in Fig. 2C, the inhibitory effect of wild-type p53 on PXR was decreased by the R175H mutant and completely rescued when larger amounts of p53 R175H were transfected, indicating that the p53 R175H mutant functions in a dominant negative manner. The dominant negative effect of p53 R175H mutant prompted us to determine whether it interacts with PXR. As shown in Fig. 3A, p53 R175H mutant interacts with PXR similarly to wild-type p53, suggesting that p53 R175H abolishes the inhibitory effect of wild-type p53 by blocking it from interacting with PXR.
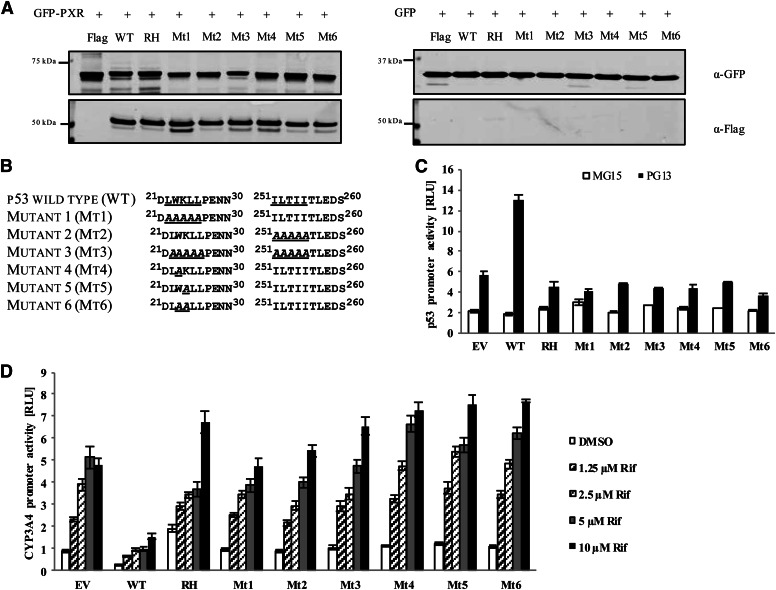
Fig. 3.
Neither LXXLL nor IXXII NR-interacting motif on p53 is required for p53-PXR interaction. (A) U2OS cells stably expressing GFP-PXR or GFP were transiently transfected with FLAG-tagged wild-type p53 (WT) or one of the FLAG-tagged p53 mutants (RH, Mt1, Mt2, Mt3, Mt4, Mt5, and Mt6) for 48 hours. Cell lysates were immunoprecipitated using GFP-specific beads and resolved using SDS-PAGE and then immunoprobed using anti-GFP (for GFP-PXR) and anti-FLAG (for FLAG-p53) antibodies. (B) FLAG-tagged p53 mutants were generated at the LWKLL and ILTII sequences by replacing one or multiple amino acids with alanine. (C) Wild-type p53 and the indicated p53 mutants were transiently transfected in HCT116 p53−/− cells along with p53 responsive luciferase reporter (PG13) and TK-Renilla luciferase as a control for 48 hours. Luciferase reporter plasmid that does not respond to p53 (MG15) was used as a control to show the specificity of the p53 promoter activity. (D) HCT116 p53−/− cells were transfected using FLAG-PXR, CYP3A4-luc, TK-Renilla luciferase plasmid, and either empty vector control (EV), wild-type p53 (WT), or mutant p53 (RH, Mt1, Mt2, Mt3, Mt4, Mt5, and Mt6) for 48 hours. Cells were treated with dimethylsulfoxide (DMSO) or with 1.25, 2.5, 5, or 10 µM rifampicin (Rif) for 24 hours before analysis using the Dual-Glo luciferase assay system. The firefly luciferase signal was normalized to the corresponding Renilla luciferase signal and reported as relative luciferase units (RLUs).
p53 Activity Is Necessary for its Inhibitory Effect on PXR.
Two unique sequences, LWKLL in the transactivation domain and ILTII in the DBD, were identified in the p53 protein that corresponds to motifs described in other coactivators and corepressors of NRs (Fig. 3B). The LXXLL motif, which is present in several coactivators of PXR, mediates protein-protein interactions to enhance the activity of the associated receptor (Plevin et al., 2005). The I/LXXI/VI corepressor NR boxes mediate binding of corepressors such as NCoR and SMRT to NRs, where they attenuate receptor activity (Perissi et al., 1999). To determine whether the LWKLL or ILTII motifs in p53 mediate the protein-protein interaction with PXR, we mutated the LWKLL or ILTII sequences individually (Mt1, Mt2) and in combination (Mt3), as indicated in Fig. 3B. Similar to the p53 R175H mutant, Mt1, Mt2, and Mt3 were unable to regulate the activity of the p53 response luciferase reporter gene PG13 (Fig. 3C). Mutation at Trp23 within the LWKLL sequence has been shown to result in a loss of p53 transactivation activity and is thus critical for p53 function (Lin et al., 1994). The loss of function was observed when either Trp23 or Lys24 within the LWKLL motif was mutated individually or together to an alanine (Mt4, Mt5, and Mt6) (Fig. 3, B and C).
In immunoprecipitation experiments from U2OS cells stably expressing GFP-PXR or GFP alone, GFP-PXR showed a similar association with all the p53 mutants that were used (Fig. 3A), suggesting that neither the LWKLL nor the ILTII motif is required for p53 to interact physically with PXR. Consistent with the results observed with the R175H mutant (Fig. 2, B and C), the inactive p53 mutants did not attenuate PXR activity (Fig. 3D) and showed a dominant negative effect on the inhibitory effect of wild-type p53 on PXR (Fig. 2C and data not shown).
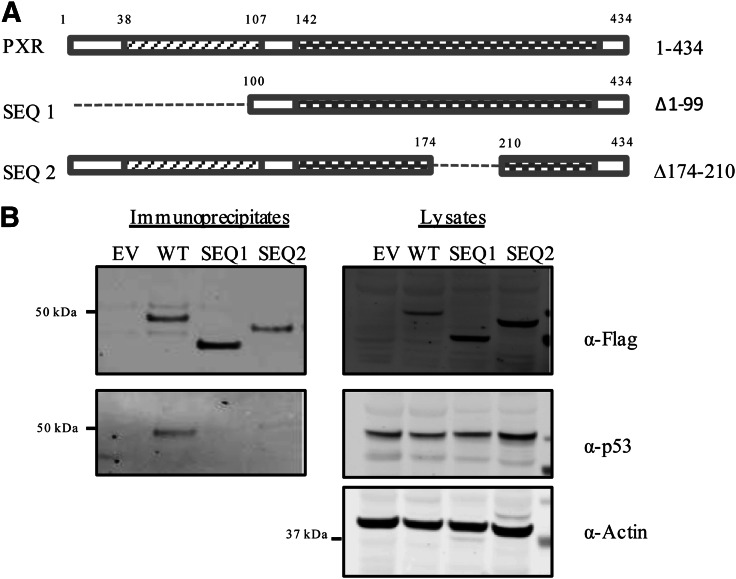
PXR.2 (PXR∆174–210) Fails to Interact with p53.
To identify which domains of PXR are responsible for interacting with p53, various truncation mutants of PXR were generated (Fig. 4A). PXR∆1–99, which contains the full LBD (amino acids 142–434) but lacks the DBD, failed to interact with p53 (Fig. 4B), suggesting that the N-terminal 99 amino acids are required for the interaction. A more refined study showed that deletion of either amino acids 2–40 (∆2–40) or amino acids 21–60 (∆21–60) at the N terminus did not noticeably affect the interaction of with p53 (Supplemental Fig. 2), suggesting that the amino acids 61–99, which are part of the DBD (amino acids 41–107), are critical for p53-PXR interaction. On the other hand, deletion of either the last 14 amino acids (∆421–434) or the last 134 amino acids (∆301–434) at the C terminus, which removes the activation function domain 2 (AF-2), also abolishes the interaction of PXR with p53 (Supplemental Fig. 2), suggesting that the AF-2 domain of PXR is critical for its interaction with p53.
Fig. 4.
The region of PXR encompassing amino acids 174–210 is important for its association with p53. (A) Schematic representation of the PXR protein and the truncation mutants that were generated: the LBD ∆1–99 (SEQ1) and ∆174–210 (SEQ2). (B) FLAG-PXR or FLAG-PXR mutants were transfected into HEK293T cells and immunoprecipitated using the FLAG-M2 beads. Left panel shows the Western blot analysis to detect FLAG-PXR using anti-FLAG (α-FLAG) and p53 using anti-p53 (α-p53) antibodies in the immunoprecipitated samples, and the protein levels in corresponding lysates are shown in the right panel. Actin levels are shown to verify equal loading of the lysates.
Most interestingly, we found that the amino acids from 174 to 210 on PXR play a significant role in preserving the p53-PXR association (Fig. 4B). This sequence is absent in the alternatively spliced isoform of PXR, also referred to as PXR.2 (Lin et al., 2009), indicating that p53 specifically associates with the more abundant isoform of PXR, PXR.1. In agreement with previously reported findings, whereas PXR.1 was robustly activated by rifampicin, PXR.2 was not (Supplemental Fig. 3, A and B). Interestingly, amino acids 174–210, which are present in PXR.1 but missing from PXR.2, are specific for PXR and are highly conserved among PXR proteins from different species identified to date (Supplemental Fig. 4). The requirement for a highly conserved and PXR-specific sequence for the interaction further indicates the specificity of the p53-PXR association.
Wild-Type p53 Attenuates PXR Binding to CYP3A4 Promoter.
As transcription factors, both PXR and p53 bind to specific sequences in the promoters of target genes to initiate transcription (Kliewer et al., 2002; Menendez et al., 2009). We used the ChIP assay to determine whether p53 modulates the binding of PXR to its target promoter in CYP3A4 in hepatocellular carcinoma cells HepG2 stably expressing FLAG-PXR, which endogenously express wild-type p53 (Puisieux et al., 1993). Anti-FLAG immunoprecipitations from HepG2 cells overexpressing wild-type p53 showed significantly less PXR recruited to the DR3 region of the CYP3A4 promoter than in immunoprecipitated samples from cells expressing only the control empty vector (Fig. 5, A and B). In contrast, overexpression of the p53 R175H mutant did not attenuate but enhanced the PXR association with CYP3A4 promoter (Fig. 5, A and B), consistent with its dominant negative effect (Fig. 2C). The levels of endogenous as well as transfected p53 are indicated in Fig. 5C.
Discussion
PXR is a master xenobiotic sensor whose activity is upregulated by binding to agonists, and the agonist-mediated receptor activity is modulated by the association of PXR with various protein partners. We showed in this study, for the first time, that p53 is one such PXR protein partner whose physical association leads to attenuation of PXR activity. We also show that the decrease in the binding of PXR to its target promoter contributes to such attenuation of receptor activity.
Indisputably, p53 is one of the most important tumor suppressors, as evidenced by the fact that more than 50% of tumors carry a p53 mutation (Hainaut and Hollstein, 2000). Most tumors carry a mutation in the p53 protein that affects either its transactivation function or its ability to bind specific DNA sequences at the promoter of p53 target genes (Soussi and Wiman, 2007). R175 is one of the hotspots for p53 missense mutations. p53 has been shown to interact with various proteins, including NRs, with various functional outcomes. In the case of p53-ER association, p53 enhances the recruitment of ER to its target promoter (Angeloni et al., 2004). In contrast, in our study, the p53-PXR association results in decreased binding of PXR to its target promoter in CYP3A4 gene, suggesting that the functional outcome of the protein-protein interaction depends on the specific protein partner involved. The effect of p53 on PXR may also vary with different PXR target promoters.
Since p53 interacts with multiple protein partners with different functional outcomes, it is important to determine whether such an interaction is specific and how this specificity is achieved. Interestingly, we found that the interaction between p53 and PXR is specific to the more abundant PXR isoform PXR.1 (434 amino acids) and does not occur with the alternatively spliced form of the protein PXR.2 (397 amino acids, with amino acids 174–210 deleted). The segment of 37 amino acids (174–210) missing from PXR.2 is unique to PXR and is not present in any other protein. It is also highly conserved among PXR proteins from different species, indicating that the interaction between p53 and PXR.1 is specific and unique. Although the physiologic function of PXR.2 has not been fully defined, such function is expected to be insensitive to p53 regulation.
PXR functions as a xenobiotic sensor mainly due to its ability to accommodate structurally diverse xenobiotics. Whereas ligand binding to PXR can alter the interaction of PXR with corepressors such as SMRT and NCoR (Ding and Staudinger, 2005; Johnson et al., 2006), it does not affect the p53-PXR.1 interaction, suggesting that the cellular levels of p53 may affect both the basal and agonist-induced activities of PXR. However, the observation that rifampicin does not alter the p53-PXR.1 association in vitro does not exclude the possibility that in vivo rifampicin could affect such an association in a subcellular localization-specific manner. Although PXR.2 can retain certain corepressor associations similar to those of PXR.1, PXR.2 fails to activate PXR target genes upon ligand treatment. The deletion of 37 amino acids (174–210) in the LBD is predicted to make PXR.2 unable to retain certain ligands, thereby altering the ligand and protein partner profile (Lin et al., 2009). It is thus not surprising that the integrity of the PXR protein structure is important to retain certain protein-protein interactions such as PXR-p53 binding, as revealed by the loss of interaction with p53 when the AF-2 domain (∆421–434) or the N-terminal 99 amino acids of PXR (∆1–99) were deleted. The N-terminal deletion mutant ∆1–99, but not the ∆2–40 or ∆21–60, abolished the interaction, suggesting that the major part of the DBD (41–107), is required for PXR to interact with p53 and is in agreement with the observation that p53 can attenuate the binding of PXR to the CYP3A4 promoter through its DBD.
As we showed in this report, p53 association is not always sufficient to attenuate the activity of the target protein since p53 wild-type and mutant forms can bind to PXR, but only the wild-type form can downregulate the activity of PXR. p53 mutants such as R175H, although inactive, have been shown to attain gain-of-function properties, which can initiate novel protein-protein interactions and transactivation of new target genes, contributing to tumor progression and chemoresistance (Blandino et al., 1999). All the transcriptionally inactive p53 mutants examined in our study maintained their association with PXR, although they all failed to attenuate PXR-mediated transactivation of target genes such as CYP3A4. As expected, these inactive p53 mutants, including p53-R175H, were shown to function in a dominant-negative manner by decreasing the interaction of wild-type p53 and PXR, thereby releasing the inhibitory effect of wild-type p53 on PXR. The dominant-negative effect of the p53-R175H mutant was also observed in the ChIP assay when ectopically expressed from a plasmid; whereas wild-type p53 decreased the binding of PXR to the CYP3A4 promoter, the R175H mutant actually enhanced the binding, possibly by replacing the endogenous wild-type p53 from the CYP3A4 promoter. Although it is unclear why binding of p53 R175H to PXR did not attenuate the binding of PXR to the CYP3A4 promoter, it is possible that the gain-of-function of p53 mutants, including the R175H mutant, contributes to the altered functional outcome of protein-protein interactions. Trp23 is necessary for the transactivation of p53 as well as its association with Mdm2 (Lin et al., 1994; Picksley et al., 1994). Interestingly, Trp23 falls within one of the sequences (LXXLL) used by coactivators to bind NRs. Although p53 also harbors the corepressor association sequence (IXXII), neither motif contributes to PXR binding.
In colon cancer cells, expression of wild-type p53 induces apoptosis (Shaw et al., 1992), whereas activation of PXR induces colon tumor growth and malignancy (Wang et al., 2011; Mani et al., 2013; Pondugula and Mani, 2013). In this study, we showed that in colon cancer cell line HCT116, wild-type p53 downregulates PXR activity. It is conceivable that in colon cancers with wild-type p53, decreased PXR activity as a result of inhibition by p53 might contribute to p53-mediated growth inhibition. Both the status of p53 and the levels of PXR will be crucial in determining their effect on tumor cell growth and chemosensitivity.
The regulation of a master xenobiotic sensor by a master tumor suppressor raises some interesting questions and exciting possibilities. As the key regulator of the drug metabolism pathway, PXR regulates drug efficacy and contributes to drug resistance in tumors (Chen et al., 2007; Wang et al., 2012). On the other hand, loss of wild-type p53 activity contributes greatly to radioresistance and chemoresistance in tumors containing p53 mutations (Fojo, 2002; El-Deiry, 2003). Our finding that wild-type p53 interacts with and attenuates the activity of PXR suggests that the status of p53 might affect the efficacy of drugs, partially through affecting PXR-regulated pathways. Further investigations, including a comprehensive study on the effect of various naturally occurring p53 mutants on the activity of PXR and correlation of such effects to clinical observations will help establish the role of p53 in systematic drug metabolism.
Supplementary Material
Acknowledgments
The authors thank Dr. Bert Vogelstein (Johns Hopkins University) for cell lines, the St. Jude Proteomics & Mass Spectrometry Shared Resource for mass spectrometric analysis, Asli Goktug for technical assistance, other members of the Chen research laboratory, Dr. Gerard Zambetti (St. Jude) for valuable discussions, and David Galloway (Department of Scientific Editing, St. Jude) for editing of the manuscript.
Abbreviations
- AF-2
activation function domain 2
- ChIP
chromatin immunoprecipitation
- DBD
DNA binding domain
- ER
estrogen receptor
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- HCT-116
human colon carcinoma cell line
- HEK293T
human embryonic kidney cell line
- HepG2
liver hepatocellular carcinoma cell line
- LBD
ligand binding domain
- NCoR
nuclear receptor corepressor
- NR
nuclear receptor
- PCR
polymerase chain reaction
- PXR
pregnane X receptor
- RH
p53 R175H mutant
- SMRT
silencing mediator for retinoid and thyroid receptors
- TBS
Tris-buffered saline
- U2OS
human osteosarcoma cell line
Authorship Contributions
Participated in research design: Elias, Chen.
Conducted experiments: Elias, Wu.
Performed data analysis: Elias, Chen.
Wrote or contributed to the writing of the manuscript: Elias, Chen.
Footnotes
This work was supported by the American Lebanese Syrian Associated Charities; St. Jude Children’s Research Hospital; the National Institutes of Health National Institute of General Medical Sciences [Grant GM086415]; and the National Institutes of Health National Cancer Institute [Grant P30-CA21765].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Alimirah F, Panchanathan R, Chen J, Zhang X, Ho SM, Choubey D. (2007) Expression of androgen receptor is negatively regulated by p53. Neoplasia 9:1152–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeloni SV, Martin MB, Garcia-Morales P, Castro-Galache MD, Ferragut JA, Saceda M. (2004) Regulation of estrogen receptor-alpha expression by the tumor suppressor gene p53 in MCF-7 cells. J Endocrinol 180:497–504 [DOI] [PubMed] [Google Scholar]
- Blandino G, Levine AJ, Oren M. (1999) Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene 18:477–485 [DOI] [PubMed] [Google Scholar]
- Blumberg B, Kang H, Bolado J, Jr, Chen H, Craig AG, Moreno TA, Umesono K, Perlmann T, De Robertis EM, Evans RM. (1998a) BXR, an embryonic orphan nuclear receptor activated by a novel class of endogenous benzoate metabolites. Genes Dev 12:1269–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Sabbagh W, Jr, Juguilon H, Bolado J, Jr, van Meter CM, Ong ES, Evans RM. (1998b) SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev 12:3195–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497–1501 [DOI] [PubMed] [Google Scholar]
- Chen Y, Tang Y, Wang MT, Zeng S, Nie D. (2007) Human pregnane X receptor and resistance to chemotherapy in prostate cancer. Cancer Res 67:10361–10367 [DOI] [PubMed] [Google Scholar]
- Ding X, Staudinger JL. (2005) Repression of PXR-mediated induction of hepatic CYP3A gene expression by protein kinase C. Biochem Pharmacol 69:867–873 [DOI] [PubMed] [Google Scholar]
- Dotzlaw H, Leygue E, Watson P, Murphy LC. (1999) The human orphan receptor PXR messenger RNA is expressed in both normal and neoplastic breast tissue. Clin Cancer Res 5:2103–2107 [PubMed] [Google Scholar]
- El-Deiry WS. (2003) The role of p53 in chemosensitivity and radiosensitivity. Oncogene 22:7486–7495 [DOI] [PubMed] [Google Scholar]
- Fan S, el-Deiry WS, Bae I, Freeman J, Jondle D, Bhatia K, Fornace AJ, Jr, Magrath I, Kohn KW, O’Connor PM. (1994) p53 gene mutations are associated with decreased sensitivity of human lymphoma cells to DNA damaging agents. Cancer Res 54:5824–5830 [PubMed] [Google Scholar]
- Fan S, Smith ML, Rivet DJ, 2nd, Duba D, Zhan Q, Kohn KW, Fornace AJ, Jr, O’Connor PM. (1995) Disruption of p53 function sensitizes breast cancer MCF-7 cells to cisplatin and pentoxifylline. Cancer Res 55:1649–1654 [PubMed] [Google Scholar]
- Fojo T. (2002) p53 as a therapeutic target: unresolved issues on the road to cancer therapy targeting mutant p53. Drug Resist Updat 5:209–216 [DOI] [PubMed] [Google Scholar]
- Goodwin B, Redinbo MR, Kliewer SA. (2002) Regulation of cyp3a gene transcription by the pregnane x receptor. Annu Rev Pharmacol Toxicol 42:1–23 [DOI] [PubMed] [Google Scholar]
- Hainaut P, Hollstein M. (2000) p53 and human cancer: the first ten thousand mutations. Adv Cancer Res 77:81–137 [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. (1991) p53 mutations in human cancers. Science 253:49–53 [DOI] [PubMed] [Google Scholar]
- Iwano S, Shibahara N, Saito T, Kamataki T. (2006) Activation of p53 as a causal step for atherosclerosis induced by polycyclic aromatic hydrocarbons. FEBS Lett 580:890–893 [DOI] [PubMed] [Google Scholar]
- Johnson DR, Li CW, Chen LY, Ghosh JC, Chen JD. (2006) Regulation and binding of pregnane X receptor by nuclear receptor corepressor silencing mediator of retinoid and thyroid hormone receptors (SMRT). Mol Pharmacol 69:99–108 [DOI] [PubMed] [Google Scholar]
- Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. (1992) Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256:827–830 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Goodwin B, Willson TM. (2002) The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr Rev 23:687–702 [DOI] [PubMed] [Google Scholar]
- Kodama S, Koike C, Negishi M, Yamamoto Y. (2004) Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol 24:7931–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba V, Yasuda K, Lamba JK, Assem M, Davila J, Strom S, Schuetz EG. (2004) PXR (NR1I2): splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol Appl Pharmacol 199:251–265 [DOI] [PubMed] [Google Scholar]
- Levine AJ, Hu W, Feng Z. (2006) The P53 pathway: what questions remain to be explored? Cell Death Differ 13:1027–1036 [DOI] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. (1994) Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev 8:1235–1246 [DOI] [PubMed] [Google Scholar]
- Lin W, Wu J, Dong H, Bouck D, Zeng FY, Chen T. (2008) Cyclin-dependent kinase 2 negatively regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. J Biol Chem 283:30650–30657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Yasuda K, Assem M, Cline C, Barber J, Li CW, Kholodovych V, Ai N, Chen JD, Welsh WJ, et al. (2009) The major human pregnane X receptor (PXR) splice variant, PXR.2, exhibits significantly diminished ligand-activated transcriptional regulation. Drug Metab Dispos 37:1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Cunningham M, Kim S, Burn T, Lin J, Sinz M, Hamilton G, Rizzo C, Jolley S, Gilbert D, et al. (2002) CYP3A4 induction by drugs: correlation between a pregnane X receptor reporter gene assay and CYP3A4 expression in human hepatocytes. Drug Metab Dispos 30:795–804 [DOI] [PubMed] [Google Scholar]
- Maeda Y, Seidel SD, Wei G, Liu X, Sladek FM. (2002) Repression of hepatocyte nuclear factor 4alpha tumor suppressor p53: involvement of the ligand-binding domain and histone deacetylase activity. Mol Endocrinol 16:402–410 [DOI] [PubMed] [Google Scholar]
- Mani S, Dou W, Redinbo MR. (2013) PXR antagonists and implication in drug metabolism. Drug Metab Rev 45:60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez D, Inga A, Resnick MA. (2009) The expanding universe of p53 targets. Nat Rev Cancer 9:724–737 [DOI] [PubMed] [Google Scholar]
- Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. (1999) Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev 13:3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picksley SM, Vojtesek B, Sparks A, Lane DP. (1994) Immunochemical analysis of the interaction of p53 with MDM2;—fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene 9:2523–2529 [PubMed] [Google Scholar]
- Plevin MJ, Mills MM, Ikura M. (2005) The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem Sci 30:66–69 [DOI] [PubMed] [Google Scholar]
- Pondugula SR, Mani S. (2013) Pregnane xenobiotic receptor in cancer pathogenesis and therapeutic response. Cancer Lett 328:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puisieux A, Galvin K, Troalen F, Bressac B, Marcais C, Galun E, Ponchel F, Yakicier C, Ji J, Ozturk M. (1993) Retinoblastoma and p53 tumor suppressor genes in human hepatoma cell lines. FASEB J 7:1407–1413 [DOI] [PubMed] [Google Scholar]
- Raynal C, Pascussi JM, Leguelinel G, Breuker C, Kantar J, Lallemant B, Poujol S, Bonnans C, Joubert D, Hollande F, et al. (2010) Pregnane X receptor (PXR) expression in colorectal cancer cells restricts irinotecan chemosensitivity through enhanced SN-38 glucuronidation. Mol Cancer 9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Bovey R, Tardy S, Sahli R, Sordat B, Costa J. (1992) Induction of apoptosis by wild-type p53 in a human colon tumor-derived cell line. Proc Natl Acad Sci USA 89:4495–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk JL, Fisher CJ, Chen SY, Zhou XF, Tillman K, Shemshedini L. (2001) p53 represses androgen-induced transactivation of prostate-specific antigen by disrupting hAR amino- to carboxyl-terminal interaction. J Biol Chem 276:38472–38479 [DOI] [PubMed] [Google Scholar]
- Shirley SH, Rundhaug JE, Tian J, Cullinan-Ammann N, Lambertz I, Conti CJ, Fuchs-Young R. (2009) Transcriptional regulation of estrogen receptor-alpha by p53 in human breast cancer cells. Cancer Res 69:3405–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T, Wiman KG. (2007) Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell 12:303–312 [DOI] [PubMed] [Google Scholar]
- Wang H, Venkatesh M, Li H, Goetz R, Mukherjee S, Biswas A, Zhu L, Kaubisch A, Wang L, Pullman J, et al. (2011) Pregnane X receptor activation induces FGF19-dependent tumor aggressiveness in humans and mice. J Clin Invest 121:3220–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YM, Ong SS, Chai SC, Chen T. (2012) Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin Drug Metab Toxicol 8:803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RE, Davis-Searles PR, Lambert MH, Redinbo MR. (2003) Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J Mol Biol 331:815–828 [DOI] [PubMed] [Google Scholar]
- Yu C, Yap N, Chen D, Cheng S. (1997) Modulation of hormone-dependent transcriptional activity of the glucocorticoid receptor by the tumor suppressor p53. Cancer Lett 116:191–196 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.