Abstract
The pancreatic islet is necessary for maintaining glucose homeostasis. Within the pancreatic islet, the homeodomain protein Nkx2.2 is essential for the differentiation of all insulin-producing β cells and a subset of glucagon-producing α cells (1). Mice lacking Nkx2.2 have relatively normal sized islets, but a large number of cells within the mutant islet fail to produce any of the four major islet hormones. In this study we demonstrate that Nkx2.2 mutant endocrine cells have been replaced by cells that produce ghrelin, an appetite-promoting peptide predominantly found in the stomach. Intriguingly, normal mouse pancreas also contains a small population of ghrelin-producing cells, defining a new islet “ε” cell population. The expansion of ghrelin-producing cells at the expense of β cells may be a general phenomenon, because we demonstrate that Pax4 mutant mice display a similar phenotype. We propose that insulin and ghrelin cells share a common progenitor and that Nkx2.2 and Pax4 are required to specify or maintain differentiation of the β cell fate. This finding also suggests that there is a genetic component underlying the balance between insulin and ghrelin in regulating glucose metabolism.
Keywords: islet, Nkx2.2, Pax4, cell-type specification
The pancreas plays a central role in energy balance and nutrient regulation through the function of two distinct populations of cells: the exocrine cells secrete digestive enzymes through a ductal system into the gastrointestinal tract, and the endocrine cells secrete hormones into the bloodstream. The islets of Langerhans are the functional units of the endocrine pancreas and are traditionally thought to be comprised of four distinct cell types: α, β, δ, and pancreatic polypeptide (PP) cells (reviewed in ref. 2). The mature cell types are defined by their unique hormone expression: glucagon, insulin, somatostatin, and PP, respectively. The β cells are the most abundant cell type, constituting >75% of the islet, and are centrally located within the islet. α, δ, and PP cells are intermixed at the periphery of the islet, with α cells representing the majority of the remaining cell types. More recently, a fifth peptide hormone has been identified in the human islet. The hormone ghrelin is produced mainly in the stomach and functions to increase secretion of growth hormone and regulate food intake and energy balance (3). Expression of ghrelin in the human islet remains somewhat controversial, because it has variably been reported to be in the α cells (4), β cells (5), or in a unique islet cell type (6). The function of ghrelin within the islet is also unknown, but it may have a paracrine role in regulating insulin secretion.
Extensive analyses of the pancreas have led to a tremendous understanding of islet morphology, islet function, and the physiological modulation of islet function and insulin secretion. More recently, there has been increasing interest in understanding the molecular mechanisms underlying embryonic pancreas development as a way to develop therapies to treat diabetes or to discover diagnostic tools for pancreatic cancer. This has led to a surge in the identification of many of the signaling pathways and transcription factors that are critical for the specification, differentiation, and maintenance of the developing pancreas (reviewed in refs. 7 and 8). Although mouse knockout studies have allowed significant progress in identifying many of the intrinsic factors regulating the specification and differentiation of each islet cell type, including Pax4, Pax6, Nkx6.1, Ngn3, Hlxb9, Pdx1, Nkx2.2, and NeuroD, the molecular mechanisms underlying the individual factors and their respective roles in cell type specification remain, for the most part, poorly understood (1, 9–18). Furthermore, islet cell lineage and the roles of individual transcription factors in directing the lineage remain obscure. It is likely that a combinatorial code of transcription factor expression (selective maintenance, activation, or repression) in the different cell types will determine islet cell type identity.
Of the transcription factors known to be involved in islet cell development, Nkx2.2, a member of the NK2 class of homeodomain transcription factors, plays a unique role in pancreatic β cell differentiation. Nkx2.2 is expressed throughout the epithelium at the onset of pancreas development, between embryonic day 8.5 (e8.5) and e9.0. As organogenesis proceeds, Nkx2.2 becomes progressively restricted to all β cells and a subset of α and PP cells. Targeted disruption of the Nkx2.2 gene in mice resulted in a complete lack of insulin-producing β cells and reduced numbers of α and PP cells (1). Notably, this is the only transcription factor knockout to eliminate 100% of the insulin-producing β cells. Furthermore, the mutant islets accumulated a large population of endocrine cells that did not produce any of the four major islet hormones.
To gain a greater understanding of the molecular function of Nkx2.2 in the islet, we undertook a comparative microarray analysis to characterize the large population of non-insulin-producing endocrine cells and to potentially identify downstream targets of Nkx2.2 in the pancreas. These studies allowed us to determine that Nkx2.2 and Pax4 are independently required to specify β cell fate in the islet and that, in the absence of either gene, β cells are replaced by cells producing the peptide hormone ghrelin. Furthermore, we demonstrate that the ghrelin cells define a unique population of cells within the normal mouse islet.
Methods
Animals. Nkx2.2 and Pax4 heterozygous mice were generated by homologous recombination as described (1, 9). Nkx2.2+/- animals were maintained on a Swiss Black background, and Pax4+/- animals were maintained on an NMRI background. Genotyping of mice and embryos was performed as described (1, 9).
Microarray Analysis. Pancreas tissue was collected from staged embryos at e12.5–e13.5, e15.5, and e18.5. For each stage, total pancreata were collected, stored individually in RNase Later (Ambion), and genotyped by PCR (1). For each embryonic stage, equal numbers of wild-type and mutant tissues were individually pooled: e12.5–e13.5, 32 -/- and 35 +/+ or +/-; e15.5, 25 -/- and 25 +/+ or +/-; e18.5, 4 -/- and 5 +/+ or +/-. Extensive analyses have revealed no phenotype associated with the heterozygous Nkx2.2 mice; therefore, for the purpose of this study, heterozygous (+/-) mice were included in the “wild-type” pools. Samples of total RNA were processed for microarray analysis by using the Mu19k A gene chip according to manufacturer's instructions (Affymetrix). Duplicate samples were analyzed for the e12.5 time point. Chip performance, background levels, and presence/absence calls were assessed by using microarray suite software (Affymetrix).
Isolation of m46 cDNA. Pancreata were dissected from ≈25 e12.5–e13.5 outbred Swiss Black wild-type embryos. RNA was prepared by using an RNAqueous Kit (Ambion). A pool of double-stranded cDNA was generated by using the SMART cDNA Kit (Clontech). PCR primers flanking the m46 cDNA (5′ATGCTGTCTTCAGGCACCATC3′ and 5′TGTCCGTGGTTACTTGTCAG3′) were used to PCR-amplify full-length cDNA from the cDNA pool. The PCR product was cloned into a Topo II Blunt vector (Invitrogen) and sequence-verified.
Processing of Embryos and Pancreatic Tissue. Tissues of dissected embryos or pancreata of newborn mice were fixed overnight by immersion in 4% paraformaldehyde at 4°C. Tissues were subjected to cryoprotection in 30% sucrose in PBS overnight at 4°C and were then embedded in tissue-freezing medium (OCT, VWR International). Tissues were cut into 8- to 10-μm sections with a cryostat. At least three embryos per stage were analyzed. For postnatal analysis, n = 7.
Immunohistochemistry. Frozen sections were used for all immunohistochemical assays. Primary antibodies used were rabbit anti-amylase (Sigma, 1:1,000), guinea pig anti-insulin (Linco Research, St. Charles, MO; 1:5,000), guinea pig anti-glucagon (Linco Research; 1:10,000), rabbit anti-mouse ghrelin and rabbit anti-human ghrelin (Phoenix Pharmaceuticals, Belmont, CA; 1:500), goat anti-ghrelin (Santa Cruz Biotechnology; 1:1,000), rabbit anti-chromogranin A (Zymed; 1:100), mouse anti-somatostatin (DAKO; 1:500), rabbit anti-PP (Zymed; 1:200), rabbit anti-islet amyloid polypeptide/amylin (Phoenix Pharmaceuticals; 1:200), rabbit anti-Pdx1 (Chemicon; 1:1,000), rabbit anti-Nkx6.1 (from J. Jansen, University of Colorado Health Sciences Center; 1:500), rabbit anti-Pax6 (Chemicon; 1:250), and guinea pig anti-Nkx2.2 (from B.S.-P., 1:100). Secondary antibodies used at 1:400 were CY2-donkey anti-goat, Texas red goat anti-rabbit, and Texas red goat anti-guinea pig (Jackson ImmunoResearch), Alexa 488 goat anti-rabbit, and Alexa 594 goat anti-rabbit (Molecular Probes). Initial experiments with each antibody were performed with and without the inclusion of primary antibody. Due to the controversy associated with ghrelin expression in the human islet, all expression was confirmed by RNA in situ analysis. Images were obtained with a BX51 microscope (Olympus, Melville, NY) and a Penguin 600CL camera (Pixera, Los Gatos, CA). photoshop 7.0 (Adobe Systems, Mountain View, CA) was used to process the images.
RNA in Situ Analysis. RNA was analyzed at the following stages: e12.5 (n = 3), e14.5 (n = 2), e15.5 (n = 5), e16.5 (n = 2), e18.5 (n = 4), neonates (n = 7). Nonradioactive in situ hybridization was performed on 8- to 10-μm frozen sections as described (19) with the following modifications. Prehybridization was performed in a humidified chamber at 55°C for 1–2 h with 500 μl of prehybridization buffer (50% formamide/5× SSC/50 μg/ml yeast tRNA/1% SDS/50 μg/ml heparin) per slide for 1–2 h. The hybridization solution was prepared by adding 1 ng of digoxigenin-labeled cRNA per 100 μl of prehybridization buffer. The sections were covered with 20- × 40-mm coverslips and not sealed. Posthybridization, slides were washed as described and then incubated in 1× MAB (100 mM maleic acid/150 mM NaC1, pH 7.5) for 5 min at room temperature before blocking [2% blocking reagent (Roche)/10% heat-inactivated sheep serum/0.1% Tween 20 in 1× MAB] at room temperature for 1 h. Blocking solution was replaced with fresh blocking solution containing α-digoxigenin antibody (Fab fragments; Roche) at a concentration of 1:5,000 and incubated overnight at 4°C. The slides were washed three times in 1× MAB with 0.1% Tween 20 for 15 min at room temperature, then washed in ddH2O with 0.1% Tween 20 for 20 min at room temperature. The bound probes were visualized with an alkaline phosphatase-conjugated anti-digoxigenin Fab fragment (Roche).
Results
Microarray Analysis of Nkx2.2-/- Pancreas. Our previous studies demonstrated that the Nkx2.2 homeodomain transcription factor is required for islet cell development and differentiation. Nkx2.2-null mice completely lack insulin-producing β cells and have reduced numbers of α cells (1) (compare Fig. 1 a and b with f and g). Furthermore, we found that the mutant islets accumulate a large population of endocrine cells (as defined by synaptophysin and chromogranin A expression), hereafter referred to as ε cells, which do not produce any of the four islet hormones (1). To characterize the gene expression profile of ε cells, we used the Affymetrix Mu19k A gene chip set to compare Nkx2.2 mutant and wild-type pancreas at three embryonic ages: e12.5, e15.5, and e18.5 (see Methods). From this analysis, we discovered a gene, m46 gastric peptide (GenBank accession no. AJ243503), that was up-regulated at least 20- to 30-fold in the Nkx2.2 mutant pancreas at all ages tested. Remarkably, m46 encodes mouse ghrelin, a recently identified 28-aa growth hormone peptide secreted predominantly from the gastric pits of the stomach to stimulate appetite (3).
Fig. 1.
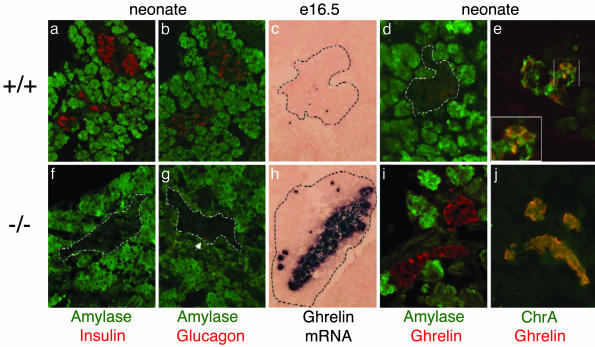
Ghrelin mRNA and protein are up-regulated in the islets of Nkx2.2 mutant mice. Immunohistochemical analysis of sectioned wild-type pancreas (a–e) or mutant pancreas (f–j) is shown. All data are from neonatal pancreata except c and h, which show pancreata from e16.5 embryos. Dashed lines delineate the pancreas at e16.6 (c and h) and an islet in neonates (d, f, and g). (a and f) Expression of amylase (green) and insulin (red). (b and g) Expression of amylase (green) and glucagon (red). (c and h) RNA in situ analysis of ghrelin mRNA in e16.5 pancreas. Incubation time for wild-type and mutant samples was identical. (d and i) Expression of ghrelin protein (red) and amylase (green). (e and j) Expression of ghrelin (red) and neuroendocrine marker chromogranin A (green).
To verify the microarray, we cloned full-length ghrelin cDNA from an embryonic cDNA library (see Methods). RNA in situ analysis using the full-length cRNA was performed on tissue sections of wild-type and mutant mice at several embryonic and postnatal stages. As early as e12.5, and at all subsequent ages tested, we confirmed that ghrelin mRNA was greatly up-regulated in the islets of the Nkx2.2 mutant mice (compare Fig. 1 c and h; data not shown). Ghrelin protein levels are correspondingly up-regulated in neonatal Nkx2.2 mutant pancreata (Fig. 1i), which showed an overabundance of ghrelin-producing cells; >90% of the mutant mature islet cells produce ghrelin (n = 7). Thus, we have uncovered the identity of the ε cells in the Nkx2.2 mutant islet, where it appears that ghrelin-producing cells have replaced the insulin-producing β cells and perhaps subsets of α and PP cells.
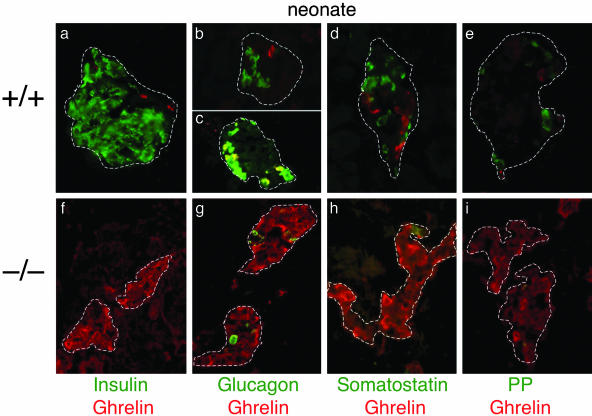
Ghrelin Cells Represent a Previously Undescribed Cell Population Within the Islet. At the time of our discovery, ghrelin had only recently been identified as a gastric peptide important for appetite regulation; very little was known about its expression and function in other tissues. Our initial analysis of ghrelin mRNA and protein expression revealed ghrelin to be normally expressed in a small population of cells within the wild-type islet (Fig. 1 c and d). To further characterize ghrelin expression within the wild-type islet and to determine whether these cells represent an islet cell population, we analyzed both RNA and protein expression at several developmental stages. These studies determined that ghrelin is expressed in the pancreas as early as e10.5 and appears to peak around e15.5 (Figs. 1, 2, 3 and data not shown). In neonates, ghrelin can be detected in a small number of cells located around the periphery of the islet and is absent from amylase-expressing exocrine cells (Fig. 1d). Ghrelin continues to be expressed at very low levels in cells at the periphery of the mouse islet into adulthood (data not shown). Coexpression of islet ghrelin with chromogranin A confirms that ghrelin cells are typical endocrine islet cells (Fig. 1 e and j). Consistent with this observation, pancreatic ghrelin cells appear to be derived from the Ngn3-dependent endocrine cell lineage (G. Gradwohl, personal communication). To further characterize ghrelin expression in the wild-type islet, we examined the expression of ghrelin with respect to the four islet hormones (Fig. 2 a–e). Due to the controversy associated with ghrelin expression in the human islet, we performed double immunofluorescence (Fig. 2), as well as single immunofluorescence, on adjacent sections (data not shown) using three different antibodies to confirm ghrelin expression in the mouse islet. From our analysis it is clear that mouse ghrelin is not coexpressed with insulin, somatostatin, or PP (Fig. 2 a, d, and e). Interestingly, in neonatal pancreata ≈30–33% of the ghrelin cells coexpress glucagon, whereas two-thirds (67%) of the ghrelin cells represent a unique islet cell population (n = 4 animals; average of 15 sections per animal). Approximately 80% of glucagon-producing α cells do not express ghrelin. We detected ghrelin single-positive cells and glucagon/ghrelin-coproducing cells in the pancreas as early as e10.5 (Fig. 3b). Even at these earliest stages of pancreas development, there was no evidence of insulin/ghrelin-coproducing cells.
Fig. 2.
Ghrelin-expressing cells define a unique subset of islet cells in the mouse that do not express the four islet hormones; in the absence of Nkx2.2, ghrelin-expressing cells replace insulin- and glucagon-expressing cell populations. Shown is immunofluorescence analysis of sectioned wild-type (a–e) and mutant (f–i) neonatal pancreas. Dashed lines delineate the islets. Red, ghrelin; green, islet hormones. (a and f) Insulin. (b, c, and g) Glucagon. (d and h) Somatostatin. (e and i) PP.
Fig. 3.
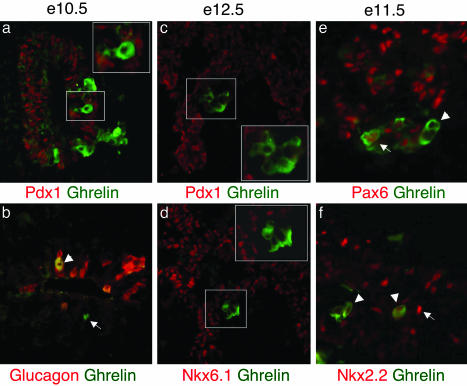
Transcription factor profile of ghrelin-positive cells in wild-type islets. Immunofluorescence analysis of sectioned embryonic pancreas shows ghrelin (green), transcription factors, and glucagon (red). (a and b) Ghrelin expression in wild-type pancreas at e10.5. (a) Ghrelin is not coexpressed with Pdx1. (b) Ghrelin-positive, glucagon-positive cell (white arrowhead) and ghrelin-positive, glucagon-negative cell (white arrow). (c and d) e12.5: ghrelin is not expressed with Pdx1 (c) or Nkx6.1 (d). (e and f) e11.5: Pax6 is expressed in some, but not all, ghrelin cells (e). Nkx2.2 is expressed at low levels or not at all in ghrelin-positive cells (arrowhead) (f). Nkx2.2-high-expressing cells do not coexpress ghrelin (white arrowhead).
The Ghrelin (ε) Cell Type Represents the Majority of Cells Within the Nkx2.2-/- Islet. To characterize the expanded ghrelin population in the Nkx2.2 mutant islet with respect to glucagon and the other islet hormones, we performed immunohistochemical analysis of the mutant islets. Similar to the wild-type islet, ghrelin-producing cells in the Nkx2.2 mutant islet do not coexpress insulin, somatostatin, or PP (Fig. 2 f, h, and i). However, unlike its expression in wild-type islets, none of the ghrelin-producing cells in the Nkx2.2 mutant coexpress glucagon (Fig. 2g). This would suggest that the ghrelin (ε) cell type is expanded, whereas the glucagon/ghrelin double-positive cells are absent from the Nkx2.2 mutant islet. Therefore, we conclude that, in normal islets, a population of glucagon-expressing α cells coexpress ghrelin, but approximately two-thirds of ghrelin-expressing cells define a new endocrine islet ε cell population. Moreover, in the Nkx2.2 mutant islet, the ghrelin-producing ε cell population has been drastically expanded at the expense of insulin- and glucagon-producing cells.
Transcription Factor Profile of Ghrelin Cells in the Islet Is Different from β Cells. The identification of ghrelin-producing ε cells as a unique cell population within the islet prompted us to characterize their normal transcription factor profile to determine their relative position in the islet cell lineage and to establish whether ghrelin cells are regulated by a similar complement of transcription factors as the other islet cell types. Pdx1 is required for pancreatic bud expansion and insulin gene transcription (16, 17, 20, 21); it is detected throughout the pancreatic epithelium at the onset of pancreas formation but becomes restricted predominantly to β cells upon endocrine cell differentiation (22, 23). Nkx6.1 is also widely expressed in the pancreatic epithelium but becomes restricted exclusively to β cells (11, 24). In e10.5 wild-type islets, when immature β cells first form, and at e12.5, when a wave of mature β cell production occurs, ghrelin-producing cells are present, but they do not express Pdx1 or Nkx6.1 (Fig. 3 a, c, and d and data not shown). This would suggest that ghrelin cells normally have a transcription factor profile that is distinct from β cells. Furthermore, in the Nkx2.2 mutant islet, Nkx6.1 expression becomes extinguished and Pdx1 expression is present but is down-regulated (ref. 1 and data not shown). This would indicate that the ε cells expressing ghrelin in the mutant islet are unlike β cells and may result from the expansion of a unique cell-type population.
Pax6 is normally expressed in all endocrine cell types and is positively regulated by Nkx2.2. Microarray analysis of Nkx2.2 mutant islets showed that Pax6 mRNA is decreased at all three ages analyzed (C.L.P. and L.S., unpublished data), and we have recently demonstrated that Pax6 protein is absent from the Nkx2.2 mutant islet (25). Interestingly, in wild-type mice at all ages tested there appear to be two populations of ghrelin cells with respect to Pax6 expression: the majority of ghrelin cells express Pax6, whereas a small number are negative for Pax6 (Fig. 3e). Due to the unavailability of appropriate antibodies, we were unable to determine how Pax6 expression distributes between the three glucagon/ghrelin cell types.
Nkx2.2 Is Normally Down-Regulated in Differentiated Ghrelin Cells. We have demonstrated that Nkx2.2 is highly expressed in β cells and is required for their differentiation (1). In this study we demonstrate that, in the absence of Nkx2.2, there is a loss of insulin-producing β cells and a corresponding increase in ghrelin-expressing ε cells. If the regulation of Nkx2.2 expression in an islet progenitor determines the choice between becoming an insulin-producing β cell (Nkx2.2-high) or a ghrelin-producing ε cell (Nkx2.2-low/off), we would expect Nkx2.2 to be down-regulated in cells that express ghrelin. Consistent with this hypothesis, between e10.5 and e14.5 in wild-type islets, ghrelin cells do not coexpress high levels of Nkx2.2, although relatively low levels of Nkx2.2 can be detected (Fig. 3f and data not shown).
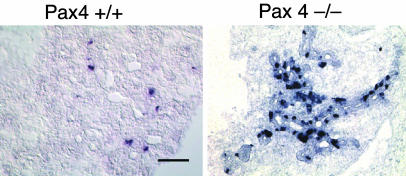
Pax4-/- Islets also Have an Increase in the Ghrelin Cell Population. The replacement of insulin-producing β cells by ghrelin-producing ε cells in the Nkx2.2 knockout mice may be indicative of a characteristic cell fate conversion that occurs whenever β cell differentiation is blocked. For example, if an active developmental switch is necessary to trigger the choice to become a β cell versus a ghrelin cell, then it might be expected that other mutants or conditions that are defective in β cell differentiation will default to a ghrelin cell fate. To determine whether or not the switch from a β cell to ghrelin cell fate is unique to the Nkx2.2 mutant, we characterized the expression of ghrelin in Pax4 mutant islets. Pax4 is expressed in differentiating β cells, and, in the absence of Pax4, 90% of insulin-producing β cells fail to form (9, 25). Similar to the Nkx2.2 knockout, Pax4 mutant islets retain a number of endocrine-committed cells that do not produce insulin, glucagon, somatostatin, or PP. Microarray analysis comparing gene expression between the Pax4 wild-type and mutant pancreas revealed a 10-fold increase in ghrelin expression in the Pax4 mutant islet (L.E. and B.S.-P., unpublished data). Additional RNA in situ and immunohistochemistry analyses confirmed that from e14.5 through birth there is a significant expansion of the number of ghrelin cells in Pax4 mutant islets (Fig. 4 and data not shown). Interestingly, ghrelin cells within the Pax4 mutant islet are not increased to the same extent seen in Nkx2.2 knockout mice, which may reflect the fact that Pax4 mutants maintain a small population of insulin-producing β cells, and there is no apparent effect on the formation of α or PP cells. In addition, unlike the Nkx2.2 mutant, Pax6 expression is unaffected in the Pax4 mutant islet; therefore, Pax6 may compensate to some extent for the loss of Pax4 (25). In the Nkx6.1 mutant, there is a 90% reduction in β cells, but, unlike the Nkx2.2 and Pax4 mutants, β cell differentiation appears to be blocked at a later developmental stage, during the secondary transition. In these mice, islet size is significantly decreased, and there is no evidence of ε-type endocrine cells (11). Consistently, there is no observable expansion of ghrelin cells in the Nkx6.1 mutant islets (M. Sander, personal communication).
Fig. 4.
Pax4 mutant islets also have increased levels of ghrelin-expressing ε cells. Shown is nonradioactive in situ analysis of ghrelin mRNA on sections of e14.5 pancreas. (a) A small number of ghrelin-expressing cells are normally expressed in the wild-type pancreas at e14.5. (b) The number of ghrelin-expressing cells is greatly increased in Pax4 mutant islets, where there is a concomitant 90% reduction of insulin-producing cells (ref. 25 and data not shown).
Discussion
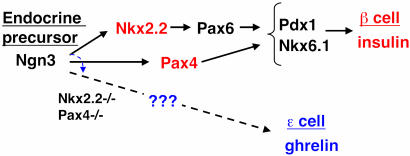
Our findings from this study suggest a model in which high levels of both Nkx2.2 and Pax4 are independently required to specify or maintain the islet β cell fate (schematized in Fig. 5). In the absence of either factor, β cells fail to form and become replaced with ghrelin-producing ε cells, a cell type within the embryonic mouse islet. Furthermore, it has recently been demonstrated that loss of Pax6 results in a similar phenotype: reduction of β cells and a corresponding expansion of ghrelin-producing ε cell numbers (P. Serup, personal communication). Because Pax6 expression depends on Nkx2.2 (25), it is likely that Nkx2.2 acts upstream of Pax6 in the same pathway to regulate the β and ε cell fate choice. The importance of these findings points to the need for extensive lineage analysis to determine whether β cells and ghrelin-producing ε cells arise from a common precursor that becomes diverted to the ghrelin cell fate in the absence of Nkx2.2 or Pax4. Alternatively, Nkx2.2 and Pax4 may promote β cell differentiation, and the absence of β cell formation causes a non-cell-autonomous effect on the expansion of ghrelin-producing ε cells from an independent progenitor population. Experiments are ongoing to resolve these issues. In the Nkx2.2 mutant, it is also possible that some ghrelin cells may be diverted from an α or PP cell fate.
Fig. 5.
Schematic of the transcription factor pathway specifying insulin-producing β cells. In the absence of Nkx2.2 or Pax4, the progenitor cells are switched to a ghrelin-producing ε cell fate.
It is conceivable that the loss of insulin-producing cells and the expansion ghrelin-producing cells are merely due to reciprocal gene expression changes of the two peptides (repression of insulin and activation of ghrelin) within the β cells. However, there are several lines of evidence to suggest that this is not occurring. First, there is currently little evidence to suggest that Nkx2.2 is an activator of insulin gene expression; rather, it may function to repress insulin gene expression (26). In addition, the characterized ghrelin promoter element does not appear to contain Nkx2.2 binding sites (27), and comparison of mouse and human ghrelin 5′ upstream sequences failed to identify conserved Nkx2.2 binding sites (C. Chao and L.S., unpublished data). Finally, the amplified population of ghrelin cells in the Nkx2.2 mutant does not express Nkx6.1 and has reduced levels of Pdx1, two canonical β cell transcription factors. This would suggest that the amplified cell population does not represent β cells but rather represents ghrelin-producing ε cells. Immunohistochemical and ultrastructural analyses are ongoing to further characterize this cell type in wild-type and Nkx2.2 mutant islets.
In summary, we have provided an excellent example of how global gene profiling analysis of knockout mouse tissue can lead to a significant leap in our understanding of cell differentiation pathways in an essential organ. These studies have allowed us to uncover two findings regarding pancreatic endocrine cell type differentiation that will have an impact on many future studies of pancreatic development, function, and regeneration. First, we have determined that ghrelin-producing ε cells represent an endocrine population within the embryonic mouse islet. Second, we have demonstrated that, in two mouse models, an early block in the differentiation of insulin-producing β cells leads to an enormous increase in ghrelin-producing ε cells, perhaps through a cell fate switch. Although the physiological role of ghrelin remains controversial, several studies have demonstrated that ghrelin functions to regulate insulin activity (28–30). Therefore, it is remarkable to find a genetic link between these two hormones. Our discoveries point to a role for ghrelin in the pancreatic islet, perhaps as a local regulator of glucose metabolism and energy balance. Furthermore, the data we present in this study provide the interesting possibility that the ghrelin-producing ε cells within the islet are derived from the same precursor that gives rise to insulin-producing β cells. Therefore, discovery of the ghrelin cell type will have a major impact on the generation of pure populations of β cells from stem or progenitor cells for cell-based therapy of diabetes. Ghrelin-producing ε cells or their precursors may now be a potential source of insulin-producing β cells for therapeutic purposes.
Acknowledgments
We thank T. Williams, H. Ford, C. V. E. Wright, and members of the Sussel laboratory for critical reading of the manuscript; J. Jensen for providing the Nkx6.1 antibody; and P. Serup, G. Gradwohl, and M. Sander for sharing unpublished data. This work was supported in part by Barbara Davis Center Diabetes & Endocrinology Research Center Core Grant NIH P30 DK5716, University of Colorado Health Sciences Center Cancer Center Microarray core grants, an American Diabetes Association Career Development Award (to L.S.), the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (L.S.), a Barbara Davis Center Blum-Kovler Fellowship (to A.E.P.-B.), Grant DK42502 from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (to B.S.-P.), and the American Lebanese Syrian Associated Charities (L.E.).
Abbreviations: PP, pancreatic polypeptide; en, embryonic day n.
References
- 1.Sussel, L., Kalamaras, J., Hartigan-O'Connor, D. J., Meneses, J. J., Pedersen, R. A., Rubenstein, J. L. & German, M. S. (1998) Development (Cambridge, U.K.) 125, 2213-2221. [DOI] [PubMed] [Google Scholar]
- 2.Slack, J. M. (1995) Development (Cambridge, U.K.) 121, 1569-1580. [DOI] [PubMed] [Google Scholar]
- 3.Kojima, M., Hosoda, H. & Kangawa, K. (2001) Horm. Res. 56, Suppl. 1, 93-97. [DOI] [PubMed] [Google Scholar]
- 4.Date, Y., Nakazato, M., Hashiguchi, S., Dezaki, K., Mondal, M. S., Hosoda, H., Kojima, M., Kangawa, K., Arima, T., Matsuo, H., et al. (2002) Diabetes 51, 124-129. [DOI] [PubMed] [Google Scholar]
- 5.Volante, M., Allia, E., Gugliotta, P., Funaro, A., Broglio, F., Deghenghi, R., Muccioli, G., Ghigo, E. & Papotti, M. (2002) J. Clin. Endocrinol. Metab. 87, 1300-1308. [DOI] [PubMed] [Google Scholar]
- 6.Wierup, N., Svensson, H., Mulder, H. & Sundler, F. (2002) Regul. Pept. 107, 63-69. [DOI] [PubMed] [Google Scholar]
- 7.Kim, S. K. & Hebrok, M. (2001) Genes Dev. 15, 111-127. [DOI] [PubMed] [Google Scholar]
- 8.Kemp, D. M., Thomas, M. K. & Habener, J. F. (2003) Rev. Endocr. Metab. Disorders 4, 5-17. [DOI] [PubMed] [Google Scholar]
- 9.Sosa-Pineda, B., Chowdhury, K., Torres, M., Oliver, G. & Gruss, P. (1997) Nature 386, 399-402. [DOI] [PubMed] [Google Scholar]
- 10.St-Onge, L., Sosa-Pineda, B., Chowdhury, K., Mansouri, A. & Gruss, P. (1997) Nature 387, 406-409. [DOI] [PubMed] [Google Scholar]
- 11.Sander, M., Sussel, L., Conners, J., Scheel, D., Kalamaras, J., Dela Cruz, F., Schwitzgebel, V., Hayes-Jordan, A. & German, M. (2000) Development (Cambridge, U.K.) 127, 5533-5540. [DOI] [PubMed] [Google Scholar]
- 12.Gradwohl, G., Dierich, A., LeMeur, M. & Guillemot, F. (2000) Proc. Natl. Acad. Sci. USA 97, 1607-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, H., Arber, S., Jessell, T. M. & Edlund, H. (1999) Nat. Genet. 23, 67-70. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, K. A., Thaler, J., Pfaff, S. L., Gu, H. & Kehrl, J. H. (1999) Nat. Genet. 23, 71-75. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson, J., Carlsson, L., Edlund, T. & Edlund, H. (1994) Nature 371, 606-609. [DOI] [PubMed] [Google Scholar]
- 16.Offield, M. F., Jetton, T. L., Labosky, P. A., Ray, M., Stein, R. W., Magnuson, M. A., Hogan, B. L. & Wright, C. V. (1996) Development (Cambridge, U.K.) 122, 983-995. [DOI] [PubMed] [Google Scholar]
- 17.Ahlgren, U., Jonsson, J. & Edlund, H. (1996) Development (Cambridge, U.K.) 122, 1409-1416. [DOI] [PubMed] [Google Scholar]
- 18.Naya, F. J., Huang, H. P., Qiu, Y., Mutoh, H., DeMayo, F. J., Leiter, A. B. & Tsai, M. J. (1997) Genes Dev. 11, 2323-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaeren-Wiemers, N. & Gerfin-Moser, A. (1993) Histochemistry 100, 431-440. [DOI] [PubMed] [Google Scholar]
- 20.Ohlsson, H., Karlsson, O. & Edlund, T. (1988) Proc. Natl. Acad. Sci. USA 85, 4228-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohlsson, H., Karlsson, K. & Edlund, T. (1993) EMBO J. 12, 4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guz, Y., Montminy, M. R., Stein, R., Leonard, J., Gamer, L. W., Wright, C. V. & Teitelman, G. (1995) Development (Cambridge, U.K.) 121, 11-18. [DOI] [PubMed] [Google Scholar]
- 23.Serup, P., Petersen, H. V., Pedersen, E. E., Edlund, H., Leonard, J., Petersen, J. S., Larsson, L. I. & Madsen, O. D. (1995) Biochem. J. 310, 997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oster, A., Jensen, J., Serup, P., Galante, P., Madsen, O. D. & Larsson, L. I. (1998) J. Histochem. Cytochem. 46, 707-715. [DOI] [PubMed] [Google Scholar]
- 25.Wang, J., Elghazi, L., Parker, S. E., Kizilocak, H., Asano, M., Sussel, L. & Sosa-Pineda, B. (2004) Dev. Biol. 266, 178-189. [DOI] [PubMed] [Google Scholar]
- 26.Cissell, M. A., Zhao, L., Sussel, L., Henderson, E. & Stein, R. (2003) J. Biol. Chem. 278, 751-756. [DOI] [PubMed] [Google Scholar]
- 27.Kishimoto, M., Okimura, Y., Nakata, H., Kudo, T., Iguchi, G., Takahashi, Y., Kaji, H. & Chihara, K. (2003) Biochem. Biophys. Res. Commun. 305, 186-192. [DOI] [PubMed] [Google Scholar]
- 28.Colombo, M., Gregersen, S., Xiao, J. & Hermansen, K. (2003) Pancreas 27, 161-166. [DOI] [PubMed] [Google Scholar]
- 29.Reimer, M. K., Pacini, G. & Ahren, B. (2003) Endocrinology 144, 916-921. [DOI] [PubMed] [Google Scholar]
- 30.Egido, E. M., Rodriguez-Gallardo, J., Silvestre, R. A. & Marco, J. (2002) Eur. J. Endocrinol. 146, 241-244. [DOI] [PubMed] [Google Scholar]