Abstract
Drosophila is a powerful model for molecular studies of hematopoiesis and innate immunity. However, its use for functional cellular studies remains hampered by the lack of single-cell assays for hemocytes (blood cells). Here we introduce a generic method combining fluorescence-activated cell sorting and nonantibody probes that enables the selective gating of live Drosophila hemocytes from the lymph glands (larval hematopoietic organ) or hemolymph (blood equivalent). Gated live hemocytes are analyzed and sorted at will based on precise quantitation of fluorescence levels originating from metabolic indicators, lectins, reporters (GFP and β-galactosidase) and antibodies. With this approach, we discriminate and sort plasmatocytes, the major hemocyte subset, from lamellocytes, an activated subset present in gain-of-function mutants of the Janus kinase and Toll pathways. We also illustrate how important, evolutionarily conserved, blood-cell-regulatory molecules, such as calcium and glutathione, can be studied functionally within hemocytes. Finally, we report an in vivo transfer of sorted live hemocytes and their successful reanalysis on retrieval from single hosts. This generic and versatile fluorescence-activated cell sorting approach for hemocyte detection, analysis, and sorting, which is efficient down to one animal, should critically enhance in vivo and ex vivo hemocyte studies in Drosophila and other species, notably mosquitoes.
Studies focusing on hematopoiesis and innate immunity in the model organism Drosophila melanogaster have identified extensive homologies between Drosophila hemocytes (blood cells) and mammalian leukocytes. Whole-animal functional studies have suggested that Drosophila hemocytes participate in similar activities to mammalian leukocytes, including phagocytosis/encapsulation of pathogens, release of reactive oxygen species (ROS) and reactive nitrogen species and antimicrobial peptides, activation of humoral serine protease cascades, scav-enging of dead bodies, wound repair, and extracellular matrix deposition (1–6). Molecular genetic studies have unravelled important evolutionarily conserved regulatory elements, including transcription factors of the Runt/acute myelogenous leukemia (7), GATA (8), and Polycomb (9) families and integral transduction cascades, including the immune deficiency/tumor necrosis receptor (2), Toll/IL-1 receptor (2), Janus kinase (10, 11), mitogen-activated protein kinase (12), Notch (13), steroid (14), and vascular endothelial growth factor (15) pathways.
Compared to mammalian species, Drosophila is particularly well suited to study the molecular genetics of blood cell development and function, thanks to the existence of a well annotated genome database, assorted genetic tools, and large mutant collections (16). By contrast, the lack of single-cell assays for Drosophila hemocytes severely restricts the scope of cellular studies (10, 11). Accordingly, our knowledge of Drosophila hemocyte subsets and functions remains very limited. In mammals, the use of fluorescence-activated cell sorting (FACS) has driven much of the progress in subset discrimination and functional analysis of leukocytes (17). Current three-laser, “multidimensional,” FACS machines enable up to 14 simultaneous single-cell measurements, namely 2 light scatters and 12 fluorescent surface/intracellular markers (18–20). FACS also enables the sorting of subsets of interest and their further use in in vitro and in vivo assays.
So far, FACS has been applied only once to freshly harvested Drosophila hemocytes, featuring one-parameter analysis of hemolymph (blood surrogate) cells for surface antibody reactivity (21). No FACS analysis of hemocytes from lymph glands, the larval hematopoietic organ (6, 14, 22), and no cell sorting of fresh hemocytes from either the lymph glands or hemolymph have been reported. Clearly, the ability to perform single-cell analyses and sorts on freshly harvested hemocytes and to use sorted hemocyte fractions with existing molecular tools for further in vitro or in vivo studies would provide great experimental opportunities, most notably for functional genomics.
In this paper, we introduce a generic FACS method that enables the detection and multidimensional analysis of live Drosophila hemocyte subsets from the hemolymph and lymph glands down to one animal. GFP and β-galactosidase (LacZ) reporters, which are widely used in Drosophila mutagenesis and transgenesis (23), can be precisely quantified within live hemocytes. Evolutionarily conserved regulatory molecules, such as Ca2+ and glutathione (GSH), can also be investigated functionally within live hemocytes. We also report (i) the successful sorting of hemocyte fractions, (ii) the successful in vivo transfer of sorted hemocytes, and (iii) the successful reanalysis of transferred cells from single hosts.
Methods
Drosophila Stocks. Stocks used in this study include y, w67 (control), Tum-l [Janus kinase gain-of-function mutant (24)], and Toll10B [Toll gain-of-function mutant (25)]. The Tum-l/11707 line was generated by crossing the Tum-l line and the LacZ enhancer-trap line, 11707 (26). The GAL4-e33c upstream activating sequence (UAS)-gfp strain was generated by crossing flies carrying the GAL4-e33c enhancer trap (27) to flies carrying the gfp transgene under control of the UAS (GAL4 response element), thus achieving constitutive GFP expression in hemolymph and lymph glands hemocytes. For in vivo transfers, we used two GFP-expressing lines: His::GFP [ubiquitous expression of a fusion protein between histone His2AvD and GFP (28)] and Tum-l; His::GFP (generated by standard crossing). Stocks were fed standard cornmeal, molasses, yeast, and agar medium and were maintained at 25°C. Late wandering third instar larvae were used for all experiments because they show maximal hemocyte numbers in lymph glands and hemolymph (6, 14).
Hemocyte Collection. Hemolymph cells were collected by rupturing the larval cuticle with a pair of fine forceps. For the collection of lymph glands cells, lymph glands were carefully dissected out, rinsed, and ruptured by repeated pipetting with siliconized tips. Cells were collected in ice-cold Schneider's medium (Invitrogen/GIBCO) containing 1× complete mini protease inhibitor mixture (Roche Applied Science) to prevent melanization, clump formation, and autolysis and kept on ice until incubation with FACS probes. Most analyses were performed with cells from 5–10 animals. However, several analyses were also performed with cells from one animal to validate single-animal hemocyte assays with both hemolymph- and lymph glands-derived hemocytes.
FACS Probes and Staining Procedures. The main probes validated so far are listed in Table 1, which is published as supporting information on the PNAS web site. Staining conditions (buffer, pH, temperature, and incubation time) were first optimized as monochlorobimane (MCB) was titrated on larval hemocyte suspensions, mbn-2 cells, and S2 cells. Other probes were titrated by using the same optimized staining conditions. Possible biochemical/optical interactions between probes were carefully checked by comparing fluorescences measured with stains applied either alone or in combination with one another. We found the conditions below to yield excellent viability of cells and reproducibility of measured signals while minimizing washing steps. Cells were incubated with probes for 20 min at room temperature, in the dark, at the concentrations indicated in Table 1, and in exactly 1,000 μl of buffer. Staining medium was Schneider's medium, pH 6.5, with 2.5 mM probenecid (Sigma) to limit active probe efflux (29). Cells were washed once with 10 ml of ice-cold staining medium and centrifuged for 5 min at 4°C at 1,500 rpm (LX-130 centrifuge; Tomy, Tokyo). Supernatant was discarded, and the pellet was kept on ice in the dark. On some occasions, antibody-staining was performed subsequently. For this purpose, H2, antilamellocyte antibody (L1a), and antiplasmatocyte antibody (P1b) hybridoma supernatants (21) were first fluorescently labeled by using the Alexa 594-Zenon reagents (Molecular Probes) according to the manufacturer's guidelines. Cells were resuspended in 90 μl of staining medium, and 10 μl of the final antibody solutions were added for a 30-min incubation on ice in the dark. Controls were incubated with medium only. After incubation, antibody-stained and control cells were washed once with 3 ml of ice-cold staining medium and centrifuged for 5 min at 4°C at 1,500 rpm. Supernatant was discarded and the pellet was kept on ice in the dark. Just before acquisition on the FACS machine (<1 h after last incubation step), cell pellets were resuspended in 100 μl of staining medium with 2 μg/ml propidium iodide (PI; Sigma) to label dead cells.
FACS Analyses and Sorts. Multidimensional analyses and sorts were performed on a modified FACStar Plus (Pharmingen) equipped with 3 lasers (a krypton laser at 407 nm, an argon laser at 488 nm, and a dye laser at 595 nm) and 13 detectors (11 fluorescences plus forward and side scatters) connected to MoFlo electronics (DakoCytomation, Fort Collins, CO) supplemented with home-built electronics (Stanford Shared FACS Facility, Stanford University School of Medicine). All analyses and sorts were repeated at least two or three times. The purity of sorted fractions was checked visually and by FACS reanalysis. Images of sorted cell fractions were obtained with a Eclipse E800 microscope (Nikon). Data were compensated, analyzed, and presented by using flowjo software (Tree Star, Ashland, OR). Details on the implementation of multidimensional FACS assays (building of customized staining combinations, compensation, and analysis) are available in refs. 18 and 19. The FACS machine was standardized with fluorochrome-containing beads, and fluorescence-reading in each channel was automatically adjusted to a constant value to ensure that data obtained on different days were comparable.
In Vivo Transfer of Sorted Hemocytes. The nozzle, sheath, and sample lines of the FACS machine were sterilized with alcohol for 15 min. GFP+ cells were sorted from suspensions of hemolymph cells of His::GFP and Tum-l; His::GFP animals. Sorted cells were pelleted, resuspended in fresh Schneider's medium without probenecid at 2,000–5,000 cells per μl, and kept on ice until in vivo transfer. For transfer, ≈50 nl was injected into the hemocoele of late third instar GFP- y, w67 hosts by using a Picospritzer III (Parker Hannifin, Cleveland, OH). GFP+ transferred cells were visualized in vivo (five repeats) with a MZ FLIII fluorescence stereomicroscope (Leica, Deerfield, IL) and reanalyzed by FACS after cell retrieval from single hosts (two repeats).
In Vivo Bacterial Injection. About 50 nl of Alexa594-labeled heat-killed Escherichia coli (Molecular Probes) was injected into the hemocoele of GAL4-e33c, UAS-gfp larvae by using the Picospritzer III. Hemolymph cells were collected 6 h after infection and analyzed by FACS as described above (three repeats).
Results
Detecting Drosophila Hemocytes Through Intracellular Glutathione. In cell suspensions from the hemolymph and lymph glands (see Methods), hemocytes are outnumbered by dietary yeast and debris of cuticle and fat body. To discriminate live hemocytes from contaminants, we first focused our attention on MCB. MCB enters cells freely and reacts with intracellular GSH, yielding fluorescent glutathione-S-bimane (GSB) adducts (29). GSH is an evolutionarily conserved antioxidant that is crucial to the function of immune cells (30). By FACS, live human leukocytes can be gated within whole blood as a PI (marking dead cells)-negative, GSB-high (PI- GSBhi) fraction (31). To test this method in Drosophila, we took advantage of the enhancer-trap line GAL4-e33c to drive the expression of UAS-gfp in hemocytes (27). Once stained with PI and MCB, the PI- GFP+ (live hemocytes) and PI- GSBhi fractions were found to be identical for both hemolymph and lymph glands suspensions (Fig. 1A). Thus, hemocytes are efficiently and selectively gated as a PI- GSBhi fraction within suspensions from both pools. By contrast, live Drosophila hemocytes could not be efficiently gated based on PI staining, forward scatter, and side scatter only (data not shown).
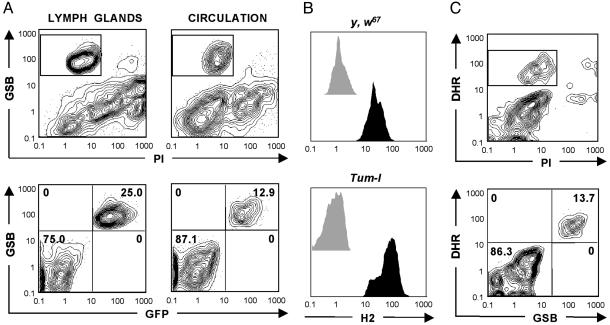
Fig. 1.
Generic methods for FACS-based hemocyte detection. (A) Live hemocytes from the lymph glands and hemolymph of GAL4-e33c UAS-gfp animals can be gated from contaminants as the PI- GSBhi fraction (Upper). Indeed, once the PI- fraction is gated, GSBhi events are identical to GFP+ events, which comprise all hemocytes (Lower). (B) PI- GSBhi live hemocytes from the hemolymph of control (y, w67) and Tum-l animals were analyzed for staining with the proposed pan-hemocyte antibody H2 (black histograms). Unstained controls are also presented (gray histograms). All control and Tum-l hemolymph-derived hemocytes were H2+.(C) Live hemolymph-derived hemocytes from y, w67 animals (Upper) can be gated as a PI- DHRhi fraction (Upper). Once the PI- fraction is gated, DHRhi events are identical to GSBhi events, which comprise all hemocytes (Lower).
By cell sorting (Fig. 2A), we found that the GSB-based detection method is efficient and selective for hemocytes, regardless of the strain and physiological conditions, including infection (see Fig. 3B), heat-shock, and partial GSH depletion (data not shown). As a further validation of the GSB-based method, we observed that PI- GSBhi hemocytes costained with the H2 antibody [a potential pan-hemocyte marker (21)], as illustrated in Fig. 1B for y, w67 control animals and Janus kinase mutants (Tum-l). Tum-l animals bear a dominant gain-of-function mutation of the Janus kinase hopscotch, resulting in cell hyperproliferation and, in the appearance of an activated subset, lamellocytes in the hemolymph (10, 24, 27).
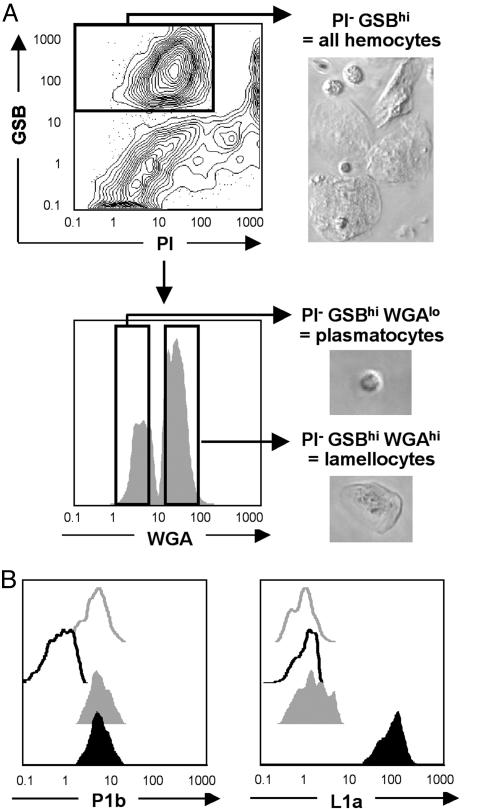
Fig. 2.
Hemocyte subset discrimination and sorting. (A) The lectin WGA can be used to discriminate and sort plasmatocytes (WGAlo) from lamellocytes (WGAhi) within PI- GSBhi live hemolymph-derived hemocytes from Tum-l animals. Sorted plasmatocytes and lamellocytes show typical round and flattened shapes, respectively. (B) Simultaneous staining with antiplasmatocyte (P1b) and antilamellocyte (L1a) antibodies confirms the identity of the Tum-l PI- GSBhi WGAlo (closed gray histograms) and PI- GSBhi WGAhi (closed black histograms) fractions as plasmatocytes and lamellocytes, respectively. Antibody-stained y, w67 (open gray histograms) and unstained Tum-l (open black histograms) PI- GSBhi total hemolymph fractions are also shown. P1b stains y, w67 and Tum-l plasmatocytes and Tum-l lamellocytes, whereas L1a only stains the latter.
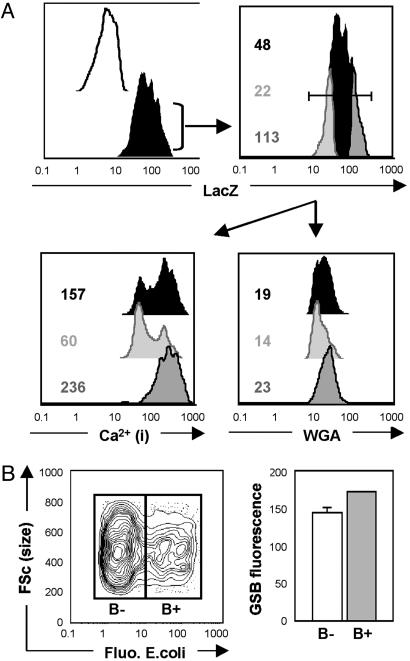
Fig. 3.
Multidimensional analysis of hemocytes. (A) Hemolymph-derived lamellocytes from the Tum-l/11707 strain were gated as a PI- GSBhi WGAhi fraction (see Fig. 2), and intracellular LacZ reporter levels were measured in a fourth fluorescence channel (Upper Left) in a sample stained with the cell-permeable LacZ substrate C12RG (filled black histogram) and an unlabeled control (open black histogram). FACS allows precise quantitative measurement of reporter activity within cells (Upper Right), thus allowing discrete lamellocyte subsets based on LacZ expression levels (whole population, lower 20%, and upper 20% shown as filled black, light gray, and dark gray histograms, respectively) to be further discriminated and analyzed for intracellular Ca2+ levels (Lower Left) and WGA-binding levels (Lower Right). Numerical values indicated are median fluorescence intensities recorded for the given subsets. (B) Live PI- GSBhi hemocytes collected from the hemolymph of GALe33c, UAS-gfp animals 6 h after injection of fluorescent (Fluo.) E. coli were separated as bacteria-negative (B-) and positive (B+) fractions (Left). Median GSB fluorescence (FSc) was then measured (Right, in arbitrary units) in both fractions and found to be significantly higher in the B+ than in the B- fraction (171.8 ± 0.4 vs. 144.0 ± 6.8, respectively; three independent experiments; P = 0.015; ANOVA).
Detecting Drosophila Hemocytes Through Intracellular ROS. One caveat of the GSB-based hemocyte detection method is that it requires two different lasers to excite PI and GSB adducts (Table 1). A valid alternative to this method, requiring only one laser, involves the combined use of PI and dihydrorhodamine 123 (DHR). DHR enters cells freely, reacts with ROS, and localizes in active mitochondria (32). Like GSH, intracellular ROS and mitochondrial activity are crucial to immune cell function (2). In all the strains and conditions tested, PI- GSBhi and PI- DHRhi fractions were identical (Fig. 1C). Hence, live hemocytes can be gated by using either the GSB- or DHR-based generic detection methods. In practice, simple hemocyte studies can rely on basic one-laser FACS machines by using the DHR-based method and a maximum of three colors.
Hemocyte Subset Discrimination and Sorting. More powerful three-laser FACS machines, as used here, enable 6–12 color assays, thus surpassing by far the capabilities of other live-cell analytical methods. With this method, hemocyte subsets can be discriminated and sorted (Fig. 2) based on staining with custom combinations of metabolic indicators, lectins, reporters (Table 1), and antibodies. As a proof of concept, we show that plasmatocyte and lamellocyte subsets present in the hemolymph of Tum-l mutants can be successfully discriminated and sorted (Fig. 2 A) based on their differential reactivity with the lectin wheat germ agglutinin (WGA) (33). We confirm the identity of plasmatocytes and lamellocytes (as defined by WGA staining) by costaining with the subset-specific P1a and L1b antibodies (21) (Fig. 2B). Identical results were obtained with plasmatocyte and lamellocyte subsets present in the hemolymph of the Toll mutant (25) Toll10B (data not shown).
Multidimensional Analysis of Hemocytes by Using Reporter Genes and Metabolic Markers. With regard to GFP (or GFP variants) and LacZ reporters, FACS is superior to microscopy in that it allows more sensitive detection and easy quantitation of expression levels (17). To illustrate this notion, we used the Tum-l/11707 strain (Fig. 3). In this strain, the enhancer-trap LacZ reporter is in the misshapen gene (part of the Jun kinase cascade) and shows strong expression in lamellocytes (26). Multidimensional FACS analysis demonstrates a 5-fold difference in LacZ expression levels between the lower 20% and upper 20% of the lamellocyte fraction (Fig. 3A). Compared with the lower 20%, the upper 20% displayed no change in size (as evaluated by forward scatter; data not shown) but showed increased surface WGA-binding (by 2-fold) and a clear shift toward increased intracellular Ca2+ (median increased by 4-fold), both of which are consistent with increased activation, as shown in mammalian leukocytes in refs. 34 and 35. Hence, discrete lamellocyte subsets can be distinguished by FACS within the Tum-l/11707 strain, showing a positive correlation between LacZ expression, WGA-binding, and intracellular Ca2+.
Besides Ca2+, several other intracellular metabolites of hemocytes can be detected easily by FACS (Table 1), including ROS/reactive nitrogen species, which are given strong attention for their role in innate immunity (36). GSH, itself, while serving as a detection marker, can also be quantified in hemocytes after MCB-staining (29). For example, we show in Fig. 3B that 6 h after injection of fluorescent heat-killed E. coli, the median GSB fluorescence is significantly increased by ≈20% in bacteria-positive compared with bacteria-negative plasmatocytes. This finding is consistent with data on mammalian macrophages that show intracellular GSH levels increasing during phagocytosis (37). Thus, Drosophila hemocytes, with the help of FACS, may be used to explore the complex in vivo regulation of GSH levels, which in turn exert pivotal control on gene transcription and activation and the apoptosis of immune cells (30).
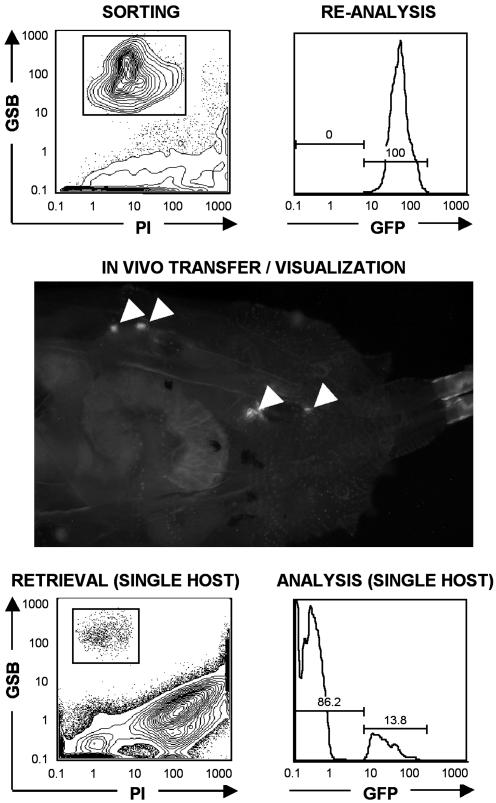
In Vivo Transfer of Sorted Hemocytes and Reanalysis from Single Host. One important asset of FACS-sorting is the ability to use live sorted fractions for further in vitro or in vivo assays. To illustrate this ability, we sorted GFP+ hemocytes from His::GFP (28) and Tum-l; His::GFP animals and used them for in vivo transfer into GFP- y, w67 hosts (Fig. 4). Transferred GFP+ cells were visible within GFP- hosts several days after transfer. Remarkably, our approach also allowed for the successful retrieval and reanalysis of transferred cells from single hosts. Such single-animal assays (also validated for lymph glands; data not shown) are enabled by the numerous yeast cells present in suspensions. Yeast cells are excellent “carriers” for hemocytes, because they minimize hemocyte loss during staining/acquisition and are efficiently excluded at analysis.
Fig. 4.
In vivo transfer of sorted hemocytes and reanalysis from a single host. A pure fraction of live GFP-labeled hemocytes was sorted from the hemolymph of Tum-l; His::GFP animals (Top Left), reanalyzed (Top Right), and visualized in vivo (arrows) 3 d after transfer into a GFP- y, w67 host (Middle). After FACS analysis of PI- GSBhi live hemocytes retrieved from the hemolymph of a single host (Bottom Left), donor-derived (GFP+) and host-derived (GFP-) fractions were successfully discriminated (Bottom Right). Note the high fraction of events outside the PI- GSBhi gate in the y, w67 host (3 d after transfer; animal close to pupal stage; Bottom Left), compared with the Tum-l donor (third instar larva; Top Left), reflecting the higher amount of debris present in the hemolymph as animals approach the pupal stage (6).
Discussion
Collectively, our results establish the feasibility of multidimensional FACS analysis, sorting, and in vivo transfer of Drosophila hemocytes. We introduced an array of FACS assays that enable the selective detection of hemolymph and lymph glands hemocytes down to one animal and yield efficient single-cell measurement of multiple surface and intracellular molecules of interest, including GFP and LacZ reporters. In essence, FACS-based analyses and sorts will allow more elaborate and precise phenotypic and functional analyses of freshly harvested hemocytes.
Our results also illustrate how nonantibody probes can be used advantageously for hemocyte studies focusing on cellular function, most notably with regard to conserved metabolic pathways (e.g., the Ca2+, GSH, ROS, and reactive nitrogen species pathways) and reporter constructs. By using the C12RG (595-nm excitation) LacZ substrate, LacZ and GFP reporters can even be quantified simultaneously within hemocytes. Hence, this approach will facilitate the functional screening of existing GFP- and/or LacZ-transgenic line collections (23). These generic methods for hemocyte detection, analysis, and sorting will likely apply to other species, including mosquitoes, for which FACS-based hemocyte assays could help study important disease-related issues (38).
Of course, it is also desirable that more antihemocyte antibodies are developed. For example, “phosphoantibodies” to activated Drosophila kinases would be advantageous for single-cell FACS analyses of intracellular signaling, as shown with mammalian leukocytes (20). New antibodies may help tailor specific FACS assays for less prominent subsets: e.g., crystal cells, implicated in coagulation reactions and prohemocytes, which correspond to hematopoietic stem/precursor cells (4, 10). Antibodies may also help resolve discrete subsets of plasmatocytes (as suggested in ref. 26) and lamellocytes (as suggested in this study).
The ability to transfer sorted cells, rather than whole tissues [e.g., lymph glands or pseudotumors (24)] and to trace their fate after transfer into single hosts should facilitate developmental studies of hemocyte subsets and pools. In particular, sequential in vivo transfers, as performed in mammals, could help characterize Drosophila hematopoietic stem/precursor cells. Genomic (39, 40) and proteomic in vitro studies of hemocyte development and function will also considerably gain in accuracy by using pure, sorted (rather than mixed), native fractions. In addition, hemocyte-sorting could lead to useful in vitro assays of fresh cells, such as clonal, secretory, and differentiation assays, as used routinely in mammalian immunology. Sorted fractions may also give rise to novel hemocyte cell lines, of which only a few are currently available (41). Finally, the successful application of FACS technology to Drosophila hemocytes could pave the way for a more medically oriented use of this model: e.g., for high-throughput screening (42) of candidate drugs with potential effects on normal hematopoiesis, hyperproliferation, and innate immune responses.
Supplementary Material
Acknowledgments
We thank Leonore A. Herzenberg, M. Krasnow, D. Parks, M. Scott, and R. Khush for advice; D. Schneider for instruction on and the use of the cell injection setup; I. Ando for the gift of H2, L1a, and P1b hybridoma supernatants; U. Theopold for the gift of mbn-2 cells; and the Bloomington Stock Center for fly stocks. This work was supported by National Institutes of Health Grants R01 CA56509 and R01 CA90307 (to J.S.L.) and R01 CA85949 (to L.A.H.).
Abbreviations: DHR, dihydrorhodamine 123; FACS, fluorescence-activated cell sorting; GSB, glutathione-S-bimane; GSH, glutathione; LacZ, β-galactosidase; MCB, monochlorobimane; PI, propidium iodide; ROS, reactive oxygen species; UAS, upstream activating sequence; WGA, wheat germ agglutinin.
References
- 1.Lavine, M. D. & Strand, M. R. (2002) Insect Biochem. Mol. Biol. 32, 1295-1309. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann, J. A. & Reichhart, J. M. (2002) Nat. Immunol. 3, 121-126. [DOI] [PubMed] [Google Scholar]
- 3.Boutros, M., Agaisse, H. & Perrimon, N. (2002) Dev. Cell 3, 711-722. [DOI] [PubMed] [Google Scholar]
- 4.Meister, M. & Lagueux, M. (2003) Cell Microbiol. 5, 573-580. [DOI] [PubMed] [Google Scholar]
- 5.Fessler, L. I., Nelson, R. E. & Fessler, J. H. (1994) Methods Enzymol. 245, 271-294. [DOI] [PubMed] [Google Scholar]
- 6.Lanot, R., Zachary, D., Holder, F. & Meister, M. (2001) Dev. Biol. 230, 243-257. [DOI] [PubMed] [Google Scholar]
- 7.Lebestky, T., Chang, T., Hartenstein, V. & Banerjee, U. (2000) Science 288, 146-149. [DOI] [PubMed] [Google Scholar]
- 8.Fossett, N. & Schulz, R. A. (2001) Differentiation 69, 83-90. [DOI] [PubMed] [Google Scholar]
- 9.Remillieux-Leschelle, N., Santamaria, P. & Randsholt, N. B. (2002) Genetics 162, 1259-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathey-Prevot, B. & Perrimon, N. (1998) Cell 92, 697-700. [DOI] [PubMed] [Google Scholar]
- 11.Dearolf, C. R. (1998) Biochim. Biophys. Acta 1377, M13-M23. [DOI] [PubMed] [Google Scholar]
- 12.Han, Z. S., Enslen, H., Hu, X., Meng, X., Wu, I. H., Barrett, T., Davis, R. J. & Ip, Y. T. (1998) Mol. Cell. Biol. 18, 3527-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duvic, B., Hoffmann, J. A., Meister, M. & Royet, J. (2002) Curr. Biol. 12, 1923-1927. [DOI] [PubMed] [Google Scholar]
- 14.Sorrentino, R. P., Carton, Y. & Govind, S. (2002) Dev. Biol. 243, 65-80. [DOI] [PubMed] [Google Scholar]
- 15.Cho, N. K., Keyes, L., Johnson, E., Heller, J., Ryner, L., Karim, F. & Krasnow, M. A. (2002) Cell 108, 865-876. [DOI] [PubMed] [Google Scholar]
- 16.Celniker, S. E. & Rubin, G. M. (2003) Annu. Rev. Genomics Hum. Genet. 4, 89-117. [DOI] [PubMed] [Google Scholar]
- 17.Herzenberg, L. A., Parks, D., Sahaf, B., Perez, O. & Roederer, M. (2002) Clin. Chem. 48, 1819-1827. [PubMed] [Google Scholar]
- 18.Baumgarth, N. & Roederer, M. (2000) J. Immunol. Methods 243, 77-97. [DOI] [PubMed] [Google Scholar]
- 19.De Rosa, S. C., Herzenberg, L. A. & Roederer, M. (2001) Nat. Med. 7, 245-248. [DOI] [PubMed] [Google Scholar]
- 20.Perez, O. D. & Nolan, G. P. (2002) Nat. Biotechnol. 20, 155-162. [DOI] [PubMed] [Google Scholar]
- 21.Kurucz, E., Zettervall, C. J., Sinka, R., Vilmos, P., Pivarcsi, A., Ekengren, S., Hegedus, Z., Ando, I. & Hultmark, D. (2003) Proc. Natl. Acad. Sci. USA 100, 2622-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holz, A., Bossinger, B., Strasser, T., Janning, W. & Klapper, R. (2003) Development (Cambridge, U.K.) 130, 4955-4962. [DOI] [PubMed] [Google Scholar]
- 23.Tickoo, S. & Russell, S. (2002) Curr. Opin. Pharmacol. 2, 555-560. [DOI] [PubMed] [Google Scholar]
- 24.Hanratty, W. P. & Ryerse, J. S. (1981) Dev. Biol. 83, 238-249. [DOI] [PubMed] [Google Scholar]
- 25.Qiu, P., Pan, P. C. & Govind, S. (1998) Development (Cambridge, U.K.) 125, 1909-1920. [DOI] [PubMed] [Google Scholar]
- 26.Braun, A., Lemaitre, B., Lanot, R., Zachary, D. & Meister, M. (1997) Genetics 147, 623-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison, D. A., Binari, R., Nahreini, T. S., Gilman, M. & Perrimon, N. (1995) EMBO J. 14, 2857-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarkson, M. & Saint, R. (1999) DNA Cell Biol. 18, 457-462. [DOI] [PubMed] [Google Scholar]
- 29.Anderson, M. T., Roederer, M., Tjioe, I., Herzenberg, L. A. & Herzenberg, L. A. (1996) in Handbook of Experimental Immunology, eds. Herzenberg, L. A., Weir, D. M., Herzenberg, L. A. & Blackwell, C. (Blackwell Scientific, Boston), Vol. 2, pp. 54.1-54.9. [Google Scholar]
- 30.Sies, H. (1999) Free Radic. Biol. Med. 27, 916-921. [DOI] [PubMed] [Google Scholar]
- 31.Scott, R. B., Collins, J. M., Matin, S., White, F. & Swerdlow, P. S. (1990) J. Clin. Lab. Anal. 4, 324-327. [DOI] [PubMed] [Google Scholar]
- 32.Vowells, S. J., Sekhsaria, S., Malech, H. L., Shalit, M. & Fleisher, T. A. (1995) J. Immunol. Methods 178, 89-97. [DOI] [PubMed] [Google Scholar]
- 33.Rizki, T. M. & Rizki, R. M. (1983) Science 220, 73-75. [DOI] [PubMed] [Google Scholar]
- 34.Dewitt, S., Laffafian, I., Morris, M. R. & Hallett, M. B. (2003) Methods Mol. Biol. 225, 47-59. [DOI] [PubMed] [Google Scholar]
- 35.Timoshenko, A. V., Andre, S., Kayser, K. & Gabius, H. J. (2001) Anticancer Res. 21, 3453-3456. [PubMed] [Google Scholar]
- 36.Nappi, A. J. & Ottaviani, E. (2000) Bioessays 22, 469-480. [DOI] [PubMed] [Google Scholar]
- 37.Seres, T., Knickelbein, R. G., Warshaw, J. B. & Johnston, R. B., Jr. (2000) J. Immunol. 165, 3333-3340. [DOI] [PubMed] [Google Scholar]
- 38.Dimopoulos, G. (2003) Cell Microbiol. 5, 3-14. [DOI] [PubMed] [Google Scholar]
- 39.Asha, H., Nagy, I., Kovacs, G., Stetson, D., Ando, I. & Dearolf, C. R. (2003) Genetics 163, 203-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Gregorio, E., Spellman, P. T., Rubin, G. M. & Lemaitre, B. (2001) Proc. Natl. Acad. Sci. USA 98, 12590-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gateff, E. (1997) in Molecular Mechanisms of Immune Responses in Insects, eds. Brey, P. T. & Hultmark, D. (Chapman & Hall, London), pp. 189-210.
- 42.Wedemeyer, N. & Potter, T. (2001) Clin. Genet. 60, 1-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.