Abstract
Wnt signaling regulates many aspects of development by increasing the signaling activity of β-catenin. Axin is a negative regulator of the Wnt signaling pathway, and it is responsible for the formation of the β-catenin degradation complex. Genetic studies with Drosophila suggest that Axin promotes cytoplasmic localization of β-catenin independent of Axin's known role of enhancing degradation of β-catenin. Here, we show that Axin is a nuclear-cytoplasmic shuttling protein. Nuclear export of Axin depends on the chromosome maintenance region 1 nuclear receptor; treatment with the chromosome maintenance region 1 inhibitor leptomycin B induces nuclear accumulation of ectopically expressed or endogenous Axin. Functional nuclear localization and nuclear export signals have been mapped within Axin. Significantly, overexpression of an Axin fragment shifts coexpressed stabilized β-catenin to the cytoplasm, and this effect requires shuttling of Axin between the cytoplasm and the nucleus. Our results suggest that Axin functions as a molecular chaperone for β-catenin and that nuclear-cytoplasmic shuttling of Axin regulates the nuclear-cytoplasmic distribution of β-catenin.
The Wnt family glycoproteins play pivotal roles in diverse developmental processes, mainly through control of the signaling activity of β-catenin (1, 2), and aberrant Wnt signaling can lead to cancer (3). Genetic experiments in Drosophila and biochemical studies in Xenopus and mammalian cells have established a framework for the Wnt signaling pathway (4). In cells that are receiving a Wnt signal, cytoplasmic β-catenin is bound to a multiprotein β-catenin destruction complex that contains several proteins including Axin, adenomatous polyposis coli (APC), and glycogen synthase kinase 3 (GSK3). In this complex, β-catenin is phosphorylated at a cluster of Ser and Thr residues near its N terminus by GSK3. Phosphorylated β-catenin is then recognized by βTrCP, a component of the SCFβTrCP ubiquitin-protein ligase complex, and is degraded by the ubiquitin-proteasome pathway. Wnt signaling disassembles the β-catenin destruction complex, stabilizing β-catenin. Accumulated β-catenin then enters the nucleus, binds to lymphoid enhancer factor/T cell factor family transcription factors, and activates the expression of β-catenin target genes.
Although most studies have focused on the mechanism by which Wnt induces the stabilization of β-catenin, it is not clear whether Wnt signaling also affects β-catenin in other important ways. In certain developmental events, Wnt protein appears to promote nuclear localization of β-catenin without changing the level of β-catenin (5–7). Furthermore, a recent study suggests that Wnts can potentiate β-catenin signaling without affecting the stability of β-catenin (8). The Wnt signal can still be transmitted in Zw3 (Drosophila GSK3)-null Drosophila embryos harboring a hypomorphic Armadillo (Drosophila β-catenin) allele, apparently by promoting nuclear localization of β-catenin (8).
Axin is a key negative regulator of the Wnt signaling pathway. Axin facilitates the formation of the β-catenin degradation complex and increases phosphorylation and degradation of cytoplasmic β-catenin. Genetic inactivation of Axin has been reported in various types of human tumors (9–12). It has been suggested that the endogenous level of Axin is very low and that the output of Wnt signaling is highly sensitive to the turnover rate of Axin (13). Indeed, it has been shown that Wnt signaling destabilizes Axin (8, 14, 15).
Recent experiments indicate that Axin also promotes the cytoplasmic localization of β-catenin (16). Armadillo is found in both nuclear and cytoplasmic locations in Drosophila Zw3-null embryos, whereas it is only nuclear in Axin-null embryos (16). Because GSK3 and Axin are genetically equivalent in promoting β-catenin phosphorylation and degradation, Axin must have an additional role in facilitating the cytoplasmic localization of β-catenin. How Axin achieves this function is currently unclear, although it has been proposed that Axin functions as a cytoplasmic anchor for β-catenin (16). Drosophila APC1 and APC2 have been implicated in promoting the cytoplasmic localization of β-catenin (17, 18), and it has been suggested that APC increases the cytoplasmic localization of β-catenin by exporting β-catenin from the nucleus (19–21). Whether Axin promotes the cytoplasmic localization of β-catenin through a mechanism analogous to that of APC is unclear.
In this study we have demonstrated that Axin constantly shuttles between the nucleus and the cytoplasm. Active nuclear import and export signals of Axin have been identified, and nuclear export of Axin depends on the chromosome maintenance region 1 (CRM1) nuclear receptor. Our data also suggest that nuclear import/export of Axin is required for the Axin-induced cytoplasmic shift of β-catenin. Thus, although Axin might serve as a cytoplasmic anchor for β-catenin, our data suggest that Axin also functions as a molecular chaperone to promote nuclear export of β-catenin.
Materials and Methods
Cell Culture and Transfection. 293 cells, COS7 cells, and 293T cells were grown at 37°C in DMEM supplemented with 10% FBS (HyClone). 293 cells were transfected with FuGENE 6 (Roche). Drosophila S2 cells were grown at room temperature in Schneider's Drosophila medium supplemented with 10% FBS (GIBCO). S2 cells were transfected with CellFECTIN (Invitrogen). Cells were analyzed 36 h after transfection. In some experiments, cells were treated with 5 ng/ml leptomycin B (LMB; Sigma) for the indicated time. A sequence encoding a GFP-Axin fusion protein was cloned into pBabe-puro retroviral vector (22). Nonreplicative virus was produced by transfection of 293T cells with GFP-Axin-pBabe-puro and helper plasmids encoding vesicular stomatitis virus glycoprotein envelope and Moloney leukemia virus Gag-Pol and was used to infect COS7 cells. COS7 cells were then selected with puromycin to generate cells stably expressing GFP-Axin.
Plasmids. Mouse form I Axin cDNA was kindly provided by Frank Costantini (Columbia University, New York) (23). The first conserved methionine between mouse and human Axin, methionine 129 in the original published sequence (23), was used as the start codon in our Axin constructs. However, to be consistent with published literature, our nomenclature for Axin is based on the original published sequence (23). GFP-tagged Axin and hemagglutinin (HA)-tagged β-catenin S37A were cloned into mammalian expression vectors under control of the cytomegalovirus promoter. The DIX domain of Axin (residues 873–956) was deleted from Axin to form AxinΔDIX. Site-directed mutagenesis of Axin was performed by using two-step PCR. All constructs were verified by DNA sequencing. A modified nuclear localization signal (NLS) of SV40 large T antigen (DPKKNRK) was fused to the C terminus of the AxinΔDIX NLS mutant to form AxinΔDIX NLSSV40. HA-tagged Drosophila Axin (DAxin) was cloned into pPac 5.1 under the control of the Drosophila actin promoter. pDM128, RevΔ3NI, and RevΔ3NI-RexNES were kindly provide by Thomas Hope (University of Illinois at Chicago, Chicago) (24, 25). A fragment of Axin (residues 433–558) was fused to the C terminus of RevΔ3NI to form RevΔ3NI-AxinNES. The last two hydrophobic residues of Axin nuclear export signal (NES) (Val-545 and Met-547) were changed to Ala in RevΔ3NI-AxinNESm.
Rev Complementation Assay. 293 cells were transfected with pDM128, plasmids encoding various Rev mutants, and internal control cytomegalovirus-Renilla. Assays were done in triplicate in six-well plates. Cell were lysed 36 h after transfection. Chloramphenicol acetyltransferase (CAT) activities were determined by using the CAT Enzyme Assay System (Promega) and normalized for Renilla activities.
Subcellular Fractionation and Immunoblotting. 293 cells were scraped into cold PBS and collected by centrifugation at 1,000 × g for 5 min. The cells were washed once with PBS and centrifuged, and the cell pellet was resuspended into hypotonic buffer (10 mM Hepes-KOH/10 mM NaCl/1 mM KH2PO4/5 mM NaHCO3/1 mM CaCal2/0.5 mM MgCl2). After a 5-min incubation on ice, cells were homogenized with 20 strokes in a Dounce homogenizer, and efficient cell lysis was verified by trypan blue staining. Cells were spun at 1,000 × g for 5 min. The supernatants from this low-speed centrifugation were spun at 100,000 × g for 90 min at 4°C to generate the cytosolic fraction. The pellet of the low-speed centrifugation was washed twice and resuspended with nuclear isolation buffer (10 mM Tris, pH 7.5/300 mM sucrose/0.1% Nonidet P-40). The pellet was homogenized 20 times by using a Dounce homogenizer and spun down. The pellet was then resuspended with nuclear isolation buffer with 1% Triton X-100 to generate the nuclear fraction. Proteins were separated by SDS/PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat dry milk for 1 h at room temperature and incubated with appropriate primary antibodies overnight at 4°C. Membranes were then washed and incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h. Membranes were washed extensively and developed with an ECL kit (Amersham Pharmacia). Antibodies used include rabbit anti-Axin polyclonal antibodies (Zymed), mouse anti-α-tubulin monoclonal antibodies (Sigma), and goat anti-lamin B polyclonal antibodies (Santa Cruz Biotechnology).
Immunofluorescence Microscopy. Thirty-six hours posttransfection, cells were fixed with 2% paraformaldehyde/PBS for 20 min and then permeabilized with 0.1% Triton X-100/PBS for 5 min. Samples were preblocked with 1% goat serum/PBS, incubated with anti-HA (HA.11) monoclonal antibodies (Covance, Berkeley, CA), and washed in PBS. Cells were then stained with Texas red-conjugated goat anti-mouse Ig antibodies (Jackson ImmunoResearch), and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were mounted with Vectorshield (Vector) and examined by fluorescence microscopy (Leica). The distribution of β-catenin in GFP-positive cells was scored as cytoplasmic (C) if >75% of β-catenin appeared to localize in the cytoplasm, and nuclear (N) if >75% of β-catenin appeared to localize in the nucleus. The rest of cells were scored as cytoplasmic and nuclear. Cells only transfected with GFP fusion constructs were fixed, permeabilized, stained with DAPI, and directly mounted. S2 cells transiently expressing HA-tagged DAxin were fixed, immunostained with anti-HA antibodies and FITC-conjugated goat anti-mouse secondary antibodies (Sigma), and examined by laser scanning microscopy (Zeiss).
Results
Nuclear-Cytoplasmic Shuttling of Axin. Because DAxin promotes the cytoplasmic localization of β-catenin, we considered the possibility that Axin might shuttle between the nucleus and the cytoplasm to accelerate nuclear export of β-catenin. The CRM1-dependent nuclear export pathway is a major pathway that cells use for transporting molecules out of the nucleus (26, 27).
To examine potential nuclear-cytoplasmic shuttling of Axin, 293 cells were transiently transfected with a plasmid encoding GFP-tagged full-length mouse Axin (form I) and were treated with LMB, a highly specific inhibitor of CRM1 (28). Overexpressed Axin appeared to localize in irregular cytoplasmic dots, and the pattern of localization was not affected by LMB (Fig. 1A). We have found that, unlike full-length Axin, an Axin mutant lacking the DIX domain (AxinΔDIX) was diffusely localized in the cytoplasm (Fig. 1 A). Because the DIX domain is responsible for the self-association of Axin (29, 30), the punctate appearance of full-length Axin in our assays might reflect protein aggregation, possibly resulting from protein overexpression. We hypothesized that nuclear-cytoplasmic shuttling might be inhibited when full-length Axin forms large protein aggregates upon overexpression. Therefore, we examined whether LMB could affect the subcellular localization of AxinΔDIX. Interestingly, LMB caused a dramatic redistribution of AxinΔDIX from the cytoplasm to the nucleus (Fig. 1 A), suggesting that the AxinΔDIX mutant shuttles between the cytoplasm and the nucleus via a CRM1-dependent pathway.
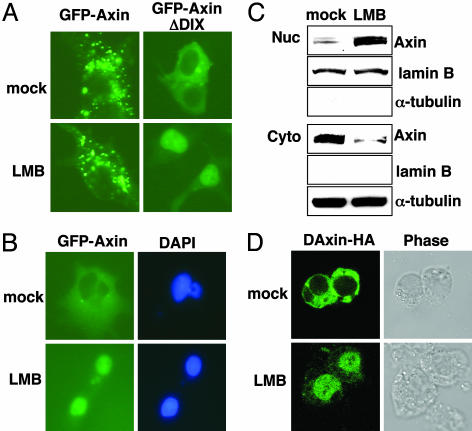
Fig. 1.
Nuclear-cytoplasmic shuttling of Axin. (A) 293 cells were transiently transfected with plasmids encoding GFP-Axin or GFP-AxinΔDIX. Thirty-six hours after transfection, cells were mock-treated or treated with 5 ng/ml LMB for 3 h and examined by fluorescence microscopy. (B) COS cells were infected with a retrovirus encoding GFP-Axin. Cells were mock-treated or treated with LMB for 3 h and examined by fluorescence microscopy. Nuclei were counterstained by 4′,6-diamidino-2-phenylindole. (C) 293 cells were mock-treated or treated with LMB for 3 h and subjected to subcellular fractionation. The levels of endogenous Axin in the nuclear and cytoplasmic fractions were determined by immunoblotting with anti-Axin antibodies. The relative purity of the nuclear and cytoplasmic fractions was confirmed by sequential probing for the nuclear marker lamin B and the cytoplasmic marker α-tubulin. (D) Drosophila S2 cells were transiently transfected with a plasmid expressing DAxin-HA. Cells were mock-treated or treated with LMB for 3 h, immunostained with anti-HA monoclonal antibodies and FITC-conjugated secondary antibodies, and examined by confocal microscopy. Cell shapes were examined by phase contrast microscopy.
To test whether the full-length Axin also shuttles between the cytoplasm and the nucleus when its level is low, a gene encoding GFP-Axin was introduced into COS7 cells through retroviral infection using a Moloney leukemia virus vector. In these cells, most of the GFP-Axin was located diffusely in the cytoplasm but accumulated in the nucleus after treatment with LMB (Fig. 1B). To test whether endogenous Axin also shuttles between nucleus and cytoplasm, 293 cells were treated with LMB, and a subcellular fractionation assay and an immunoblotting analysis were performed to measure nuclear and cytoplasmic Axin. LMB treatment resulted in a significant shift of Axin from the cytoplasmic fraction to the nuclear fraction (Fig. 1C), indicating that endogenous Axin also undergoes nucleocytoplasmic shuttling.
Because DAxin promotes the cytoplasmic localization of Armadillo (16), we tested whether DAxin also shuttles between the nucleus and the cytoplasm in Drosophila cells. Ectopically expressed HA-tagged DAxin had a mostly diffuse cytoplasmic location in Drosophila S2 cells but accumulated in the nucleus upon LMB treatment (Fig. 1D). These data suggest that DAxin, like its mammalian counterpart, undergoes nuclear-cytoplasmic shuttling. In this setting, transiently expressed DAxin shuttles between nucleus and cytoplasm, even when it contains the DIX domain, possibly reflecting a low level of expression.
Identification of NES and NLS of Axin. CRM1 functions as a receptor for leucine-rich NESs. This type of NES is characterized by the presence of four critically spaced, large-group hydrophobic residues, most often represented by leucine (31). Visual examination of the protein sequence of mouse Axin identified a potential NES (residues 538–547) in the GSK3 binding domain that resembles the known leucine-rich NES sequences (Fig. 2B).
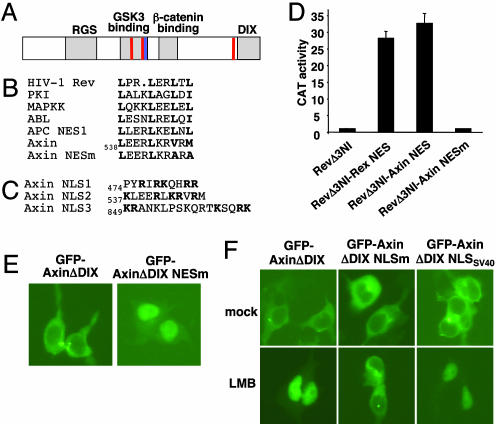
Fig. 2.
Characterization of NES and NLS sequences of Axin. (A) Schematic representation of the structure of Axin. Functional domains of Axin are labeled. The potential NES sequence is indicated by a blue bar, and proposed NLS sequences are indicated by red bars. (B) Alignment of selected NES sequences from known nuclear-cytoplasmic shuttling proteins with the Axin NES. The critical hydrophobic residues are shown in boldface. The last two conserved hydrophobic residues (Val-545 and Met-547) were substituted with Ala in the mutant form of Axin NES (Axin NESm). (C) Sequences of three potential NLSs of Axin. The basic residues in NLS1, NLS2, and bipartite NLS3 are shown in boldface. (D) Complementation of the nuclear export activity of Rev by the Axin NES. Rev mutants were coexpressed with pDM128 indicator construct and cytomegalovirus-Renilla control in 293 cells. Nuclear export and expression of unspliced CAT mRNA require the nuclear export activity of Rev. Controls include RevΔ3NI (NES-mutated Rev) and RevΔ3NI fused with the NES of human T-lymphotropic virus I Rex. The experiments were performed in triplicate, and CAT activities were normalized to Renilla luciferase activities. (E) Nuclear accumulation of NES-mutated AxinΔDIX. 293 cells were transiently transfected with plasmids expressing the wild-type or NES-mutated GFP-AxinΔDIX and examined by fluorescence microscopy. (F) Identification of NLS sequences of Axin. 293 cells were transiently transfected with plasmids encoding various forms of GFP-AxinΔDIX, mock-treated or treated with LMB for 3 h, and examined by fluorescence microscopy. Mutating three stretches of positively charged residues of GFP-AxinΔDIX abolished nuclear import of the protein upon LMB treatment. Nuclear import of this mutant protein was restored by adding to its C terminus a modified NLS from SV40 large T antigen.
To examine the functionality of this NES, we tested whether it can substitute for the NES of HIV-1 Rev protein. HIV-1 Rev protein binds to unspliced HIV-1 RNA through its RNA binding domain and exports it from nucleus via the CRM1-dependent nuclear export pathway. Rev mutants were coexpressed with a pDM128 reporter in 293 cells through transient transfection. The transcript produced by pDM128 harbors a single intron containing the CAT coding sequence, which is removed when the RNA is spliced. There is a HIV-1 Rev response element located between intact 5′ and 3′ splice sites in the transcript; hence, expression of the unspliced CAT mRNA requires the nuclear export function of Rev (24). As a positive control, fusion of the NES sequence of human T-lymphotropic virus I Rex protein (25) to a NES-mutated Rev (RevΔ3NI) significantly increased Rev-dependent expression of CAT (Fig. 2D). A fragment of Axin (residues 443–558) containing the potential NES sequence was fused to the C terminus of RevΔ3NI to form RevΔ3NI-AxinNES, and this chimeric protein significantly increased the expression of CAT (Fig. 2D), suggesting that the Axin fragment containing the potential NES sequence can restore the nuclear export function of Rev. Furthermore, mutation of the last two hydrophobic residues of Axin NES (V545A, M547A) within this Axin fragment abolished the stimulatory effect of RevΔ3NI-AxinNES on the expression of CAT (Fig. 2D). These data suggest that the proposed NES sequence of Axin is a functional NES.
Next, we tested whether this NES is functional within the context of the Axin sequence. GFP-tagged AxinΔDIX is cytoplasmic, and a double mutant of AxinΔDIX with the NES sequence mutated (V545A and M547A) became accumulated in the nucleus (Fig. 2E). These data strongly support the functionality of the NES sequence as a nuclear exporting signal and suggest that active nuclear export is required for maintaining the cytoplasmic localization of Axin.
Nuclear accumulation of Axin upon inhibition of nuclear export implies the existence of an NLS in Axin protein. It is more difficult to recognize an NLS than the leucine-rich type of NES. There is a bipartite NLS consensus (residues 849–865) in the C-terminal region of mouse Axin (form I) (ref. 23 and Fig. 2C). However, mutating this potential NLS did not affect the subcellular localization of AxinΔDIX in the presence of LMB (data not shown). Thus, to identify a potential NLS in Axin protein, various fragments of Axin protein were fused to the C terminus of a GFP-GST chimeric protein, and the subcellular locations of these chimeric proteins were examined in 293 cells with or without LMB treatment (data not shown). In this analysis, GST-GFP-Axin 443–558 was normally cytoplasmic and became nuclear when cells were treated with LMB (data not shown), suggesting the existence of both an NLS and an NES in this fragment. Indeed, this Axin fragment contains the previously identified NES, and mutation of the NES led to nuclear accumulation of this chimeric protein (data not shown) as it did for GFP-AxinΔDIX (Fig. 2E).
There are two stretches of basic amino acid residues (residues 474–483 and 537–547) in this Axin fragment (Fig. 2C); mutation of either one reduced nuclear localization, and mutation of both completely abolished the nuclear localization of GST-GFP-Axin 443–558 upon LMB treatment (data not shown). Mutation of both potential NLS sequences (R476Q, R478Q, K479Q, R482Q, R483Q, K543Q, and R544Q) in the context of AxinΔDIX significantly decreased its nuclear localization upon LMB treatment (data not shown). Mutating the C-terminal bipartite NLS consensus sequence (R859Q, K861Q, R864Q, and K865Q) in the context of AxinΔDIX with mutated central NLS sequences largely abolished nuclear localization of Axin after LMB treatment (Fig. 2F). Therefore, there seem to be at least three NLS sequences that contribute to the nuclear localization of Axin protein. The second NLS (residues 537–547) appears to be the strongest among the three NLS sequences; mutation of it has the most significant effect on AxinΔDIX localization upon LMB treatment (data not shown). Of interest, the second NLS and the NES of Axin overlap.
The AxinΔDIX mutant with all three putative NLS sequences mutated was denoted AxinΔDIX NLSm. To restore nuclear import of Axin, a modified NLS from SV40 large T antigen was fused to the C terminus of AxinΔDIX NLSm to form AxinΔDIX NLSSV40. Like AxinΔDIX, this mutant was mainly cytoplasmic and did not form aggregates, and it became entirely nuclear upon LMB treatment (Fig. 2E).
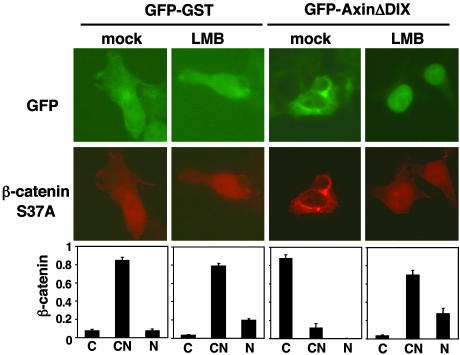
Requirement of Nuclear-Cytoplasmic Shuttling of Axin for Axin-Induced Cytoplasmic Shifting of β-Catenin. Experiments with Drosophila have suggested a role for Axin in promoting cytoplasmic localization of β-catenin (8, 16). Because Axin shuttles between the nucleus and the cytoplasm and physically interacts with β-catenin, we tested whether nuclear-cytoplasmic shuttling of Axin could increase the proportion of β-catenin in the cytoplasm. GFP-GST or GFP-AxinΔDIX was coexpressed with HA-tagged β-catenin S37A in 293 cells, and cells were either mock-treated or treated with LMB. The subcellular location of β-catenin was determined by indirect immunofluorescence staining with anti-HA antibodies and scored only in GFP-positive cells (Fig. 3). In GST-GFP-positive cells, HA-tagged β-catenin was found in both the cytoplasm and the nucleus. In contrast, coexpression of GFP-AxinΔDIX resulted in a significant shift of β-catenin from nucleus to cytoplasm (Fig. 3). After LMB treatment, AxinΔDIX accumulated in the nucleus, reversing the AxinΔDIX-dependent shift of β-catenin to the cytoplasm (Fig. 3). These results suggest that the steady-state nuclear-cytoplasmic distribution of AxinΔDIX determines the location of β-catenin.
Fig. 3.
AxinΔDIX induces cytoplasmic shift of β-catenin in a CRM1-dependent manner. GFP-GST or GFP-AxinΔDIX was coexpressed with HA-tagged β-catenin S37A in 293 cells. Cells were treated with vehicle or LMB for 16 h. β-Catenin was detected by indirect immunofluorescence staining with anti-HA monoclonal antibodies and Texas red-conjugated secondary antibodies. The distribution of β-catenin in GFP-positive cells was scored as cytoplasmic (C), cytoplasmic and nuclear (CN), and nuclear (N) and graphed. The results are the average of three independent experiments; at least 300 GFP-positive cells were scored for each sample in every experiment.
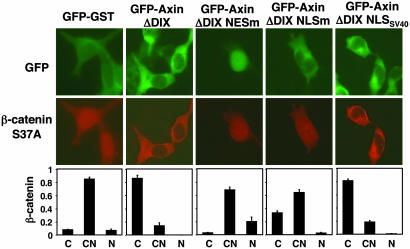
In the above experiment, AxinΔDIX may have increased the cytoplasmic localization of β-catenin by either actively shuttling β-catenin out of the nucleus or simply serving as a cytoplasmic anchor for β-catenin as proposed in ref. 16. If Axin serves as a molecular chaperone for β-catenin, nuclear import of AxinΔDIX would be necessary for the Axin-dependent cytoplasmic shift of β-catenin. By contrast, nuclear import of AxinΔDIX would be dispensable if Axin functions only as a cytoplasmic anchor. To distinguish these two possibilities, the effects of AxinΔDIX and AxinΔDIX NLSm on β-catenin localization were compared. As predicted by the chaperone model, although AxinΔDIX NLSm, like AxinΔDIX, was located mainly in the cytoplasm, its ability to induce a cytoplasmic shift of β-catenin was markedly attenuated (Fig. 4). Importantly, adding the NLS from the SV40 large T antigen to the C terminus of AxinΔDIX NLSm restored the cytoplasmic localization of β-catenin. This finding excludes the possibility that the inability of AxinΔDIX NLSm to relocate β-catenin to the cytoplasm is due to its lower binding affinity for β-catenin; this is consistent with the fact that all three NLS sequences are outside the β-catenin binding domain of Axin (residues 563–626). Together, these results suggest that nuclear-cytoplasmic shuttling of Axin is required for the Axin-dependent cytoplasmic shift of β-catenin.
Fig. 4.
Nuclear-cytoplasmic shuttling is required for AxinΔDIX-induced cytoplasmic shift of β-catenin. GFP-GST or various forms of GFP-AxinΔDIX were coexpressed with HA-tagged β-catenin S37A in 293 cells. The distributions of HA-tagged β-catenin in GFP-positive cells were scored as in Fig. 3. The expression levels of various mutants of GFP-AxinΔDIX were similar as determined by immunoblotting with anti-GFP antibodies (data not shown).
It should be noted that, compared with GST-GFP, AxinΔDIX NLSm increased β-catenin cytoplasmic localization to a certain extent (Fig. 4). This effect may mean that the ectopically expressed Axin also functions as a cytoplasmic anchor to trap β-catenin in the cytoplasm. Therefore, the Axin-dependent cytoplasmic shift of β-catenin most likely results from both nuclear export and cytoplasmic anchorage of β-catenin in this experimental setting.
Discussion
β-Catenin exerts its signaling activity only in the nucleus (32, 33). Therefore, it is important to understand how the distribution of β-catenin between the cytoplasm and the nucleus is regulated. Experiments with Drosophila have strongly supported a role for Axin in controlling the cytoplasmic-nuclear partitioning of β-catenin, independent of its ability to promote β-catenin degradation. It has been suggested that Axin promotes the cytoplasmic localization of β-catenin by means of cytoplasmic anchorage and that Wnt increases nuclear localization of stabilized β-catenin through destruction of Axin (8, 16). Although Axin has generally been considered to be a cytoplasmic protein, we have demonstrated using LMB that localization of Axin is a highly dynamic process; both Drosophila and mouse Axin constantly shuttle between the nucleus and the cytoplasm. We propose that Axin functions as a molecular chaperone to accelerate the export of β-catenin from the nucleus.
The mechanisms by which β-catenin enters and exits the nucleus have not been well established. β-Catenin contains no recognizable NLS and NES. The nuclear localization of β-catenin can be enhanced when coexpressed with the lymphoid enhancer factor/T cell factor family of transcription factors, so it has been proposed that β-catenin is imported into the nucleus by a piggyback mechanism (34–36). However, it has also been shown that β-catenin can enter the nucleus independent of transport factors such as the importins and RanGTPase (37, 38). Indeed, the armadillo repeats of β-catenin are structurally related to importin-β HEAT repeats that bind to the nuclear pore complex (39, 40). In Drosophila, an Armadillo mutant that has armadillo repeats 3–6 deleted and presumably does not bind to most of its binding partners is localized in the nucleus (41), suggesting that nuclear import of β-catenin is constitutive and that the localization of β-catenin is determined mostly by nuclear export or cytoplasmic retention.
β-Catenin can be exported from the nucleus through different mechanisms. It has been suggested that APC shuttles between the nucleus and the cytoplasm in a CRM1-dependent manner and that APC exports β-catenin out of the nucleus as a molecular chaperone (19–21, 42). However, under certain conditions, β-catenin can exit from the nucleus through a CRM1-independent mechanism. By injecting recombinant β-catenin protein into Xenopus oocytes and mammalian cells, or by using semipermeabilized SW480 colon cancer cells, it has been suggested that the nuclear export of β-catenin is mediated by the armadillo repeats of β-catenin and that it is independent of CRM1 and RanGTPase (38, 43, 44). However, it should be noted that the levels of β-catenin in these experiments were much higher than in normal situations, and it is unclear whether nuclear export of β-catenin is mediated mainly by the CRM1-independent pathway in normal cells.
On the contrary, we have found that nuclear export of endogenous β-catenin in normal cells occurs at least in part through a CRM1-dependent pathway. Consistent with previous findings (20), we have found that treatment of COS7 cells with LMB causes nuclear accumulation of endogenous β-catenin (data not shown). Moreover, stabilized or overexpressed β-catenin accumulates in the nucleus (45, 46), suggesting that CRM1-independent export of β-catenin is a relatively slow and inefficient process.
We have demonstrated that nuclear import of Axin is required for Axin-induced cytoplasmic shift of β-catenin (Fig. 4). Nuclear-cytoplasmic shuttling of Axin significantly increases the proportion of β-catenin in the cytoplasm. Therefore, CRM1-independent nuclear export of β-catenin does not seem to play a dominant role at least in our experimental setting. We propose that CRM1-independent nuclear export of β-catenin is relatively slow and that Axin accelerates nuclear export of β-catenin. In normal cells, Axin shuttles between nucleus and cytoplasm to export the low level of nuclear β-catenin. Once in the cytoplasm, β-catenin can be degraded by the β-catenin destruction complex or can form a complex with the membrane E-cadherin. This mechanism could minimize β-catenin signaling activity in cells that do not receive a Wnt signal or quickly down-regulate β-catenin signaling activity when the Wnt signal is withdrawn. Because Axin and APC interact with each other and both bind to β-catenin, it is conceivable that Axin accelerates nuclear export of β-catenin in a complex with APC. It is possible that Axin- and APC-dependent nuclear export of β-catenin plays a dominant role mainly in normal cells. In tumor cells, β-catenin might be accumulated to a level above the threshold for the regulation of Axin and APC.
Genetic studies in Drosophila have strongly suggested that Axin plays a role in promoting cytoplasmic localization of β-catenin (8, 16), e.g., as a cytoplasmic anchor for β-catenin (16). Consistent with our proposal that Axin functions as a molecular chaperone for β-catenin, we have shown that DAxin shuttles between nucleus and cytoplasm (Fig. 1D), although the potential NLS and NES in DAxin have not yet been identified. Our model does not exclude the possibility that Axin also serves as a cytoplasmic anchor for β-catenin (16). However, it has been shown that the concentration of endogenous Axin is very low (much lower than the concentration of other major components of the Wnt pathway) (13). Therefore, the level of endogenous Axin might not be high enough to trap β-catenin in the cytoplasm. It seems more likely that Axin cycles between nucleus and cytoplasm to move nuclear β-catenin to the cytoplasm and accelerate the nuclear export of β-catenin.
The nuclear-cytoplasmic shuttling of Axin suggests that Axin could also have a nuclear function. Indeed, nuclear Axin has been observed in normal and cancerous intestinal cells (47). Interestingly, Axin has been shown to interact with protein inhibitor of activated signal transducer and activator of transcription and become sumoylated (48), consistent with the fact that the majority of sumoylated proteins are in the nucleus. Whether sumoylation affects the function and subcellular localization of Axin is currently unclear. Additional experiments will be necessary to determine how Axin functions, what interacts with it in the nucleus, and whether the nuclear-cytoplasmic distribution of Axin is regulated.
Acknowledgments
We thank Frank Costantini, Thomas Hope, Richard Mann, and James Woodgett for providing reagents critical to this work and William Pao and Liang Schweizer for reading the manuscript. F.C. is supported by a postdoctoral fellowship from the Susan G. Komen Breast Cancer Foundation.
Abbreviations: APC, adenomatous polyposis coli; LMB, leptomycin B; NES, nuclear export signal; NLS, nuclear localization signal; GSK3, glycogen synthase kinase 3; CRM1, chromosome maintenance region 1; CAT, chloramphenicol acetyltransferase; DAxin, Drosophila Axin; HA, hemagglutinin.
References
- 1.Cadigan, K. M. & Nusse, R. (1997) Genes Dev. 11, 3286-3305. [DOI] [PubMed] [Google Scholar]
- 2.Wodarz, A. & Nusse, R. (1998) Annu. Rev. Cell Dev. Biol. 14, 59-88. [DOI] [PubMed] [Google Scholar]
- 3.Polakis, P. (2000) Genes Dev. 14, 1837-1851. [PubMed] [Google Scholar]
- 4.Peifer, M. & Polakis, P. (2000) Science 287, 1606-1609. [DOI] [PubMed] [Google Scholar]
- 5.Schneider, S., Steinbeisser, H., Warga, R. M. & Hausen, P. (1996) Mech. Dev. 57, 191-198. [DOI] [PubMed] [Google Scholar]
- 6.Novak, A., Hsu, S. C., Leung-Hagesteijn, C., Radeva, G., Papkoff, J., Montesano, R., Roskelley, C., Grosschedl, R. & Dedhar, S. (1998) Proc. Natl. Acad. Sci. USA 95, 4374-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logan, C. Y., Miller, J. R., Ferkowicz, M. J. & McClay, D. R. (1999) Development (Cambridge, U.K.) 126, 345-357. [DOI] [PubMed] [Google Scholar]
- 8.Tolwinski, N. S., Wehrli, M., Rives, A., Erdeniz, N., DiNardo, S. & Wieschaus, E. (2003) Dev. Cell 4, 407-418. [DOI] [PubMed] [Google Scholar]
- 9.Satoh, S., Daigo, Y., Furukawa, Y., Kato, T., Miwa, N., Nishiwaki, T., Kawasoe, T., Ishiguro, H., Fujita, M., Tokino, T., et al. (2000) Nat. Genet. 24, 245-250. [DOI] [PubMed] [Google Scholar]
- 10.Taniguchi, K., Roberts, L. R., Aderca, I. N., Dong, X., Qian, C., Murphy, L. M., Nagorney, D. M., Burgart, L. J., Roche, P. C., Smith, D. I., et al. (2002) Oncogene 21, 4863-4871. [DOI] [PubMed] [Google Scholar]
- 11.Wu, R., Zhai, Y., Fearon, E. R. & Cho, K. R. (2001) Cancer Res. 61, 8247-8255. [PubMed] [Google Scholar]
- 12.Dahmen, R. P., Koch, A., Denkhaus, D., Tonn, J. C., Sorensen, N., Berthold, F., Behrens, J., Birchmeier, W., Wiestler, O. D. & Pietsch, T. (2001) Cancer Res. 61, 7039-7043. [PubMed] [Google Scholar]
- 13.Lee, E., Salic, A., Kruger, R., Heinrich, R. & Kirschner, M. W. (2003) PLoS Biol. 1, E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willert, K., Shibamoto, S. & Nusse, R. (1999) Genes Dev. 13, 1768-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao, J., Wang, J., Liu, B., Pan, W., Farr, G. H., III, Flynn, C., Yuan, H., Takada, S., Kimelman, D., Li, L. & Wu, D. (2001) Mol. Cell 7, 801-809. [DOI] [PubMed] [Google Scholar]
- 16.Tolwinski, N. S. & Wieschaus, E. (2001) Development (Cambridge, U.K.) 128, 2107-2117. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed, Y., Nouri, A. & Wieschaus, E. (2002) Development (Cambridge, U.K.) 129, 1751-1762. [DOI] [PubMed] [Google Scholar]
- 18.Akong, K., Grevengoed, E. E., Price, M. H., McCartney, B. M., Hayden, M. A., DeNofrio, J. C. & Peifer, M. (2002) Dev. Biol. 250, 91-100. [DOI] [PubMed] [Google Scholar]
- 19.Rosin-Arbesfeld, R., Townsley, F. & Bienz, M. (2000) Nature 406, 1009-1012. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, B. R. (2000) Nat. Cell Biol. 2, 653-660. [DOI] [PubMed] [Google Scholar]
- 21.Neufeld, K. L., Nix, D. A., Bogerd, H., Kang, Y., Beckerle, M. C., Cullen, B. R. & White, R. L. (2000) Proc. Natl. Acad. Sci. USA 97, 12085-12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgenstern, J. P. & Land, H. (1990) Nucleic Acids Res. 18, 1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng, L., Fagotto, F., Zhang, T., Hsu, W., Vasicek, T. J., Perry, W. L., III, Lee, J. J., Tilghman, S. M., Gumbiner, B. M. & Costantini, F. (1997) Cell 90, 181-192. [DOI] [PubMed] [Google Scholar]
- 24.Hope, T. J., Bond, B. L., McDonald, D., Klein, N. P. & Parslow, T. G. (1991) J. Virol. 65, 6001-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, F. J., Beeche, A. A., Hunter, J. J., Chin, D. J. & Hope, T. J. (1996) Mol. Cell. Biol. 16, 5147-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattaj, I. W. & Englmeier, L. (1998) Annu. Rev. Biochem. 67, 265-306. [DOI] [PubMed] [Google Scholar]
- 27.Nakielny, S. & Dreyfuss, G. (1999) Cell 99, 677-690. [DOI] [PubMed] [Google Scholar]
- 28.Fukuda, M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M. & Nishida, E. (1997) Nature 390, 308-311. [DOI] [PubMed] [Google Scholar]
- 29.Hsu, W., Zeng, L. & Costantini, F. (1999) J. Biol. Chem. 274, 3439-3445. [DOI] [PubMed] [Google Scholar]
- 30.Kishida, S., Yamamoto, H., Hino, S., Ikeda, S., Kishida, M. & Kikuchi, A. (1999) Mol. Cell. Biol. 19, 4414-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogerd, H. P., Fridell, R. A., Benson, R. E., Hua, J. & Cullen, B. R. (1996) Mol. Cell. Biol. 16, 4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox, R. T., Pai, L. M., Miller, J. R., Orsulic, S., Stein, J., McCormick, C. A., Audeh, Y., Wang, W., Moon, R. T. & Peifer, M. (1999) Development (Cambridge, U.K.) 126, 1327-1335. [DOI] [PubMed] [Google Scholar]
- 33.Cong, F., Schweizer, L., Chamorro, M. & Varmus, H. (2003) Mol. Cell. Biol. 23, 8462-8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Behrens, J., von Kries, J. P., Kuhl, M., Bruhn, L., Wedlich, D., Grosschedl, R. & Birchmeier, W. (1996) Nature 382, 638-642. [DOI] [PubMed] [Google Scholar]
- 35.Huber, O., Korn, R., McLaughlin, J., Ohsugi, M., Herrmann, B. G. & Kemler, R. (1996) Mech. Dev. 59, 3-10. [DOI] [PubMed] [Google Scholar]
- 36.Molenaar, M., van de Wetering, M., Oosterwegel, M., Peterson-Maduro, J., Godsave, S., Korinek, V., Roose, J., Destree, O. & Clevers, H. (1996) Cell 86, 391-399. [DOI] [PubMed] [Google Scholar]
- 37.Fagotto, F., Gluck, U. & Gumbiner, B. M. (1998) Curr. Biol. 8, 181-190. [DOI] [PubMed] [Google Scholar]
- 38.Yokoya, F., Imamoto, N., Tachibana, T. & Yoneda, Y. (1999) Mol. Biol. Cell 10, 1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malik, H. S., Eickbush, T. H. & Goldfarb, D. S. (1997) Proc. Natl. Acad. Sci. USA 94, 13738-13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kutay, U., Izaurralde, E., Bischoff, F. R., Mattaj, I. W. & Gorlich, D. (1997) EMBO J. 16, 1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orsulic, S. & Peifer, M. (1996) J. Cell Biol. 134, 1283-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosin-Arbesfeld, R., Cliffe, A., Brabletz, T. & Bienz, M. (2003) EMBO J. 22, 1101-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiechens, N. & Fagotto, F. (2001) Curr. Biol. 11, 18-27. [DOI] [PubMed] [Google Scholar]
- 44.Eleftheriou, A., Yoshida, M. & Henderson, B. R. (2001) J. Biol. Chem. 276, 25883-25888. [DOI] [PubMed] [Google Scholar]
- 45.Simcha, I., Shtutman, M., Salomon, D., Zhurinsky, J., Sadot, E., Geiger, B. & Ben-Ze'ev, A. (1998) J. Cell Biol. 141, 1433-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harada, N., Tamai, Y., Ishikawa, T., Sauer, B., Takaku, K., Oshima, M. & Taketo, M. M. (1999) EMBO J. 18, 5931-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson, C. B., Neufeld, K. L. & White, R. L. (2002) Proc. Natl. Acad. Sci. USA 99, 8683-8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rui, H. L., Fan, E., Zhou, H. M., Xu, Z., Zhang, Y. & Lin, S. C. (2002) J. Biol. Chem. 277, 42981-42986. [DOI] [PubMed] [Google Scholar]