Abstract
The aryl hydrocarbon receptor (AhR) is a ligand-mediated basic helix-loop-helix transcription factor of the Per/Arnt/Sim family that regulates adaptive and toxic responses to a variety of chemical pollutants, including polycyclic aromatic hydrocarbons and halogenated aromatic hydrocarbons, most notably 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Ligand activation leads to AhR nuclear translocation and binding to a xenobiotic response element (XRE) in association with the Arnt to regulate gene expression. Several recent genome-wide transcriptional studies identified numerous AhR target genes that lack the canonical XRE recognition site in the promoter regions. Characterization of one such target gene, the plasminogen activator inhibitor 1, identified a novel nonconsensus XRE (NC-XRE) that confers TCDD responsiveness independently of the Arnt protein. Studies reported here show that the NC-XRE is a recognition site for the AhR and a new binding partner, the Kruppel-like factor (KLF) family member KLF6. In vivo chromatin immunoprecipitations and in vitro DNA binding studies demonstrate that the AhR and KLF6 proteins form an obligatory heterodimer necessary for NC-XRE binding. Mutational analyses show that the protein-protein interactions involve the AhR C terminus and KLF6 N terminus, respectively. Moreover, NC-XRE binding depends on the 5′ basic region in KLF6 rather than the previously characterized zinc finger DNA binding domain. Collectively, the results unmask a novel AhR signaling mechanism distinct from the canonical XRE-driven process that will enrich our future understanding of AhR biology.
Introduction
The aryl hydrocarbon receptor (AhR), a member of the Per/Arnt/Sim (PAS) family of transcription factors, is a known mediator of the toxic response to the potent and persistent toxicant 2,3,7,8-tetracholodibenzo-p-dioxin (TCDD), and regulates the expression of multiple genes involved in toxicant metabolism, such as CYP1A1, in addition to modulating cell proliferation (Puga et al., 2000; Santini et al., 2001; Huang and Elferink, 2005; Mitchell et al., 2006, 2010; Mitchell and Elferink, 2009). In steady-state conditions, the AhR is sequestered in the cytoplasm bound to heat shock protein 90 (HSP90) and the chaperonin AhR interacting protein (ARA9/AIP/XAP2) via its basic helix-loop-helix (bHLH) and PAS domains (Perdew, 1988; Carver and Bradfield, 1997; Ma and Whitlock, 1997; Carver et al., 1998) Upon ligand binding to the PAS B domain, the nuclear localization sequence in the N terminus is revealed, resulting in translocation to the nucleus. Once in the nucleus and shed of its chaperonins, in the classic characterization of AhR activity, the AhR heterodimerizes with another member of the PAS family, the aryl hydrocarbon receptor nuclear translocator (Arnt), through both the bHLH and PAS domains (Hoffman et al., 1991; Prokipcak and Okey, 1991; Probst et al., 1993; Ikuta et al., 1998; Lees and Whitelaw, 1999). The AhR-Arnt heterodimer binds to canonical DNA sequence (5′TnGCGTG-3′), referred to as the xenobiotic response element (XRE), in the promoter region of target genes, inducing their transcription (Probst et al., 1993; Elferink and Whitlock, 1994; Hankinson, 1995). Given the wide array of functions attributed to activated AhR from toxicant metabolism to cell proliferation, speculation about alternate mechanisms of action abounds.
Apart from the AhR complex binding to XRE, recent studies have identified a novel and specific DNA sequence in the plasminogen activator inhibitor 1 (PAI-1) promoter, referred to as nonconsensus XRE (NC-XRE), to which AhR binds in a TCDD-inducible manner (Huang and Elferink, 2012). However, the XRE and NC-XRE share no sequence homology. AhR binding to the NC-XRE involves a 5′-GGGA-3′-tetranucleotide motif, a marked departure from the 5′-GCGTG-3′ core consensus motif identifying the XRE. Likewise, the protein-DNA complexes that form at the XRE and NC-XRE are biochemically distinct, with the latter not requiring the Arnt protein. These data suggest that AhR binding to the NC-XRE involves novel protein interactions.
While the XRE and NC-XRE are distinct entities, the NC-XRE shares marked homology with the DNA binding sequence of the Kruppel-like factor (KLF) family. The KLF family is a recently identified and growing superfamily of transcription factors related to specificity protein 1 (Sp1) and characterized by three zinc finger domains in their C termini that confer binding to a “GC-box” linked to diverse target genes (Philipsen and Suske, 1999; Bieker, 2001). Supporting a potential interaction between the AhR and a KLF family member, two separate KLF family members have been associated with the established AhR target gene CYP1A1: KLF9 can bind to regions in the rat CYP1A1 promoter and modulate its expression (Imataka et al., 1992), and KLF4 inhibits CYP1A1 induction via an interaction with Sp1 (Zhang et al., 1998). However, neither KLF9 nor KLF4 are expressed in the liver, the site of AhR-dependent PAI-1 induction involving the NC-XRE. KLF6 is a member of the KLF family of transcription factors expressed in the liver and characterized as a tumor suppressor (Ratziu et al., 1998). Reminiscent of the AhR-regulated p21 (WAF1/CIP1) gene expression, KLF6 has also been shown to induce transcription of p21 (Kojima et al., 2000; Narla et al., 2001, 2007), suggestive of an overlapping or complementary role for both proteins in cell cycle regulation. Results of this study indicate that AhR interacts with KLF6 at NC-XRE in the PAI-1 promoter in a dioxin-dependent manner. Furthermore, sequential deletion studies demonstrate the importance of the C terminus of the AhR and the N-terminal domains of KLF6 in this interaction. In addition, KLF6 DNA binding is dependent on key arginine residues located outside the zinc finger domain.
Materials and Methods
Animals and TCDD Treatment.
All care and procedure conditions were approved by the guidelines set by the Institutional Animal Care and Use Committee at the University of Texas Medical Branch at Galveston. C57BL/6 mice (8- to 10-week-old females, weighing 22–25 g) were purchased from The Jackson Laboratories (Bar Harbor, ME). TCDD (Cerilliant, Round Rock, TX) was dissolved in anisole and diluted to 2 µg/ml in peanut oil or to 200 nM in dimethylsulfoxide, whereas the vehicle control consisted of peanut oil with a corresponding amount of anisole or dimethylsulfoxide alone for in vivo and in vitro experiments, respectively. Animals were administered via gavage with either vehicle or TCDD (20 μg/kg) 2 hours before sacrifice by isoflurane overdose followed by cervical dislocation. In vitro transcribed and translated proteins were treated with vehicle or 20 nM TCDD for 1 hour at 30°C or 20 minutes at 37°C for coimmunoprecipitation and electrophoretic mobility shift assay (EMSA) studies, respectively.
AhR and KLF6 Constructs.
Full-length human AhR was cloned into the pSport vector using KpnI and SalI sites, was Sp6 promoter driven, and was a gift from Dr. William Chan (University of the Pacific, Stockton, CA). The C-terminal deletion of human AhR was created by restriction enzyme digestion of full-length human AhR with BstY I (New England BioLabs, Ipswich, MA). Human KLF6 full-length and mutant constructs were provided by Dr. Scott Friedman (Mount Sinai School of Medicine, New York, NY) in the pCS2+MT vector under the T7 promoter. Due to poor expression levels, the human KLF6 constructs were subcloned into the pSport vector via SalI and KpnI sites. Using the full-length human KLF6/pCS2+MT construct as a template, polymerase chain reaction (PCR) was performed with the primers listed in Table 1. Mouse AhR constructs were in the pcDNA1/neo vector under the T7 promoter, and were a gift from Dr. Oliver Hankinson (UCLA, Los Angeles, CA). Mouse KLF6 constructs were generated using the same strategy as for the human KLF6 constructs, and were subcloned into the pSport vector. The cDNA reverse transcribed from liver RNA using the primer 5′ CTCTTTTAGCCTACAGGATTCGTC-3′ was used as a template. Primers designed were based on the mouse KLF6 sequence (Inuzuka et al., 1999) and are listed in Table 1.
TABLE 1.
Oligonucleotides used in PCR and ChIP
| Oligonucleotides (Forward/Reverse) | DNA Sequence (5′→3′) |
|---|---|
| Cloning primers | |
| WT hKLF6 | GGTACCATGGATTACAAGGATGACGACGATAAGATGGACGTGCTCCCCATGTGCAG |
| GTCGACTCAGAGGTGCCTCTTCATGTGCAG | |
| Δ27 hKLF6 | GGTACCATGGATTACAAGGATGACGACGATAAGCTGGAGGAGTACTGGCAACAGACC |
| GTCGACTCAGAGGTGCCTCTTCATGTGCAG | |
| Δ128 hKLF6 | GGTACCATGGATTACAAGGATGACGACGATAAGCCCATTGGCGAAGTTTTGGTCAGC |
| GTCGACTCAGAGGTGCCTCTTCATGTGCAG | |
| Δ178 hKLF6 | GGTACCATGGATTACAAGGATGACGACGATAAGACTTCGGGGAAGCCAGGTGACAAG |
| GTCGACTCAGAGGTGCCTCTTCATGTGCAG | |
| Δ202-283 hKLF6 | GGTACCATGGATTACAAGGATGACGACGATAAGATGGACGTGCTCCCCATGTGCAG |
| GTCGACTCACCGGTGCACCCTCCTCCTGCCGTC | |
| WT mKLF6 | GGTACCATGGATTACAAGGATGACGACGATAAGATGAAACTTTCACCTGCGCTCCCGGGAACA |
| GTCGACTCAGAGGTGCCTCTTCATGTGCAG | |
| Δ34 mKLF6 | GGTACCATGGATTACAAGGATGACGACGATAAGATGGATGTGCTCCCAATGTGTAGCATCTTC |
| GTCGACTCAGAGGTGCCTCTTCATGTGCAG | |
| Δ61 mKLF6 | GGTACCATGGATTACAAGGATGACGACGATAAGCTGGAGGAATATTGGCAACAGACC |
| GTCGACTCAGAGGTGCCTCTTCATGTGCAG | |
| Δ128 mKLF6 | GGTACCATGGATTACAAGGATGACGACGATAAGTTTAATTATAACTTAGAGACCAATAGCCTG |
| GTCGACTCAGAGGTGCCTCTTCATGTGCAG | |
| Δ212-318 mKLF6 | GGTACCATGGATTACAAGGATGACGACGATAAGATGAAACTTTCACCTGCGCTCCCGGGAACA |
| GTCGACTCAACTTCGAACCTTCCCAGGTGAGGGCAGGTC | |
| ChIP primers | |
| PAI-1 | GTCCCAGCAAGTCACTGGGAGG |
| CTGGAGGCGGGTGTGCGGCG | |
| CYP1A1 | CTATCTCTTAAACCCCACCCCAA |
| CTAAGTATGGTGGAGGAAAGGGTG |
WT, wild type.
In Vitro Transcription/Translation.
AhR and KLF6 constructs were expressed using the TnT coupled reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer’s protocol. For the 50-µl reaction, 2 µg of each construct was added, and expression was driven by the T7 promoter for the mouse AhR constructs or by the Sp6 promoter for human AhR, human KLF6, and mouse KLF6 constructs.
Nuclear Extract Preparation.
Following sacrifice, livers were blanched with ice-cold phosphate-buffered saline, removed, finely minced, and transferred to a Dounce homogenizer with 1.6 ml of sterile phosphate-buffered saline and 4 ml of 2.2-M sucrose solution [2.2 M sucrose, 10 mM HEPES, 15 mM KCl, 2 mM EDTA (pH 8.0), and added just prior to use 0.15 mM spermine, 0.5 mM spermidine, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 5 µl/ml protease inhibitor cocktail]. Samples were homogenized on ice and filtered through cheesecloth. An additional 3 ml of the 2.2-M sucrose solution was added to each sample and mixed gently. This homogenate was layered onto 4 ml of a 2.05-M sucrose cushion (same composition as the 2.2-M sucrose solution, but with 2.05 M sucrose) and centrifuged at 30,000g in a Beckman Coulter (Brea, CA) MLS-50 rotor for 1 hour at 4°C. The white nuclear pellets were resuspended in 1 ml of nuclear lysis buffer [10 mM HEPES, 102 mM KCl, 0.1 mM EDTA (pH 8.0), 11.4% glycerol, 0.15 mM spermine, 0.5 mM spermidine, 1 mM NaF, 1 mM Na2VO4, 1 mM ZnSO4, and 5 µl/ml protease inhibitor cocktail] on ice, and washed twice with nuclear lysis buffer by centrifugation at 1500g. After the final wash, the pellet was resuspended in 200 µl of nuclear lysis buffer. An equal volume of 2X NUN buffer (2 M urea, 2% NP40, 650 mM NaCl, 50 mM HEPES, and 2 mM DTT) was added, drop by drop. Samples were incubated on ice for 20 minutes and centrifuged at 55,000g for 20 minutes at 4°C (TLA 55; Beckman Instruments). The supernatant was collected, and nuclear protein concentration was measured with a Bio-Rad (Hercules, CA) protein assay kit according to manufacturer’s instructions.
Coimmunoprecipitation and Western Blotting.
Nuclear extracts or in vitro transcribed/translated proteins were suspended in ice-cold TGH buffer [50 mM HEPES (pH 7.4), 150 mM NaCl, 10% glycerol, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 1 mM PMSF, 10 mM NaF, 1 mM Na3VO4, 5 µl/ml protease inhibitor cocktail, and 1 µg/ml bovine serum albumin]. Nuclear extracts were incubated overnight at 4°C with either a rabbit anti-AhR (Enzo Life Sciences, Farmingdale, NY) or goat anti-KLF6 (Santa Cruz Biotechnology, Dallas, TX) antibody, followed by precipitation with protein A/G PLUS-agarose beads (Santa Cruz Biotechnology). The immunoprecipitated proteins were separated on 10% SDS-PAGE gels, transferred, and fixed with 5% milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T). Filters were probed with goat anti-KLF6 or rabbit anti-AhR antibodies overnight followed by appropriate Cy3–labeled secondary antibodies (GE Healthcare Lifesciences, Pittsburgh, PA) at room temperature for 1 hour. Images were captured using the Typhoon Trio Variable Mode Imager (GE Healthcare).
Electrophoretic Mobility Shift Assay.
Either 10 µg of nuclear extract or 5 µl of proteins from in vitro transcription/translation were incubated in HEDG buffer (25 mM HEPES, 1 mM EDTA, 1 mM DTT, and 10% glycerol, pH 7.4) with 100 ng of poly(deoxyinosinic-deoxycytidylic)acid (PolydI·dC) and 100 mM KCl. Then 300 ng of 32P-NC-XRE or 32P-NC-XRE mutants M2, M3, M4, and M5 (32P-GTP end-labeled double-stranded DNA probe, 3000 mCi/mmol) were added and incubated at room temperature for an additional 15 minutes (Huang and Elferink, 2012). Samples were electrophoresed on 6% nondenaturing polyacrylamide gels in TAE (40 mM Tris, 20 mM acetic acid, 1 mM EDTA, pH8) buffer, exposed to phosphoscreens (GE Healthcare Life Sciences), and imaged on Typhoon Trio.
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation (ChIP) assays for the whole liver were performed as described previously (Huang and Elferink, 2012). In brief, following 2-hour vehicle or TCDD treatment, liver tissues from C57BL/6 female mice were isolated, finely minced, and crosslinked with 1% formaldehyde (Fisher Scientific, Houston, TX) in phosphate-buffered saline at room temperature for 10 minutes. Samples were homogenized in a Dounce homogenizer and centrifuged at 3200g for 5 minutes at 4°C. The supernatant was discarded, the pellet was resuspended in 4 ml of cell lysis buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 8.0), 85 mM KCl, 0.5% NP40, and 4 µl of protease inhibitor cocktail], and homogenized with four Dounce strokes. Samples were incubated on ice for 20 minutes, centrifuged at 3200g for 5 minutes at 4°C, and the pellet was processed using ChIP-IT Express Enzymatic Kit (Active Motif, Carlsbad, CA) according to the manufacturer’s protocol. The nuclei were sheared using Enzymatic Shearing Cocktail (Active Motif) at 37°C for 15 minutes, and the sheared chromatin was incubated overnight with protein G magnetic beads with appropriate antibodies, AhR (Abcam, Cambridge, MA), KLF6 (Santa Cruz Biotechnology), histone H3 as positive control (Abcam), and IgG as negative control (Cell Signaling Technology, Danvers, MA). Immunoprecipitated and input DNA were PCR amplified using primers specific to the NC-XRE in the PAI-1 promoter or Cyp1a1 (Table 1). PCR product was separated on 5% polyacrylamide gels, stained with SYBR Green I (Life Technologies, Grand Island, NY), and imaged on Typhoon Trio, and band intensity was measured using ImageQuant (GE Healthcare Life Sciences).
Statistical Analysis.
All statistical data are represented as the mean + S.E. Differences between the groups were considered statistically significant if the P value was <0.05. Data were analyzed by applying the t test using Sigma Plot software (Systat Software, San Jose, CA).
Results
KLF6 Interacts with the AhR In Vivo and Is Part of the NC-XRE Complex.
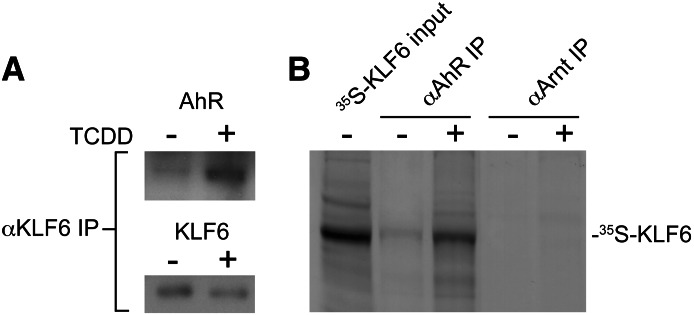
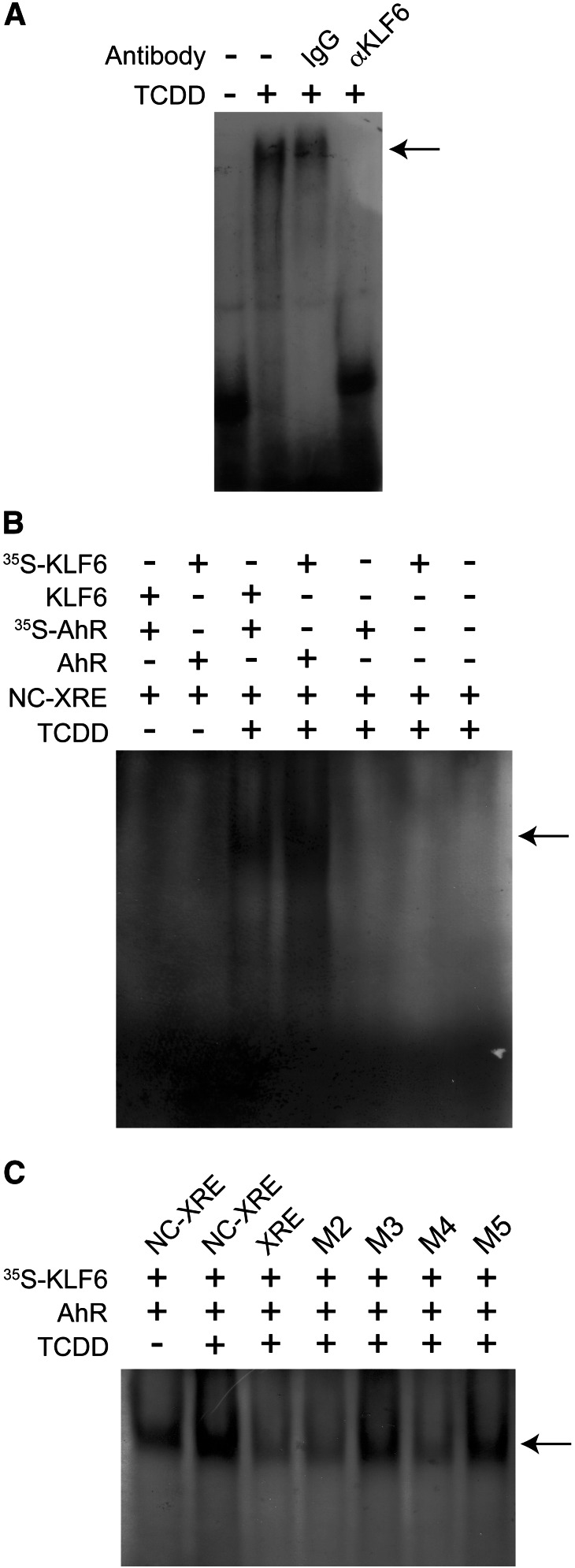
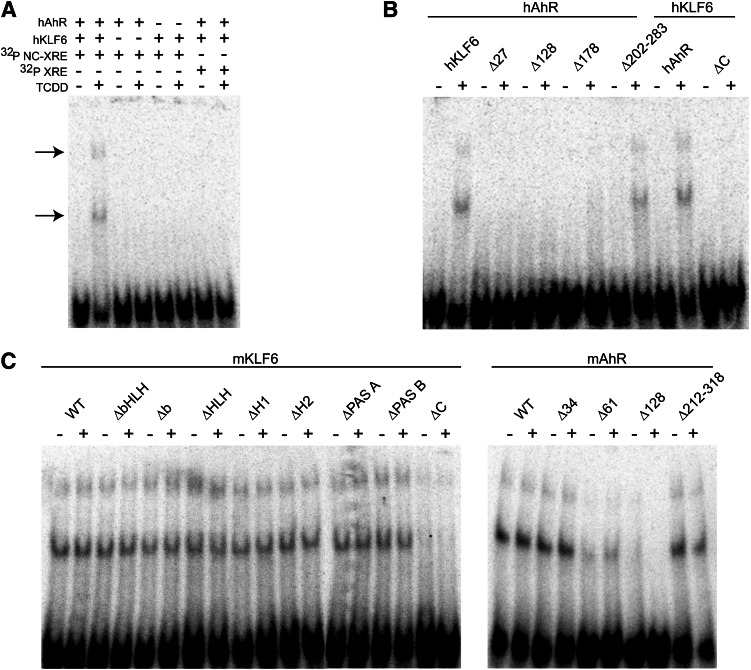
To establish the existence of an interaction between AhR and KLF6 in vivo, nuclear extracts were prepared from mice pretreated with vehicle or 20 µg/kg TCDD, 2 hours prior to sacrifice, to activate the AhR. KLF6 and its associated proteins were immunoprecipitated, and subsequent immunoblotting confirmed that the AhR was associated with KLF6 in a TCDD-dependent manner (Fig. 1A). Additionally, complementary reciprocal coimmunoprecipitation experiments using unlabeled AhR or Arnt protein and 35S-labeled KLF6 expressed by in vitro transcription and translation detected an AhR-KLF6 interaction (Fig. 1B). The evidence shows that radiolabeled KLF6 is immunoprecipitated in a TCDD-dependent manner using the AhR antibody, but not with the Arnt protein antibody. These results indicate that the AhR and KLF6 interact with each other independently of the Arnt protein, consistent with our recent ChIP finding showing that AhR binding to the PAI-1 NC-XRE does not require the Arnt protein (Huang and Elferink, 2012). To assess DNA binding, EMSA was performed using mouse liver nuclear extracts incubated with an IgG control antibody or an anti-KLF6 antibody before exposure to radioactively labeled NC-XRE (Fig. 2A). The KLF6 antibody specifically abolishes formation of the protein-DNA complex, suggesting that KLF6 is indeed a component of the NC-XRE binding complex. Disruption of the protein-DNA complex rather than formation of a super-shift product suggests that the KLF6 antibody recognizes an epitope at or near the region in KLF6 important for DNA binding. Therefore, to obtain direct evidence of AhR and KLF6 binding to the NC-XRE, we applied an EMSA strategy using 35S-radiolabeled AhR or KLF6 in conjunction with an unlabeled “cold” NC-XRE oligonucleotide (Fig. 2B). The results identified a gel shift product (Fig. 2B, lanes 3 and 4) that is TCDD-inducible and absolutely dependent on both AhR and KLF6. Moreover, using 35S-radiolabeled KLF6 to monitor direct AhR-KLF6 DNA binding to mutant NC-XRE sites (Fig. 2C) revealed the same sequence preferences previously characterized using crude mouse liver nuclear extracts (Huang and Elferink, 2012), confirming NC-XRE binding by an AhR-KLF6 heterodimer.
Fig. 1.
Protein-protein interaction between AhR and KLF6. C57BL/6 mice were gavaged with either vehicle (-) or 20 µg/kg TCDD (+) 2 hours before sacrifice and preparation of liver nuclear extracts as described in the Materials and Methods section. (A) Nuclear proteins were immunoprecipitated with an antibody against KLF6 and immunoblotted to detect the presence of KLF6 and the AhR. (B) Human recombinant 35S-labeled KLF6 and unlabeled AhR and Arnt were synthesized using the in vitro transcription and translation system. KLF6 was incubated with either the AhR or Arnt protein in the absence (-) or presence (+) of 6 nM TCDD for 2 hours prior to coimmunoprecipitation with antibodies directed against the AhR or Arnt protein. Coimmunoprecipitated KLF6 was detected by autoradiography after fractionation by SDS-PAGE. IP, immunoprecipitation.
Fig. 2.
The AhR and KLF6 bind to the NC-XRE. (A) Nuclear extracts were prepared from vehicle- and TCDD-treated mouse livers as described in Fig. 1, and were preincubated with either IgG (negative control) or αKLF6 antibodies prior to EMSA with an NC-XRE probe. The arrow denotes the TCDD-inducible NC-XRE complex. (B) EMSA was performed using TnT in vitro–generated recombinant unlabeled or 35S-radiolabeled AhR or KLF6 and unlabeled NC-XRE. Protein-DNA complexes were identified by autoradiography to detect the 35S-radiolabeled proteins. (C) EMSA was performed using TnT-expressed 35S-KLF6 and unlabeled AhR with the NC-XRE, XRE, and several site-specific NC-XRE mutants described previously (Huang and Elferink, 2012). Protein-DNA complexes were visualized by autoradiography.
The AhR and KLF6 Bind Together at the PAI-1 Promoter In Vivo in Response to TCDD.
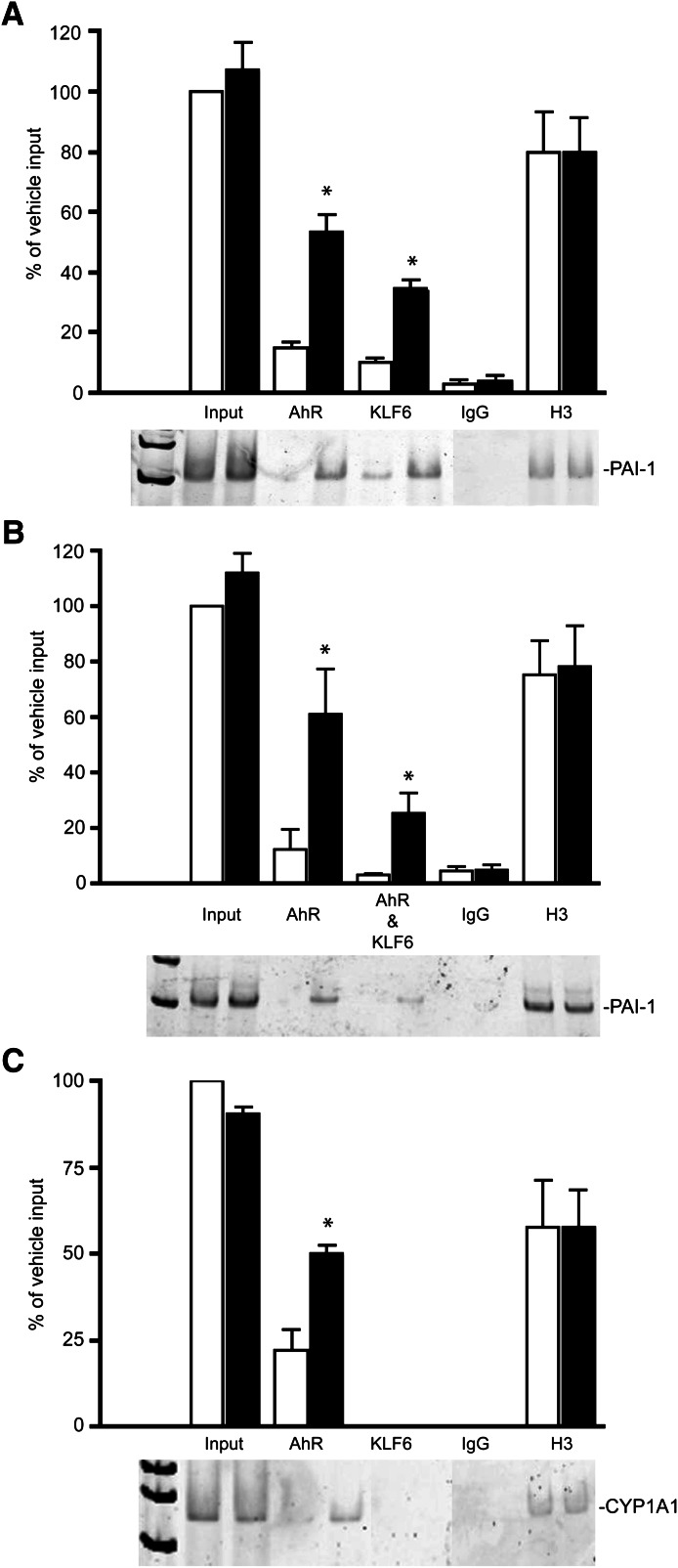
To verify the AhR and KLF6 association with the NC-XRE in vivo, ChIP assays were performed on the PAI-1 promoter using whole liver from vehicle- or TCDD-pretreated mice using antibodies against the AhR, KLF6, H3 (positive control), and IgG (negative control) as previously described (Huang and Elferink, 2012). The results revealed that both the murine AhR and KLF6 bind to the PAI-1 promoter region encompassing the NC-XRE in a TCDD-dependent manner (Fig. 3A). Furthermore, sequential chromatin immunoprecipitation of AhR and KLF6 confirms that both proteins are simultaneously bound to the PAI-1 promoter (Fig. 3B). Moreover, the TCDD-inducible KLF6 interaction appears to be specific for the NC-XRE because analysis of the Cyp1a1 promoter containing several XREs only detected the AhR interaction (Fig. 3C). Quantitation of the ChIP data relied on densitometric analyses of the PCR products from at least three independent experiments. Collectively, the evidence indicates that AhR binding to the NC-XRE both in vitro and in vivo requires KLF6, but is independent of the Arnt protein.
Fig. 3.
TCDD-dependent AhR and KLF6 binding to the PAI-1 promoter. ChIP assays were performed on mouse liver tissue isolated from vehicle- and 20 µg/kg TCDD–treated (2 hours) animals. Antibodies against the AhR, KLF6, H3 (positive control), and IgG (negative control) were used to immunoprecipitate the target proteins. PCR using primers directed against the PAI-1 or Cyp1a1 promoter was used to amplify the isolated DNA. PCR products were separated on 5% polyacrylamide gel and visualized with SYBR green. Quantitation of the PCR products generated from 3–4 independent experiments used ImageQuant (GE Healthcare Lifesciences) software, and is presented as a percentage of input DNA (average ± S.E.) isolated from vehicle-treated (open bars) and TCDD-treated (solid bars) mice. (A) PCR using primers specific to PAI-1 promoter following ChIP with individual antibodies. (B) PCR on DNA isolated in a sequential reChIP experiment using antibodies directed against the AhR, followed by KLF6. (C) PCR on the Cyp1a1 promoter following ChIP with the individual antibodies. *P < 0.05.
The C Terminus of the AhR and the N Terminus of KLF6 Are Critical for Protein-Protein Interactions In Vitro.
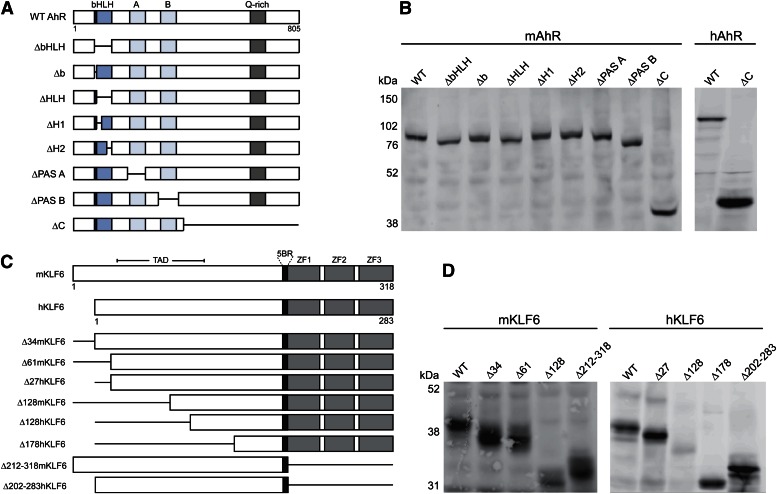
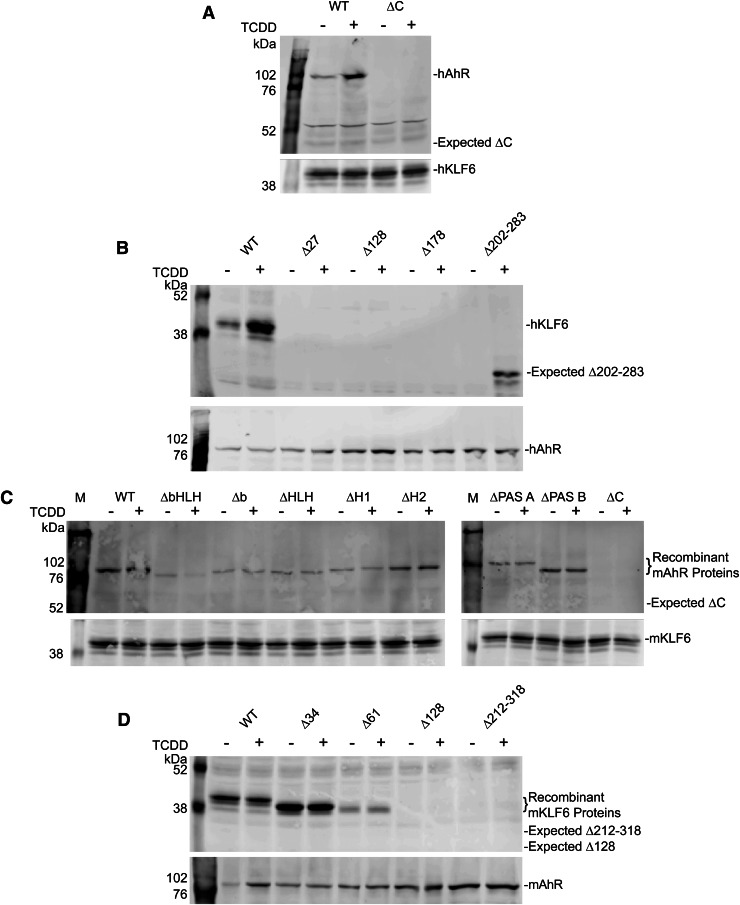
To study the AhR and KLF6 protein-protein interactions, we generated a series of AhR and KLF6 deletion constructs targeting known functional domains (Fig. 4). Constructs encoding the murine AhR harbored targeted deletions spanning the basic helix-loop-helix domain, the basic (DNA binding) domain, the PAS A and B domains, and the C terminus (containing a glutamine-rich transactivation domain). We also generated human AhR constructs encoding the full-length receptor and a C-terminal deletion (Fig. 4, A and B). We similarly generated a series of mouse KLF6 deletion constructs lacking the N-terminal 34 amino acids (Δ34), 61 amino acids (Δ61), and 128 amino acids (Δ128). It is noteworthy that the mouse KLF6 protein contains an additional 34 N-terminal residues not present in the human protein, hence the Δ34 and Δ61 murine proteins can be viewed as structurally equivalent to the human full-length and Δ27 KLF6 proteins, respectively. In addition, KLF6 constructs lacking the DNA binding domain (Δ212–318 for murine KLF6, Δ202–283 for human KLF6) were generated (Fig. 4C). All AhR and KLF6 constructs were expressed using an in vitro coupled transcription and translation system (Fig. 4, B and D, respectively).
Fig. 4.
Expression of recombinant murine and human AhR and KLF6 constructs. Murine (mAhR, mKLF6) and human (hAhR, hKLF6) full-length and deletion constructs depicted in panels (A) (AhR) and (C) (KLF6) were generated using the in vitro TnT system. Recombinant protein expression was monitored by SDS-PAGE and Western blotting against the AhR (panel B) and KLF6 (panel D) proteins. WT, wild type.
Analysis of the protein-protein interactions between the AhR and KLF6 involved reciprocal coimmunoprecipitation using in vitro synthesized proteins with antibodies against the AhR and KLF6. Coimmunoprecipitation of the recombinant human AhR with an antibody against KLF6 revealed that KLF6 binding to the AhR requires the receptor’s C-terminal region (Fig. 5A). Reciprocal coimmunoprecipitation studies with the recombinant human proteins containing an intact N terminus demonstrated that the N-terminal 27 amino acid region of KLF6 conferred AhR binding, since removal of this region completely abolished the AhR-KLF6 interaction (Fig. 5B). Using a series of recombinant murine AhR proteins lacking the well characterized functional domains spanning the bHLH and PAS regions, coimmunoprecipitation studies confirmed that these domains are not required for the receptor interaction with KLF6 (Fig. 5C). Akin to the human AhR, loss of the murine receptor’s C terminus abolished the KLF6 interaction. However, in contrast to the finding with the human proteins, under these cell-free in vitro conditions, the murine proteins do not retain the TCDD dependency. Although the basis for this species difference is unknown, it appears to be a property restricted to in vitro experimental conditions, since the TCDD dependency is clearly evident following coimmunoprecipitation of the AhR-KLF6 complex using mouse liver nuclear extracts (Fig. 1A). Evaluation of the murine KLF6 deletion constructs reveals that the region between amino acids 34 and 61 in the mouse protein is important for AhR binding, and further removal of N-terminal residues completely abolishes the protein-protein interaction (Fig. 5D). It is noteworthy that the N-terminal 27 amino acid sequence in the human KLF6 is 100% identical to residues 34–61 in mouse KLF6, indicating that this is an evolutionarily conserved AhR interaction domain. It should be noted that, in contrast to the human KLF6 protein (Δ202–283), the murine KLF6 Δ212–318 C-terminal deletion failed to coimmunoprecipitate. Conceivably, this reflects a genuine species difference or points to an experimental artifact possibly related to a conformational defect in the recombinant mouse protein.
Fig. 5.
Characterization of the AhR-KLF6 protein-protein interaction. Proteins generated as described in Fig. 4 were used in coimmunoprecipitation studies using lysates pretreated with vehicle (-) or 20 nM TCDD (+) for 20 minutes at 37°C. (A) Human KLF6 was immunoprecipitated and the lysates subjected to SDS-PAGE and Western blotting to detect coimmunoprecipitated hAhR. AhR and KLF6 protein were detected using the appropriate Cy3-labeled secondary antibodies (GE Healthcare), and images were captured using a Typhoon Trio Variable Mode Imager (GE Healthcare). (B) Reciprocal coimmunoprecipitation studies targeted the hAhR and assayed for coprecipitation of the hKLF6 proteins. (C) Murine KLF6 was used to coimmunoprecipitate the full-length mAhR as well as several deletion constructs. (D) Reciprocal coimmunoprecipitation used mAhR to pull down the mKLF6 deletion constructs.
The C Terminus of the AhR and the N Terminus of KLF6 Are Critical for Protein-NC-XRE Interactions In Vitro.
We next proceeded to explore the DNA binding properties of the AhR-KLF6 complex, including the deletion mutants, using the traditional EMSA (Fig. 6). Unlabeled recombinant human AhR and KLF6 proteins were transformed in vitro with vehicle or TCDD and subjected to EMSA using a 32P-radiolabeled NC-XRE or XRE (Fig. 6A). The EMSA detected two TCDD-inducible protein-DNA complexes with the NC-XRE probe that required the presence of both the human AhR and KLF6. No DNA binding was observed with the XRE, indicating that the complex is specific to the NC-XRE. As observed previously, this EMSA signature is specific to the NC-XRE-protein complex and distinct from the canonical XRE complex (Huang and Elferink, 2012). These data corroborate the findings in Fig. 2B and confirm that both proteins are necessary to form the protein-DNA complex. DNA binding by the AhR-KLF6 complex relied on the same protein domains shown to be necessary for the protein-protein interaction (Fig. 6B). This suggests that DNA binding is dependent upon AhR-KLF6 heterodimerization.
Fig. 6.
Characterization of the AhR-KLF6 protein-DNA complex using the wild-type and mutant recombinant proteins. The recombinant human and murine AhR and KLF6 full-length and deletion constructs described in Fig. 4 were treated with vehicle (-) or 20 nM TCDD (+) for 20 minutes at 37°C and used in EMSA with 32P-radiolabeled NC-XRE (A–C) and XRE (A) probes. (A) Protein-DNA complexes and free DNA were visualized by autoradiography. EMSA under these in vitro conditions generated two TCDD-inducible protein-DNA complexes with the NC-XRE probe only (arrows). (B) EMSA using the human AhR and KLF6 proteins. (C) EMSA using the murine AhR and KLF6 proteins.
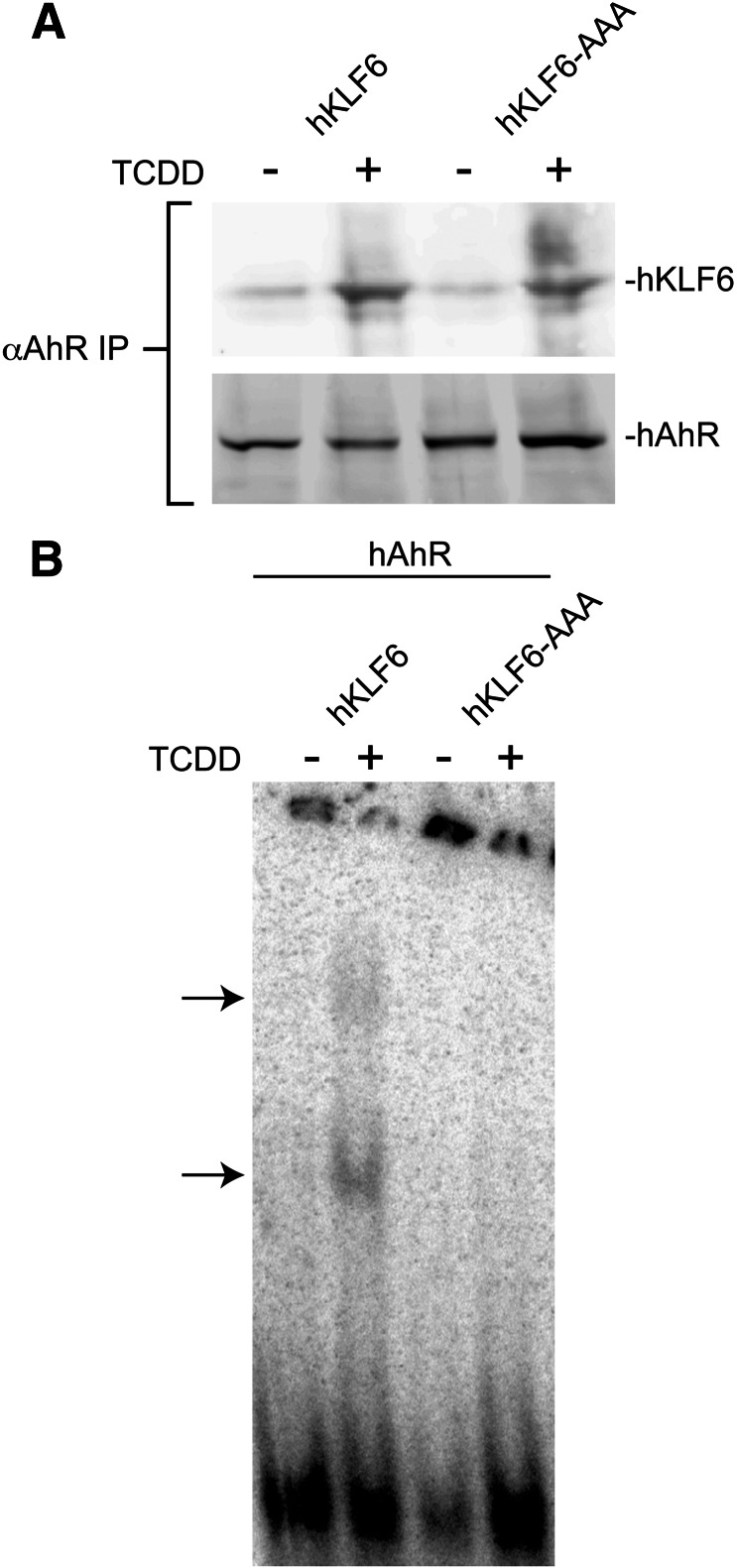
The DNA binding properties exhibited by the murine proteins demonstrated that removal of the AhR’s C terminus disrupts DNA binding, consistent with the finding obtained using the human proteins. Targeted deletions eliminating other functional domains—including the basic region, and the HLH and PAS domains—had no noticeable impact on DNA binding (Fig. 6C). The finding that removal of the basic region (Δb and ΔbHLH) required for XRE binding did not disrupt EMSA complex formation was unexpected, and suggests that AhR binding to the NC-XRE is fundamentally different from binding to the XRE. Analysis of the KLF6 deletion constructs indicated that the peptide sequence encoded by residues 34–61 required for the AhR-KLF6 protein interactions is also necessary for DNA binding (Fig. 6C). In contrast, the KLF6 C-terminal region (Δ212–318 and Δ202–283 in the murine and human proteins, respectively) encompassing the Cys2/His2 Kruppel-like zinc fingers is expendable for DNA binding (Fig. 6). Since the conserved zinc finger region defines the DNA binding domain in the KLF family (Turner and Crossley, 1999), the observation that removal of this domain did not disrupt DNA binding was intriguing. It is noteworthy that several KLF proteins contain a 5′ basic region juxtaposing the first zinc finger. Although the 5′ basic region is a putative nuclear localization signal (Shields and Yang, 1997), this was not borne out in recent studies of KLF6 (Rodriguez et al., 2010). However, biophysical studies have shown that electrostatic interactions between positively charged basic residues and the negatively charged phosphate DNA backbone are the primary mediators of nucleic acid binding (Elrod-Erickson et al., 1996). Hence, we explored the possibility that the 5′ basic region conferred DNA binding on KLF6 by replacing three consecutive arginine residues from 196–198 in hKLF6 with alanines to generate hKLF6-AAA (Fig. 7). Coimmunoprecipitation studies using in vitro–expressed hAhR and hKLF6 or hKLF6-AAA revealed that the alanine substitutions did not disrupt the TCDD-inducible AhR-KLF6 interaction, suggesting that the point mutations did not markedly affect protein structure (Fig. 7A). However, the EMSA revealed that the hKLF6-AAA protein was incapable of forming a protein-DNA complex with the AhR (Fig. 7B), suggesting that one or more of the arginine residues in the 5′ basic region are required for DNA binding. Collectively, these data indicate that the AhR-KLF6 protein-protein interaction, and indirectly the protein-DNA interaction, is dependent on amino acids located in the N terminus of KLF6 and C terminus of the AhR, respectively. In addition, KLF6 DNA binding is dependent on key arginine residues located outside the zinc finger domain.
Fig. 7.
Arginines 196–198 in the KLF6 5′ basic region are critical for NC-XRE binding. (A) Wild-type hKLF6 and a mutant protein with three consecutive arginine residues (Arg196–198) replaced by alanines (hKLF6-AAA) were expressed along with the hAhR in vitro using the TnT expression system. (A) Recombinant proteins were treated with vehicle (-) or 20 nM TCDD (+) for 20 minutes at 37°C and subjected to coimmunoprecipitation using the anti-AhR antibody. Coprecipitation of hKLF6 was evaluated by SDS-PAGE and Western blotting using an anti-KLF6 antibody. (B) EMSA was performed using the recombinant proteins and 32P-radiolabeled NC-XRE, and complex formation visualized by autoradiography (arrows). IP, immunoprecipitation.
Discussion
Previous studies support AhR binding and activity being more complex than initially thought. Apart from classic AhR-Arnt heterodimer binding to the XRE, the AhR also interacts with Sp1, pRb, and the RelA and RelB subunits of NF-κB (Ge and Elferink, 1998; Tian et al., 1999; Wang et al., 1999; Puga et al., 2000; Vogel et al., 2007). Examination of the AhR-RelB interaction identified a novel 8-nucleotide TCDD-responsive DNA binding site coined the RelBAhRE in the interleukin-8 promoter, distinct from the XRE (Vogel et al., 2007). We recently characterized a novel AhR binding site in the PAI-1 gene promoter referred to as the nonconsensus XRE (Huang and Elferink, 2012). As with the RelBAhRE, AhR binding to the NC-XRE occurs in the absence of the Arnt protein. However, the RelBAhRE bears minimal resemblance to the NC-XRE sequence found in the PAI-1 promoter. This led to the supposition that receptor binding to the NC-XRE relies on a new AhR binding partner distinct from RelB. The findings presented here build on that premise by identifying KLF6 as a hitherto unknown AhR binding partner involved in complex formation at the NC-XRE.
The rationale for examining KLF6 as a component of the NC-XRE–bound complex in the PAI-1 promoter was founded on several complementary observations: 1) sequence homology between the NC-XRE and the consensus binding site for KLF proteins, 2) prior evidence of a functional relationship between the AhR and a KLF family member in the brain and gut (Imataka et al., 1992; Zhang et al., 1998), 3) expression of KLF6 in the liver (Ratziu et al., 1998; Narla et al., 2007), 4) a role of KLF6 in cell proliferation reminiscent of the AhR’s role in cell proliferation, and 5) increased nuclear localization of KLF6 resulting in enhanced PAI-1 expression (Gehrau et al., 2010). KLF6 binds to the GC-rich region or CACCC elements of the DNA sequence in the target gene promoter (Philipsen and Suske, 1999; Bieker, 2001) and regulates expression of a large number of genes involved in differentiation, proliferation, and apoptosis. KLF6 has been shown to induce p21 (WAF1/CIP1), transforming growth factor β1, IGF1R, hINOS, and E-cadherin, as well as repress DlK1 (through recruitment of HDAC3) and MMP9 (by forming a repression complex with Sp2) (Ratziu et al., 1998; Kojima et al., 2000; Narla et al., 2001, 2007; Warke et al., 2003; Kremer-Tal et al., 2004; Li et al., 2005; Das et al., 2006; DiFeo et al., 2006). KLF6 directly activates p21 (WAF1/CIP1) in a p53-independent manner, leading to inhibition of cell cycle progression in the prostate and decreased hepatocyte proliferation (Narla et al., 2001, 2007), suggesting that it functions as a tumor suppressor. Furthermore, KLF6 loss of heterozygosity, mutations, and downregulation is associated with the development of hepatocellular, prostate, colorectal, primary non–small-cell lung carcinoma, and glioblastoma (Ito et al., 2004; Bureau et al., 2009).
As the tumor suppressor characteristics of KLF6 were being identified, it became evident that the subcellular localization varies between noncancerous and cancerous tissues, with KLF6 remaining confined to the cytoplasm in carcinomas (Gehrau et al., 2010). Similar to all members of the KLF superfamily, KLF6 is characterized by three highly conserved Cys2/His2 zinc fingers and an N-terminal activation domain (Kaczynski et al., 2003; Suske et al., 2005). Previous work demonstrated that the zinc fingers of KLF1 through KLF4 are necessary for nuclear localization as well as DNA binding, and likewise the first zinc finger also plays an important role in KLF6 nuclear localization (Rodriguez et al., 2010). However, despite the established role for the zinc finger domain in DNA binding, our observations indicated that loss of this domain in KLF6 did not abolish NC-XRE binding (Fig. 6). Accordingly, the evidence showed that the 5′ basic region adjacent to the first zinc finger conferred binding to the NC-XRE (Fig. 7). This implies that the DNA binding domain in KLF6 extends beyond the zinc finger domain. Also, since the NC-XRE and consensus KLF recognition sequences differ from each other somewhat, it is conceivable that the 5′ basic region imparts latitude to KLF6 DNA binding not inherent in the zinc fingers.
In this study, we demonstrated a direct interaction between the AhR and KLF6, involving the AhR C terminus and KLF6 N terminus. Moreover, the presence of both proteins is essential for complex formation in vitro at the NC-XRE (Figs. 2B and 6A). In vivo, both proteins bind the PAI-1 promoter in a region containing the NC-XRE, and sequential chromatin immunoprecipitation data suggest a direct and simultaneous presence of both the AhR and KLF6 at this site (Fig. 3). Although the in vivo coimmunoprecipitation and ChIP experiments in mice established a TCDD dependency, this property could not be recapitulated via in vitro studies using the murine proteins. In contrast, the human proteins did retain agonist responsiveness in vitro. Despite this disparity, the in vitro studies using the mouse and human proteins provided several common observations. Notably, NC-XRE binding does not depend on the KLF6 zinc finger domain, deletion of the receptor’s C terminus abolishes both KLF6 binding and formation of the DNA complex, and the AhR-KLF6 interaction depends on a completely conserved 27 amino acid region in the KLF6 protein. This suggests that the observed discrepancies reflect minor species differences or artifacts attributable to the in vitro experimental conditions. For instance, it is formally possible that the species difference can be attributed to as yet unidentified cofactors involved in the AhR-KLF6 interactions at NC-XRE. Several studies have suggested a differential response in ligand binding, and to dioxins in particular, between human and murine AhR when using a yeast reporter system and in vitro–generated recombinant proteins, respectively (Kawanishi et al., 2003; Ramadoss and Perdew, 2004). Other work also suggests that human AhR (hAhR) is more responsive to indoxyl-3-sulfate, an endogenous ligand, than mouse AhR (mAhR) in hepatoma cell lines (Schroeder et al., 2010). Using XRE-driven reporter assays, recent studies have also demonstrated differences in transcription activation by human versus mouse AhR (Flaveny et al., 2008).
The functional consequences of the Arnt-independent AhR-KLF6 interaction with the NC-XRE require further exploration. Recent evidence using an Arnt protein conditional knockout mouse model provided compelling evidence that Arnt is required for AhR-mediated dioxin-induced hepatotoxicity including phase I gene expression, hepatomegaly, and hepatic inflammation and steatosis (Nukaya et al., 2010). Studies using mice expressing a hypomorphic Arnt allele also provide evidence that XRE-driven gene expression contributes to vascular development in the absence of exogenous agonists (Walisser et al., 2004). Although possible, these data do not confirm that the AhR-Arnt heterodimer is exclusively responsible for dioxin toxicity, including tumor promotion and teratogenicity. Therefore, the AhR-KLF6 complex may contribute to dioxin-mediated carcinogenesis, particularly given the documented role for both proteins in cell cycle control (Ge and Elferink, 1998; Kolluri et al., 1999; Puga et al., 2000; Narla et al., 2001, 2007; Benzeno et al., 2004; Huang and Elferink, 2005; Mitchell et al., 2006; Bureau et al., 2009). Alternatively, the AhR-KLF6 complex may serve regulatory functions unrelated to dioxin toxicity but be critically important in normal physiologic events. Hence, genome-wide assessments of the transcriptional targets controlled by this new complex offer a means to address this issue and represent promising research opportunities.
Acknowledgments
The authors thank Dr. Premnath Shetty for intellectual contributions and valuable discussion at the outset of these studies.
Abbreviations
- AhR
aryl hydrocarbon receptor
- Arnt
aryl hydrocarbon receptor nuclear translocator
- bHLH
basic helix-loop-helix
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- hAhR
human aryl hydrocarbon receptor
- hKLF6
human kruppel like factor 6
- KLF
Kruppel-like factor
- mAhR
mouse aryl hydrocarbon receptor
- mKLF6
mouse kruppel like factor 6
- NC-XRE
nonconsensus xenobiotic response element
- PAI-1
plasminogen activator inhibitor 1
- PAS
Per/Arnt/Sim
- PCR
polymerase chain reaction
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- TnT
transcription and translation protein expression system
- XRE
xenobiotic response element
Authorship Contributions
Participated in research design: Wilson, Joshi, Elferink.
Conducted experiments: Wilson, Joshi.
Contributed new reagents or analytic tools: Wilson, Joshi.
Performed data analysis: Wilson, Joshi, Elferink.
Wrote or contributed to the writing of the manuscript: Wilson, Joshi, Elferink.
Footnotes
This work was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grants R01ES007800 and P30ES006676] (to C.J.E.) and [Grant F30ES016490] (to S.R.W.).
References
- Benzeno S, Narla G, Allina J, Cheng GZ, Reeves HL, Banck MS, Odin JA, Diehl JA, Germain D, Friedman SL. (2004) Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res 64:3885–3891 [DOI] [PubMed] [Google Scholar]
- Bieker JJ. (2001) Krüppel-like factors: three fingers in many pies. J Biol Chem 276:34355–34358 [DOI] [PubMed] [Google Scholar]
- Bureau C, Hanoun N, Torrisani J, Vinel J-P, Buscail L, Cordelier P. (2009) Expression and function of kruppel like-factors (KLF) in carcinogenesis. Curr Genomics 10:353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver LA, Bradfield CA. (1997) Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. J Biol Chem 272:11452–11456 [DOI] [PubMed] [Google Scholar]
- Carver LA, LaPres JJ, Jain S, Dunham EE, Bradfield CA. (1998) Characterization of the Ah receptor-associated protein, ARA9. J Biol Chem 273:33580–33587 [DOI] [PubMed] [Google Scholar]
- Das A, Fernandez-Zapico ME, Cao S, Yao J, Fiorucci S, Hebbel RP, Urrutia R, Shah VH. (2006) Disruption of an SP2/KLF6 repression complex by SHP is required for farnesoid X receptor-induced endothelial cell migration. J Biol Chem 281:39105–39113 [DOI] [PubMed] [Google Scholar]
- DiFeo A, Narla G, Hirshfeld J, Camacho-Vanegas O, Narla J, Rose SL, Kalir T, Yao S, Levine A, Birrer MJ, et al. (2006) Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res 12:3730–3739 [DOI] [PubMed] [Google Scholar]
- Elferink CJ, Whitlock JP., Jr (1994) Dioxin-dependent, DNA sequence-specific binding of a multiprotein complex containing the Ah receptor. Receptor 4:157–173 [PubMed] [Google Scholar]
- Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. (1996) Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure 4:1171–1180 [DOI] [PubMed] [Google Scholar]
- Flaveny C, Reen RK, Kusnadi A, Perdew GH. (2008) The mouse and human Ah receptor differ in recognition of LXXLL motifs. Arch Biochem Biophys 471:215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge NL, Elferink CJ. (1998) A direct interaction between the aryl hydrocarbon receptor and retinoblastoma protein. Linking dioxin signaling to the cell cycle. J Biol Chem 273:22708–22713 [DOI] [PubMed] [Google Scholar]
- Gehrau RC, D’Astolfo DS, Dumur CI, Bocco JL, Koritschoner NP. (2010) Nuclear expression of KLF6 tumor suppressor factor is highly associated with overexpression of ERBB2 oncoprotein in ductal breast carcinomas. PLoS ONE 5:e8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson O. (1995) The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol 35:307–340 [DOI] [PubMed] [Google Scholar]
- Hoffman EC, Reyes H, Chu F-F, Sander F, Conley LH, Brooks BA, Hankinson O. (1991) Cloning of a factor required for activity of the Ah (dioxin) receptor. Science 252:954–958 [DOI] [PubMed] [Google Scholar]
- Huang G, Elferink CJ. (2012) A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol Pharmacol 81:338–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Elferink CJ. (2005) Multiple mechanisms are involved in Ah receptor-mediated cell cycle arrest. Mol Pharmacol 67:88–96 [DOI] [PubMed] [Google Scholar]
- Ikuta T, Eguchi H, Tachibana T, Yoneda Y, Kawajiri K. (1998) Nuclear localization and export signals of the human aryl hydrocarbon receptor. J Biol Chem 273:2895–2904 [DOI] [PubMed] [Google Scholar]
- Imataka H, Sogawa K, Yasumoto K-i, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. (1992) Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J 11:3663–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Wakao H, Masuho Y, Muramatsu MA, Tojo H, Nanbu-Wakao R. (1999) cDNA cloning and expression analysis of mouse zf9, a Krüppel-like transcription factor gene that is induced by adipogenic hormonal stimulation in 3T3-L1 cells. Biochim Biophys Acta 1447:199–207 [DOI] [PubMed] [Google Scholar]
- Ito G, Uchiyama M, Kondo M, Mori S, Usami N, Maeda O, Kawabe T, Hasegawa Y, Shimokata K, Sekido Y. (2004) Krüppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res 64:3838–3843 [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. (2003) Sp1- and Krüppel-like transcription factors. Genome Biol 4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanishi M, Sakamoto M, Ito A, Kishi K, Yagi T. (2003) Construction of reporter yeasts for mouse aryl hydrocarbon receptor ligand activity. Mutat Res 540:99–105 [DOI] [PubMed] [Google Scholar]
- Kojima S, Hayashi S, Shimokado K, Suzuki Y, Shimada J, Crippa MP, Friedman SL. (2000) Transcriptional activation of urokinase by the Krüppel-like factor Zf9/COPEB activates latent TGF-β1 in vascular endothelial cells. Blood 95:1309–1316 [PubMed] [Google Scholar]
- Kolluri SK, Weiss C, Koff A, Göttlicher M. (1999) p27(Kip1) induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev 13:1742–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer-Tal S, Reeves HL, Narla G, Thung SN, Schwartz M, Difeo A, Katz A, Bruix J, Bioulac-Sage P, Martignetti JA, et al. (2004) Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology 40:1047–1052 [DOI] [PubMed] [Google Scholar]
- Lees MJ, Whitelaw ML. (1999) Multiple roles of ligand in transforming the dioxin receptor to an active basic helix-loop-helix/PAS transcription factor complex with the nuclear protein Arnt. Mol Cell Biol 19:5811–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Yea S, Li S, Chen Z, Narla G, Banck M, Laborda J, Tan S, Friedman JM, Friedman SL, et al. (2005) Krüppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J Biol Chem 280:26941–26952 [DOI] [PubMed] [Google Scholar]
- Ma Q, Whitlock JP., Jr (1997) A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem 272:8878–8884 [PubMed] [Google Scholar]
- Mitchell KA, Elferink CJ. (2009) Timing is everything: consequences of transient and sustained AhR activity. Biochem Pharmacol 77:947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KA, Lockhart CA, Huang G, Elferink CJ. (2006) Sustained aryl hydrocarbon receptor activity attenuates liver regeneration. Mol Pharmacol 70:163–170 [DOI] [PubMed] [Google Scholar]
- Mitchell KA, Wilson SR, Elferink CJ. (2010) The activated aryl hydrocarbon receptor synergizes mitogen-induced murine liver hyperplasia. Toxicology 276:103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, et al. (2001) KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294:2563–2566 [DOI] [PubMed] [Google Scholar]
- Narla G, Kremer-Tal S, Matsumoto N, Zhao X, Yao S, Kelley K, Tarocchi M, Friedman SL. (2007) In vivo regulation of p21 by the Kruppel-like factor 6 tumor-suppressor gene in mouse liver and human hepatocellular carcinoma. Oncogene 26:4428–4434 [DOI] [PubMed] [Google Scholar]
- Nukaya M, Walisser JA, Moran SM, Kennedy GD, Bradfield CA. (2010) Aryl hydrocarbon receptor nuclear translocator in hepatocytes is required for aryl hydrocarbon receptor-mediated adaptive and toxic responses in liver. Toxicol Sci 118:554–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdew GH. (1988) Association of the Ah receptor with the 90-kDa heat shock protein. J Biol Chem 263:13802–13805 [PubMed] [Google Scholar]
- Philipsen S, Suske G. (1999) A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res 27:2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst MR, Reisz-Porszasz S, Agbunag RV, Ong MS, Hankinson O. (1993) Role of the aryl hydrocarbon receptor nuclear translocator protein in aryl hydrocarbon (dioxin) receptor action. Mol Pharmacol 44:511–518 [PubMed] [Google Scholar]
- Prokipcak RD, Okey AB. (1991) Downregulation of the Ah receptor in mouse hepatoma cells treated in culture with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Can J Physiol Pharmacol 69:1204–1210 [DOI] [PubMed] [Google Scholar]
- Puga A, Barnes SJ, Dalton TP, Chang C-y, Knudsen ES, Maier MA. (2000) Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J Biol Chem 275:2943–2950 [DOI] [PubMed] [Google Scholar]
- Ramadoss P, Perdew GH. (2004) Use of 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin as a probe to determine the relative ligand affinity of human versus mouse aryl hydrocarbon receptor in cultured cells. Mol Pharmacol 66:129–136 [DOI] [PubMed] [Google Scholar]
- Ratziu V, Lalazar A, Wong L, Dang Q, Collins C, Shaulian E, Jensen S, Friedman SL. (1998) Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci USA 95:9500–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez E, Aburjania N, Priedigkeit NM, DiFeo A, Martignetti JA. (2010) Nucleo-cytoplasmic localization domains regulate Krüppel-like factor 6 (KLF6) protein stability and tumor suppressor function. PLoS ONE 5:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini RP, Myrand S, Elferink C, Reiners JJ., Jr (2001) Regulation of Cyp1a1 induction by dioxin as a function of cell cycle phase. J Pharmacol Exp Ther 299:718–728 [PubMed] [Google Scholar]
- Schroeder JC, Dinatale BC, Murray IA, Flaveny CA, Liu Q, Laurenzana EM, Lin JM, Strom SC, Omiecinski CJ, Amin S, et al. (2010) The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry 49:393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields JM, Yang VW. (1997) Two potent nuclear localization signals in the gut-enriched Krüppel-like factor define a subfamily of closely related Krüppel proteins. J Biol Chem 272:18504–18507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G, Bruford E, Philipsen S. (2005) Mammalian SP/KLF transcription factors: bring in the family. Genomics 85:551–556 [DOI] [PubMed] [Google Scholar]
- Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. (1999) Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J Biol Chem 274:510–515 [DOI] [PubMed] [Google Scholar]
- Turner J, Crossley M. (1999) Mammalian Krüppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci 24:236–240 [DOI] [PubMed] [Google Scholar]
- Vogel CFA, Sciullo E, Li W, Wong P, Lazennec G, Matsumura F. (2007) RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol Endocrinol 21:2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walisser JA, Bunger MK, Glover E, Bradfield CA. (2004) Gestational exposure of Ahr and Arnt hypomorphs to dioxin rescues vascular development. Proc Natl Acad Sci USA 101:16677–16682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang W, Safe S. (1999) Regulation of constitutive gene expression through interactions of Sp1 protein with the nuclear aryl hydrocarbon receptor complex. Biochemistry 38:11490–11500 [DOI] [PubMed] [Google Scholar]
- Warke VG, Nambiar MP, Krishnan S, Tenbrock K, Geller DA, Koritschoner NP, Atkins JL, Farber DL, Tsokos GC. (2003) Transcriptional activation of the human inducible nitric-oxide synthase promoter by Kruppel-like factor 6. J Biol Chem 278:14812–14819 [DOI] [PubMed] [Google Scholar]
- Zhang W, Shields JM, Sogawa K, Fujii-Kuriyama Y, Yang VW. (1998) The gut-enriched Krüppel-like factor suppresses the activity of the CYP1A1 promoter in an Sp1-dependent fashion. J Biol Chem 273:17917–17925 [DOI] [PMC free article] [PubMed] [Google Scholar]