Abstract
The function of some multidomain proteins is regulated by interdomain communication. We use second-site suppressor cysteine mutations to test a hypothesis on how the inserted (I)-like domain in the integrin β-subunit regulates ligand binding by the neighboring I domain in the integrin α-subunit [Huth, J. R., Olejniczak, E. T., Mendoza, R., Liang, H., Harris, E. A., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 5231–5236; and Alonso, J. L., Essafi, M., Xiong, J. P., Stehle, T. & Arnaout, M. A. (2002) Curr. Biol. 12, R340–R342]. The hypothesis is that an interaction between the β I-like metal ion-dependent adhesion site (MIDAS) and an intrinsic ligand in the linker following the α I domain, Glu-310, exerts a pull that activates the α I domain. Individual mutation of αL linker residue Glu-310 or β2 MIDAS residues Ala-210 or Tyr-115 to cysteine abolishes I domain activation, whereas the double mutation of αL-E310C with either β2-A210C or β2-Y115C forms a disulfide bond that constitutively activates ligand binding. The disulfide-bonded mutant is resistant to small molecule antagonists that bind to the β I-like domain near its interface with the α I domain and inhibit communication between these domains but remains susceptible to small molecule antagonists that bind underneath the I domain α7-helix and certain allosteric antagonistic antibodies. Thus, the α7-helix and its linker are better modeled as a pull spring than a bell rope. The results suggest that αL residue Glu-310, which is universally conserved in all I domain-containing integrins, functions as an intrinsic ligand for the β I-like domain, and that when integrins are activated, the β I-like MIDAS binds to Glu-310, pulls the spring, and thereby activates the α I domain.
Integrins are a large family of adhesion receptors that regulate cell migration and tissue organization and transduce signals bidirectionally across the plasma membrane. They are the most structurally complicated adhesion molecules yet known, with noncovalently associated α- and β-transmembrane subunits containing five and eight distinctive domains, respectively, in their extracellular segments. Half of vertebrate integrin α-subunits and all β-subunits contain von Willebrand factor-type A domains, termed inserted (I) and I-like domains, respectively (1–3). Both I and I-like domains have an α/β-fold with a central β-sheet surrounded by α-helices and a metal ion-dependent adhesion site (MIDAS) at the C-terminal ends of the central β-strands, i.e., the “top” face (1, 4–6). In integrins that lack I domains, I-like domains directly mediate ligand binding: a metal at the MIDAS coordinates to an acidic residue in the ligand (7). In I domain-containing integrins such as αLβ2, the I domain binds the acidic residue of the ligand through its MIDAS (4, 8–10), whereas the I-like domain regulates binding by the I domain (11). However, the molecular mechanism of I domain regulation by the I-like domain remains unknown.
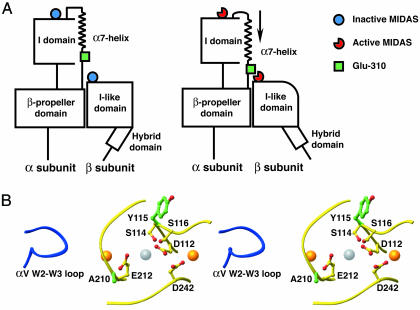
The I domain is inserted in the integrin α-subunit between blades 2 and 3 of the β-propeller domain (12). The I domain C-terminal α7-helix and the linker connecting it to the β-propeller domain are crucial for regulation of ligand binding. Downward movement of the α7-helix activates the I domain (8, 9, 13–15). Mutations in the α7-helix and linker may either activate or inactivate the I domain (16–19). A liganded crystal structure of integrin αVβ3, which lacks an α I domain, shows that the acidic Asp side chain of a ligand-mimetic peptide Arg-Gly-Asp is bound to the MIDAS of the β3 I-like domain, whereas the Arg side chain binds to loops of the αV β-propeller, at a site equivalent to where the I domain is inserted into the αL β-propeller domain (6). Because a Glu residue in the linker between the I and β-propeller domains corresponding to Glu-310 in αL is absolutely conserved in all I domain-containing integrins, and mutation of this residue in αL (16) or αM (20) abolishes I domain activation, it previously has been proposed that Glu-310 might interact with the metal in the β2 MIDAS in a way that mimics ligand binding by integrins that lack I domains (Fig. 1A) (1, 16, 20). However, a large number of explanations are possible for the negative effect of mutation of αL Glu-310, and no evidence for an interaction with the β I-like domain has been presented. In this article, by constructing second-site revertant mutations (21), we test the hypothesis that when activated, the β2 I-like domain MIDAS binds αL residue Glu-310 in the linker between the I domain and the β-propeller domain and exerts a downward pull on the α7-helix of the I domain that activates the I domain (Fig. 1 A).
Fig. 1.
The structural hypothesis. (A) Schematic. It is hypothesized that αL-Glu-310 acts as an intrinsic ligand that binds to the β2-subunit I-like domain MIDAS and, thus, axially displaces the I domain α7-helix in the C-terminal direction, reshapes the β6-α7 loop, and activates the αL I domain MIDAS (1, 16, 20). C-terminal axial displacement of the α7-helix and β6-α7 loop reshaping is known to result in a 10,000-fold increase in affinity for ligand of the αL I domain (9). Swing-out of the hybrid domain is depicted as demonstrated for αVβ3 and α5β1 (35, 36). (B) Stereo diagram of the MIDAS loops of the β2 and β3 I-like domains (yellow) and the W2-W3 loop of the αV β-propeller domain (blue). The structure shown is that of liganded αVβ3 (6); all side chains shown are identical in β2 and β3, and numbering is that of β2. Mutated residues are shown in green; MIDAS-coordinating residues are shown in yellow; O atoms are red. The ligand-induced metal-binding site (LIMBS), MIDAS, and adjacent to MIDAS (ADMIDAS) metal ions are gold, silver, and gold spheres, respectively, from left to right.
Materials and Methods
Cell Lines, Antibodies, and Small Molecule Inhibitors. cDNAs of wild-type αL and β2 were inserted into pcDNA3.1/Hygro(+) or pcDNA3.1(+) and used as the template for mutagenesis. The αL and β2 mutations were generated by using QuikChange XL Site-Directed Mutagenesis Kit (Stratagene). All constructs were verified by DNA sequencing. 293T cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. K562 cells were transfected by electroporation and selected with 1 mg/ml G418 (22). mAbs to human αL and β2 are as described (11). mAbs m24 (23) and KIM127 (24) were kind gifts of N. Hogg (Imperial Cancer Research Fund, London) and M. Robinson (Celltech, Slough, U.K), respectively. mAbs were used as 10 μg/ml purified IgG or 1:200 ascites. LFA703 (25, 26) was kindly provided by Novartis Pharma (Basel). XVA143 (27) was synthesized according to example 345 of the patent (28) and was obtained from Paul Gillespie (Roche, Nutley, NJ)
Cell Adhesion Assay. Binding of fluorescently labeled transfectants to immobilized intercellular adhesion molecule-1 (ICAM-1) was as described (22). Briefly, soluble ICAM-1 (domains 1–5) was immobilized at 10 μg/ml on microtiter plates. Binding of the 293T transient transfectants to immobilized ICAM-1 was determined in 2.5% FBS/L15 medium. Binding of K562 stable transfectants to immobilized ICAM-1 was determined in Hepes/NaCl/Glucose/BSA (20 mM Hepes, pH 7.5/140 mM NaCl/2 mg/ml glucose/1% BSA) supplemented as indicated with divalent cations and DTT. After incubation at room temperature for 30 min, unbound cells were washed off and bound cells were quantitated (22).
Binding of Soluble ICAM-1. Binding of soluble ICAM-1-IgA/Fc fusion protein complexed with affinity-purified, FITC anti-human IgA was measured by immunofluorescence flow cytometry (29).
Cell Surface Biotinylation and Immunoprecipitation. Cell surface biotinylation and immunoprecipitation were as described (29).
Results
Design and Cell Surface Expression of αLβ2 Second-Site Reversion Mutants. αL- and β2-subunits were coexpressed in transient 293T or stable K562 transfectants, and adhesion to ICAM-1 immobilized on substrates or binding to soluble multimeric fluorescent ICAM-1 was measured (Fig. 2). Mutation αL-E310C abolished binding to ICAM-1 similarly to αL-E310A, confirming a crucial role for Glu-310 in I domain activation (Fig. 2 A) (16). To search for a site in the β2-subunit where a cysteine could be introduced that would suppress the αL-E310C mutation by formation of an intersubunit disulfide bond, the liganded αVβ3 structure (7) was examined for a residue around the I-like domain MIDAS that was close to the loop between blade 2 and blade 3 of the β-propeller, where the I domain is inserted in αL (12). Metal coordinating residues and buried residues were excluded. By using these criteria, residue β2-A210 was selected, which is in the MIDAS loop that bears the metal-coordinating residue Glu-212 (Fig. 1B).
Fig. 2.
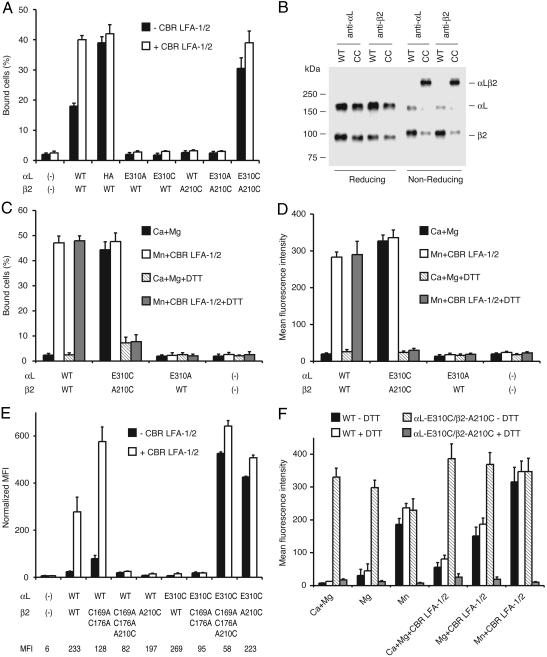
Second-site suppressor αL-E310C and β2-A210C mutations constitutively activate αLβ2 by forming a disulfide bond. (A) Binding of 293T cell transfectants to immobilized ICAM-1. Adhesion to ICAM-1 of cells transfected with the indicated αL and β2 cDNA was determined in the absence (black bars) or presence (white bars) of activating mAb CBR LFA-1/2 at 37°C. HA, high-affinity αL K287C/K294C I domain mutant (8). (B) Immunoprecipitation. K562 transfectants expressing wild-type αLβ2 (WT) and αL-E310C/β2-A210C (CC) were surface-labeled with biotin, and lysates were immunoprecipitated by either αL antibody (TS2/4) or β2 antibody (May.017). Immunoprecipitates were subjected to reducing and nonreducing SDS/7.5% PAGE, transferred to nitrocellulose, and Western-blotted with horseradish peroxidase-streptavidin and enhanced chemiluminescence. (C–F) Binding of K562 stable (C, D, and F) or 293T transient (E) transfectants to ICAM-1 substrates (C) and soluble ICAM-1 complexes (D–F). Binding was assayed in Hepes/NaCl/glucose/BSA (K562 transfectants) or 2.5% FBS/L15 medium (293T transfectants) supplemented with 1 mM CaCl2/1 mM MgCl2, 2 mM MgCl2, or 2 mM MnCl2 plus 10 μg/ml CBR LFA-1/2 or 2 mM DTT as indicated at room temperature. In E, binding of ICAM-1 by different 293T transfectants was normalized to their αLβ2 cell surface expression by multiplying by the ratio of the specific mean fluorescence intensity (MFI) of TS2/4 mAb binding to wild-type and mutant αLβ2. MFI of TS2/4 mAb before subtraction of the MFI of untransfected cells to obtain specific MFI is shown in the bottom row of E. Binding by the mock transfectant was not normalized.
We expected that αL-E310 would bind to the β2 MIDAS as part of a larger intersubunit interface, and that exposed residues in β2 MIDAS loops also would contribute to this interface. β2-A210 is nearby the MIDAS coordinating residue β2-E212, and we therefore hoped that the mutation β2-A210C might by itself inactivate αLβ2. Indeed, the β2-A210C mutation abolished binding to ICAM-1 (Fig. 2 A), showing a crucial role for a non-metal-coordinating β2 MIDAS loop residue in αLβ2 activation and suggesting that residues in the vicinity of the β2 MIDAS, including β2-Ala-210, might interact with the I domain C-terminal linker.
Formation of an Intersubunit Disulfide Bond Between αL-E310C and β2-A210C Constitutively Activates Integrin αLβ2. To directly test the hypothesis that an interaction between residues in the vicinity of αL linker residue E310 and β2 MIDAS residue A210 activates αLβ2, the αL-E310C and β2-A210C mutants were cotransfected. Despite the abolition of binding by the individual substitutions in the αL-E310C/β2 and αL/β2-A210C heterodimers, the αL-E310C/β2-A210C double mutant heterodimer was fully activated (Fig. 2 A, C, and D). By contrast, the αL-E310A/β2-A210C double mutant was inactive. Immunofluorescent flow cytometry showed all αL-E310 and β2-A210 single and double mutants were as well expressed as wild-type αLβ2 in both 293T and K562 transfectants (Fig. 2E, bottom line, and data not shown). These results demonstrate second-site reversion between mutations at residues αL-E310 and β2-A210 when each residue is mutated to cysteine. Immunoprecipitation from K562 transfectants and reducing and nonreducing SDS/PAGE demonstrated that the αL-E310C/β2-A210C heterodimer, but not the wild-type αLβ2 heterodimer, is covalently linked with a disulfide bond (Fig. 2B), with an efficiency of formation of 80%. β I-like domains contain a specificity-determining loop with disulfide-bonded cysteines that locate ≈12 Å from the I-like MIDAS and 16 Å from β2-A210 (7). To rule out any possible interaction with the engineered disulfide, these cysteines were mutated to Ala in the β2-C169A/C176A mutant. Although αL/β2-C169A/C176A/A210C was inactive, αL-E310C/β2-C169A/C176A/A210C was constitutively active in binding to ICAM-1 and bound as well as αL-E310C/β2-A210C and activated wild-type αLβ2 after correction for the lower expression of heterodimers containing the β2-C169A/C176A mutation (Fig. 2E). Furthermore, αL-E310C/β2-C169A/C176A/A210C formed a disulfide-linked heterodimer (data not shown).
Suppression between the αL-E310C and β2-A210C mutations did more than restore wild-type ligand binding, it also resulted in constitutive activation. In 293T transfectants in the absence of activation, the αL-E310C/β2-A210C double mutant was more active than wild-type αLβ2 and appeared maximally activated, as shown by lack of further activation by CBR LFA-1/2 mAb to the β2 I-EGF3 domain and comparison to αLβ2 with a mutant high-affinity (HA) αL I domain (Fig. 2 A). In K562 transfectants, wild-type αLβ2 was inactive under basal conditions in Ca2+/Mg2+, whereas the αL-E310C/β2-A210C double mutant was maximally active in Ca2+/Mg2+ both in adhesion assays and in binding of soluble multimeric ICAM-1 (Fig. 2 C and D). In contrast to wild-type αLβ2, αL-E310C/β2-A210C was active independent of whether Ca2+ plus Mg2+, Mg2+, Mn2+, or activating mAb CBR LFA-1/2 was present (Fig. 2F).
DTT was used to reduce the disulfide bond. Although reduction with 10 mM DTT at 37°C can activate β2 integrins (8, 30), 2 mM DTT at room temperature did not affect binding of wild-type αLβ2 to immobilized or soluble ICAM-1 (Fig. 2 C, D, and F). However, 2 mM DTT abolished the binding of αL-E310C/β2-A210C to ICAM-1 (Fig. 2 C, D, and F). Ligand binding by wild-type αLβ2, but not DTT-treated αL-E310C/β2-A210C, was activated by CBR LFA-1/2 mAb and Mn2+ (Fig. 2 C, D, and F). The inability of DTT-treated αL-E310C/β2-A210C to bind ligand agrees with the finding above that both αL-E310C/β2 and αL/β2-A210C heterodimers failed to bind ligand. We conclude that (i) in the active conformation of αLβ2, residues αL-310 and β2-210 are in sufficiently close proximity to form a disulfide bond when mutated to cysteine; (ii) disulfide bond formation is required for second-site reversion between the αL-E310C and β2-A210C mutations; and (iii) formation of a disulfide bond between these residues constitutively activates the αL I domain.
Susceptibility to Small Molecule Antagonists and Inhibitory Antibodies. mAbs inhibit αLβ2 function by different mechanisms. mAbs that directly, i.e., competitively block binding to ICAM-1, inhibit binding to activated wild-type αLβ2 as well as αLβ2 containing an I domain locked in the high-affinity, ligand-binding configuration with a disulfide bond (HA αLβ2) (11). By contrast, mAbs that indirectly, i.e., allosterically block binding to ICAM-1, inhibit wild-type αLβ2 but not HA αLβ2. Competitive inhibitor mAbs to the αL I domain, i.e., TS2/6, May.035, MHM24, and TS1/22, equivalently blocked wild-type αLβ2, αL-E310C/β2-A210C, and HA αLβ2 (Table 1) (8). By contrast, TS2/14 mAb, which noncompetitively inhibits ICAM-1 binding to LFA-1, blocked binding of the αL-E310C/β2-A210C mutant but not the HA αLβ2 mutant to ICAM-1, suggesting that the αL-E310C/β2-A210C mutation does not irreversibly activate the I domain. mAbs to the β2 I-like domain inhibit ICAM-1 binding allosterically, as shown by inhibition of wild-type but not HA αLβ2 (Table 1) (11). Interestingly, mAbs May.017 and MHM23, which bind to the specificity-determining loop of the I-like domain, which locates in or very near to the α I/β I-like interface, partially inhibited ligand binding by αL-E310C/β2-A210C (Table 1). By contrast, mAbs that bind distal to this interface, to the β2 I-like domain α1- and α7-helices, did not inhibit αL-E310C/β2-A210C (Table 1).
Table 1. Inhibition by αL I and β2 I-like domain antibodies of multimeric ICAM-1 binding to αLβ2 mutants.
| Inhibition, %
|
|||||
|---|---|---|---|---|---|
| mAb | Epitope | Wild-type αLβ2 | αL-E310C/β2-A210C | HA αLβ2 | |
| TS2/6 | αL I domain | 154–183 | 97 ± 2 | 96 ± 2 | 97 ± 1 |
| May.035 | αL I domain | K197, H201 | 98 ± 1 | 98 ± 0 | 97 ± 1 |
| MHM24 | αL I domain | K197 | 96 ± 2 | 97 ± 1 | 96 ± 0 |
| TS1/22 | αL I domain | Q266, S270 | 96 ± 1 | 97 ± 2 | 92 ± 1 |
| TS2/14 | αL I domain | S270, E272 | 99 ± 0 | 99 ± 0 | 14 ± 2 |
| CBR LFA-1/1* | αL I domain | 301–338 | 97 ± 2 | 2 ± 0 | 2 ± 1 |
| May.017 | β2 I-like domain | E175, ? | 98 ± 0 | 70 ± 8 | 3 ± 2 |
| MHM23 | β2 I-like domain | E175 | 97 ± 2 | 40 ± 6 | 2 ± 2 |
| TS1/18 | β2 I-like domain | R133, H332 | 98 ± 1 | 4 ± 3 | 0 ± 2 |
| YFC51 | β2 I-like domain | R133, H332 | 98 ± 0 | 2 ± 2 | 0 ± 1 |
| CLB LFA-1/.1 | β2 I-like domain | H332, N339 | 97 ± 1 | 2 ± 2 | 0 ± 0 |
Wild-type αLβ2 in K562 transfectants was activated by preincubation with mAb CBR LFA-1/2. Binding to soluble, multimeric ICAM-1 in medium containing 1 mM CaCl2 and 1 mM MgCl2 was in the presence of the indicated mAb. Results are means ± SD of three experiments. HA, high-affinity I domain mutant (8).
The epitope spans the linker including αL-E310C. Binding of CBR LFA-1/1 to the αL-E310C/β2-A210C and HA mutants was ≈50% of binding to wild-type αLβ2 (8). All other mAbs bound to αL-E310C/β2-A210C, HA αLβ2, and wild-type αLβ2 equally well (data not shown)
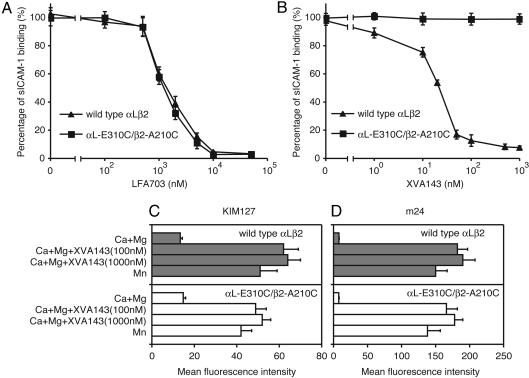
The mechanism of activation by the disulfide between the αL I linker and the β2 I-like MIDAS was investigated further with representatives of two distinct classes of small molecule antagonists, LFA703 and XVA143. Both are allosteric antagonists, as shown by lack of inhibition of HA αLβ2 (29). LFA703 binds to the hydrophobic pocket underneath the α7-helix of the αL I domain and stabilizes the low-affinity, closed conformation of the I domain (25, 26). αL-E310C/β2-A210C was as sensitive to inhibition by LFA703 as wild-type αLβ2 (Fig. 3A). XVA143 binds to the MIDAS of the β2 I-like domain and blocks its ability to communicate activation to the α I domain (27, 29). The αL-E310C/β2-A210C mutant was totally resistant to inhibition by XVA143 (Fig. 3B).
Fig. 3.
Inhibition by small molecule antagonists of binding to ICAM-1 and induction of activation epitopes. (A and B) Inhibition of binding of soluble, multimeric ICAM-1 by LFA703 (A) or XVA143 (B). Binding of wild-type αLβ2 K562 transfectants activated by preincubation with mAb CBR LFA-1/2 for 30 min or αL-E310C/β2-A210C transfectants was measured in medium containing 1 mM CaCl2 and 1 mM MgCl2.(C and D) Induction by XVA143 or Mn2+ of KIM127 (C) and m24 (D) epitopes. Transfectants in medium containing 1 mM CaCl2/1 mM MgCl2, 2 mM MnCl2, and XVA143 as indicated were stained with KIM127 or m24 mAbs and subjected to immunofluorescence flow cytometry. Expression of activation-insensitive mAb TS2/4 was not affected by XVA143 or Mn2+ (data not shown).
The global conformation of the αL-E310C/β2-A210C mutant was examined with the m24 mAb to an activation epitope on the β2 I-like domain (11, 23, 31) and the KIM127 mAb to an epitope on the β2 I-EGF2 domain that is buried in the bent integrin conformation and exposed in the extended conformation (32, 33). The disulfide connecting the αL I domain linker to the β2 I-like MIDAS did not induce exposure of either epitope, suggesting that the extended conformation is not induced and that the I-like domain remains in an inactive conformation (Fig. 3 C and D). This finding is as expected, because unlike coordination of αL-Glu-310 with the β2 MIDAS metal, the disulfide bond is not expected to alter MIDAS coordination and β2 I-like domain conformation. Similarly, mutationally stabilizing the αL I domain in the high-affinity conformation with the HA αLβ2 mutant does not lead to β2 I-like domain activation or global conformational change (11). Nonetheless, the active conformation of the β2 I-like domain detected by m24 mAb and the extended integrin conformation detected by KIM127 mAb were induced in the αL-E310C/β2-A210C mutant by Mn2+ and by XVA143 (Fig. 3 C and D). This finding shows that the αL-E310C/β2-A210C mutant is capable of undergoing global conformational change and binds to XVA143 despite lack of inhibition of ligand binding by XVA143.
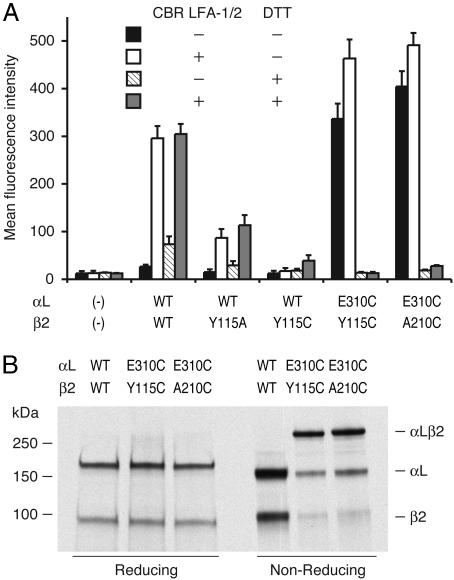
Another Second-Site Reversion αLβ2 Mutant That also Constitutively Binds ICAM-1. To obtain further evidence for an interaction of αL-Glu-310 with the β2 MIDAS, we mutated residue β2-Tyr-115 (Fig. 1B). Tyr-115 is located between the two Ser residues in the MIDAS DXSYS sequence motif. Ala-210 and Tyr-115 locate opposite one another on either side of the MIDAS metal ion (Fig. 1B). ICAM-1 binding by αLβ2 was reduced by the β2-Y115A mutation and totally abolished by the β2-Y115C mutation (Fig. 4A). The double mutant αL-E310C/β2-Y115C constitutively bound ICAM-1, exactly as observed for the αL-E310C/β2-A210C mutant (Fig. 4A). Moreover, immunoprecipitation from 293T transfectants and SDS/PAGE demonstrated that the αL-E310C and β2-Y115C subunits were covalently linked together with a disulfide bond, with an efficiency of formation of 63% (Fig. 4B). Ligand binding by αL-E310C/β2-Y115C was abolished by DTT reduction, demonstrating that the intersubunit disulfide bond was indispensable for ligand-binding activity (Fig. 4A).
Fig. 4.
Constitutive activation of αL-E310A/β2-Y115C with an intersubunit disulfide bond. (A) Binding of 293T cell transfectants to soluble ICAM-1 complexes. Binding was assayed in 2.5% FBS/L15 medium supplemented with 10 μg/ml CBR LFA-1/2 or 2 mM DTT as indicated at room temperature. All mutants were as well expressed as wild-type αLβ2 in 293T cells. (B) Immunoprecipitation. Lysates from 35S-labeled 293T transfectants were immunoprecipitated with TS2/4 mAb and subjected to SDS/7.5% PAGE and fluorography.
Discussion
To the classical technique of second-site reversion mutations (21), we have added the twists of disulfide bond formation and mutations that activate function as well as suppress loss of function. The αL mutation E310C and the β2 mutations A210C and Y115C individually abolish activation of ligand binding by αLβ2, but use of the αL mutation in combination with either of the β2 mutations constitutively induces αLβ2 activation. The formation of the intersubunit disulfide bonds and their requirement for activation of ligand binding directly demonstrate interaction between these regions of αL and β2 in the active integrin conformation. The β I-like MIDAS metal ion is centered immediately between residues Ala-210 and Tyr-115, only 5 Å from their Cα atoms. The activating crosslinks to these residues strongly support the hypothesis that interaction between αL residue Glu-310 and the metal of the β2 I-like domain MIDAS induces the high-affinity conformation of the αL I domain. Glu-310 is the only acidic residue in the αL I domain α7-helix or its linker to the β-propeller domain that is conserved in all integrin I domains, and mutation of the other αL acidic residues in the same polypeptide segment, Glu-301 (16), Asp-316, and Glu-323 (data not shown) does not abolish αLβ2 activation. In integrins that lack I domains, the β I-like domain MIDAS metal ion when activated directly coordinates an acidic residue in the ligand, e.g., the Asp of the Arg-Gly-Asp (RGD) motif common to many integrin ligands (7). Our data strongly support the concept that integrins that contain I domains are activated by a similar interaction in which an intrinsic ligand-like residue, αL-Glu-310, rather than an extrinsic ligand, binds to a metal ion at the β-subunit I-like MIDAS.
Receptor-ligand-like interaction between neighboring domains/subunits is a previously uncharacterized mechanism for signal transmission in the extracellular environment and in adhesion molecules and may turn out to have parallels in the diverse range of pro- and eukaryotic intracellular enzymes and extracellular recognition molecules in which von Willebrand factor type A, i.e., I domains, are present (34). α I domains appear in integrins late in metazoan evolution, whereas β I-like domains are present in all integrins, including those in Porifera, Drosophila, and Caenorhabditis elegans. Binding of I-like domains to an intrinsic ligand in the linker following the I domain must have evolved when I domains became inserted in integrin α-subunits, to enable regulated binding to an extrinsic ligand to be transferred from the β I-like domain to the α I domain.
Our study provides insight into the mechanics of the linkage between β I-like and α I domains. Movement of residue αL-310 to a position near the β2 I-like MIDAS is sufficient to pull down the α7-helix, reshape the β6-α7 loop, and activate the αL I domain. The conformation of the β6-α7 loop, but not that of the α7-helix, is important for transducing conformational change to the α I MIDAS (9). Allosteric inhibition of αL-E310C/β2-A210C by certain mAbs to the αL and β2 I-like domains and LFA703 shows that the segment connecting the β6-α7 loop (αL residue ≈293) to αL residue 310 remains elastic. The connecting segment thus should be viewed as a pull spring rather than a bell rope or connecting rod. A pull spring, but not the other two types of mechanical connections, has the important feature that it would enable the β6-α7 loop to assume three ratchet positions, as recently observed for the closed, intermediate, and open conformations of the αL I domain, which have low, intermediate, and high affinity for ICAM-1, respectively (9).
Acknowledgments
We thank Uli von Andrian for reviewing the manuscript. This work was supported by National Institutes of Health Grant CA31798.
Abbreviations: MIDAS, metal ion-dependent adhesion site; I, inserted; ICAM-1, intercellular adhesion molecule-1; HA, high-affinity.
References
- 1.Shimaoka, M., Takagi, J. & Springer, T. A. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 485-516. [DOI] [PubMed] [Google Scholar]
- 2.Humphries, M. J. (2000) Biochem. Soc. Trans. 28, 311-339. [PubMed] [Google Scholar]
- 3.Gahmberg, C. G., Tolvanen, M. & Kotovuori, P. (1997) Eur. J. Biochem. 245, 215-232. [DOI] [PubMed] [Google Scholar]
- 4.Lee, J.-O., Rieu, P., Arnaout, M. A. & Liddington, R. (1995) Cell 80, 631-638. [DOI] [PubMed] [Google Scholar]
- 5.Huang, C., Zang, Q., Takagi, J. & Springer, T. A. (2000) J. Biol. Chem. 275, 21514-21524. [DOI] [PubMed] [Google Scholar]
- 6.Xiong, J.-P., Stehle, T., Diefenbach, B., Zhang, R., Dunker, R., Scott, D. L., Joachimiak, A., Goodman, S. L. & Arnaout, M. A. (2001) Science 294, 339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong, J.-P., Stehle, T., Zhang, R., Joachimiak, A., Frech, M., Goodman, S. L. & Arnaout, M. A. (2002) Science 296, 151-155. [DOI] [PubMed] [Google Scholar]
- 8.Lu, C., Shimaoka, M., Ferzly, M., Oxvig, C., Takagi, J. & Springer, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 2387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimaoka, M., Xiao, T., Liu, J.-H., Yang, Y., Dong, Y., Jun, C.-D., McCormack, A., Zhang, R., Joachimiak, A., Takagi, J., Wang, J.-H. & Springer, T. A. (2003) Cell 112, 99-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emsley, J., Knight, C. G., Farndale, R. W., Barnes, M. J. & Liddington, R. C. (2000) Cell 101, 47-56. [DOI] [PubMed] [Google Scholar]
- 11.Lu, C., Shimaoka, M., Zang, Q., Takagi, J. & Springer, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 2393-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Springer, T. A. (1997) Proc. Natl. Acad. Sci. USA 94, 65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, J.-O., Bankston, L. A., Arnaout, M. A. & Liddington, R. C. (1995) Structure 3, 1333-1340. [DOI] [PubMed] [Google Scholar]
- 14.Shimaoka, M., Shifman, J. M., Jing, H., Takagi, J., Mayo, S. L. & Springer, T. A. (2000) Nat. Struct. Biol. 7, 674-678. [DOI] [PubMed] [Google Scholar]
- 15.Shimaoka, M., Lu, C., Palframan, R., von Andrian, U. H., Takagi, J. & Springer, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 6009-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huth, J. R., Olejniczak, E. T., Mendoza, R., Liang, H., Harris, E. A., Lupher, M. L., Jr., Wilson, A. E., Fesik, S. W. & Staunton, D. E. (2000) Proc. Natl. Acad. Sci. USA 97, 5231-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, L. & Plow, E. F. (1996) J. Biol. Chem. 271, 29953-29957. [DOI] [PubMed] [Google Scholar]
- 18.Oxvig, C., Lu, C. & Springer, T. A. (1999) Proc. Natl. Acad. Sci. USA 96, 2215-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupher, M. L., Jr., Harris, E. A., Beals, C. R., Sui, L., Liddington, R. C. & Staunton, D. E. (2001) J. Immunol. 167, 1431-1439. [DOI] [PubMed] [Google Scholar]
- 20.Alonso, J. L., Essafi, M., Xiong, J.-P., Stehle, T. & Arnaout, M. A. (2002) Curr. Biol. 12, R340-R342. [DOI] [PubMed] [Google Scholar]
- 21.Yanofsky, C. (2003) J. Biol. Chem. 278, 10859-10878. [DOI] [PubMed] [Google Scholar]
- 22.Lu, C. & Springer, T. A. (1997) J. Immunol. 159, 268-278. [PubMed] [Google Scholar]
- 23.Dransfield, I. & Hogg, N. (1989) EMBO J. 8, 3759-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson, M. K., Andrew, D., Rosen, H., Brown, D., Ortlepp, S., Stephens, P. & Butcher, E. C. (1992) J. Immunol. 148, 1080-1085. [PubMed] [Google Scholar]
- 25.Kallen, J., Welzenbach, K., Ramage, P., Geyl, D., Kriwacki, R., Legge, G., Cottens, S., Weitz-Schmidt, G. & Hommel, U. (1999) J. Mol. Biol. 292, 1-9. [DOI] [PubMed] [Google Scholar]
- 26.Weitz-Schmidt, G., Welzenbach, K., Brinkmann, V., Kamata, T., Kallen, J., Bruns, C., Cottens, S., Takada, Y. & Hommel, U. (2001) Nat. Med. 7, 687-692. [DOI] [PubMed] [Google Scholar]
- 27.Welzenbach, K., Hommel, U. & Weitz-Schmidt, G. (2002) J. Biol. Chem. 277, 10590-10598. [DOI] [PubMed] [Google Scholar]
- 28.Fotouhi, N., Gillespie, P., Guthrie, R., Pietranico-Cole, S. & Yun, W. (1999) PCT Int. Appl. WO0021920 (Hoffmann-La Roche, Basel).
- 29.Shimaoka, M., Salas, A., Yang, W., Weitz-Schmidt, G. & Springer, T. A. (2003) Immunity 19, 391-402. [DOI] [PubMed] [Google Scholar]
- 30.Shimaoka, M., Lu, C., Salas, A., Xiao, T., Takagi, J. & Springer, T. A. (2002) Proc. Natl. Acad. Sci. USA 99, 16737-16741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamata, T., Tieu, K. K., Tarui, T., Puzon-McLaughlin, W., Hogg, N. & Takada, Y. (2002) J. Immunol. 168, 2296-2301. [DOI] [PubMed] [Google Scholar]
- 32.Lu, C., Ferzly, M., Takagi, J. & Springer, T. A. (2001) J. Immunol. 166, 5629-5637. [DOI] [PubMed] [Google Scholar]
- 33.Beglova, N., Blacklow, S. C., Takagi, J. & Springer, T. A. (2002) Nat. Struct. Biol. 9, 282-287. [DOI] [PubMed] [Google Scholar]
- 34.Whittaker, C. A. & Hynes, R. O. (2002) Mol. Biol. Cell 13, 3369-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takagi, J., Petre, B. M., Walz, T. & Springer, T. A. (2002) Cell 110, 599-611. [DOI] [PubMed] [Google Scholar]
- 36.Takagi, J., Strokovich, K., Springer, T. A. & Walz, T. (2003) EMBO J. 22, 4607-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]