Abstract
How polyphagous herbivores cope with the diversity and unpredictability of plant defenses remains largely unknown at both the genetic and molecular levels. To examine whether generalist counterdefense enzymes are structurally more flexible and functionally more diverse, two counterdefensive allelochemical-metabolizing cytochrome P450 proteins, CYP6B1 from the specialist Papilio polyxenes, feeding on furanocoumarin-containing plants, and CYP6B8 from the generalist Helicoverpa zea, feeding on hundreds of host plant species, are compared structurally and functionally. Molecular modeling indicates that CYP6B8 has more flexible overall folding, a more elastic catalytic pocket, and one more substrate access channel than CYP6B1. Baculovirus-mediated expression of the CYP6B8 and CYP6B1 proteins demonstrates that CYP6B8 metabolizes six biosynthetically diverse plant allelochemicals (xanthotoxin, quercetin, flavone, chlorogenic acid, indole-3-carbinol, and rutin) and three insecticides (diazinon, cypermethrin, and aldrin), whereas CYP6B1 metabolizes only two allelochemicals (xanthotoxin and flavone) and one insecticide (diazinon) of those tested. These results indicate that generalist counterdefense proteins are capable of accepting a more structurally diverse array of compounds compared with specialist counterdefense proteins.
The majority of herbivorous insects are oligophagous, i.e., specialized on a relatively narrow range of host plants (three or fewer plant families) (1, 2). Such specialization is thought to reduce competition for food from other herbivores and to lower predation/parasitoid risk from generalist enemies (3). Because of the diversification of plant chemical defenses, however, this strategy may compromise a specialist's ability to make use of alternative food sources. A small proportion of insect herbivores, including some of the world's most important agricultural pests, are polyphagous, i.e., feed on a wide range of plant families. Although polyphagous species may have fewer limitations with respect to food availability, the toxicological challenge of generalized feeding is considerable in view of the tremendous diversity of plant defense compounds (allelochemicals). Because these compounds tend to be taxonomically limited in distribution, a polyphagous diet exposes an insect herbivore to a broad and unpredictable array of plant defenses. In fact, biochemical adaptation to a relatively narrow range of plant defense compounds may explain why the majority of herbivorous insects are specialists.
How generalists cope with the diversity and unpredictability of plant defenses remains largely unknown at both the genetic and molecular levels. One possibility is that counterdefense genes of generalists have multiple functions; in the case of detoxificative genes such as cytochrome P450 monooxygenases (4), individual proteins from generalists may have a much wider substrate spectrum. Greater functional flexibility could in theory allow generalists to cope with the unpredictability of plant defense (1).
Only a few counterdefense genes have been well characterized in herbivorous insects. Perhaps best characterized are allelochemical-metabolizing P450s in the CYP6B subfamily isolated from larval Lepidoptera (4–8). The black swallowtail butterfly, Papilio polyxenes, a specialist restricted to furanocoumarin-containing plants, relies on at least two P450s, CYP6B1 and CYP6B3, for protection against plant allelochemicals (4, 7). In a baculovirus-mediated expression system, CYP6B1 efficiently metabolizes xanthotoxin and other furanocoumarins (5, 8), whereas CYP6B3 metabolizes xanthotoxin only at low levels (J.-S. Chen, M.R.B., and M.A.S., unpublished observations). Related members of the CYP6B subfamily are also found in the polyphagous noctuids Helicoverpa zea (9, 10), Helicoverpa armigera (11), and Heliothis virescens (12, 13). Four CYP6B genes in H. zea, CYP6B8, CYP6B9, CYP6B27, and CYP6B28, are induced in response to a number of plant allelochemicals, including indole-3-carbinol and chlorogenic acid (14), and plant defense signaling compounds, including jasmonate and salicylate (15), strongly implicating all four of these CYP6B genes in metabolism of host plant allelochemicals.
To address whether generalist counterdefense proteins are structurally more flexible and functionally more diverse than comparable specialist counterdefense proteins, we chose to compare CYP6B1 from the specialist P. polyxenes and CYP6B8 from the generalist H. zea. Evolutionarily and functionally, the CYP6B1 and CYP6B8 proteins are related in their amino acid sequence (53% identity) (refs. 9 and 10; see Fig. 4, which is published as supporting information on the PNAS web site), their inducibility by furanocoumarins (6, 9, 14, 16), and their capacity to metabolize furanocoumarins (5, 8). Ecologically, they are expressed in herbivores that share a number of furanocoumarin-containing host plants, including parsnip (Pastinaca sativa), carrot (Daucus carota), and celery (Apium graveolens), among the preferred hosts of P. polyxenes and occasional hosts of H. zea (ref. 17; www.dallasbutterflies.com/Butterflies/html/black.html). Here, we present homology-based 3D structural models for both proteins, constructed to identify commonalities and differences in their overall folding, flexibility, and catalytic site configurations. We also present substrate metabolism profiles for both proteins, conducted to define the degree of divergence in their metabolic profiles. Together, these structural and metabolic comparisons are consistent with the hypothesis that CYP6B8 in H. zea, our representative generalist, is structurally more flexible and functionally more versatile than CYP6B1 in P. polyxenes, our representative specialist.
Materials and Methods
Homology Modeling and Molecular Dynamics (MD). Homology modeling was done by using the moe program (Chemical Computing Group, Montreal). The modeling of the CYP6B1 structure has been previously described (18), and the same procedure was followed here for modeling of CYP6B8. In brief, the Bacillus megaterium CYP102 crystal structure was selected as the template for constructing CYP6B1 and CYP6B8 models because these two CYP6B proteins share the highest degree of amino acid sequence identity (19.7% for CYP6B8, 19.8% for CYP6B1) with CYP102. The choice of CYP102 as template over rabbit CYP2C5 is based on the fact that CYP102, CYP6B1, and CYP6B8 are functionally related in their ability to metabolize xanthotoxin, whereas CYP2C5 cannot metabolize this compound. In addition, several high-energy regions in the CYP2C5 crystal structure (19) were reported to strongly bias the predicted binding sites geometry in several P450s (20). Ten models were generated and subjected to a coarse energy minimization for each target protein, with the explicit inclusion of the heme coordinates in all steps of the homology model generation. The best model with the highest moe's residue packing scores (-2.9152 for CYPB8) was selected for further full energy minimizations. Heme coordinates were duplicated from the template CYP102 crystal structure. Full energy minimizations were run by using the charmm22 force field (21) distributed in moe, using a dielectric constant of 1 with distance-dependent dielectric treatment and a cutoff between 6.5 Å and 7 Å for nonbonded interactions. The fully energy-minimized CYP6B1 and CYP6B8 models were structurally superimposed and substrate channels were identified for both CYP6B proteins by running Alpha Site Finder of the moe program.
MD simulations of the two enzymes were run by using the charmm22 force field distributed in moe, starting from the fully minimized structures in the (NVT) ensemble at a temperature of 300 K. An integration timestep of 1 fs was used. No restraints were applied to atomic positions. No explicit water molecules were added to the homology model, and a distance-dependent dielectric was used. The total simulation time was 350 ps. The average model was calculated from every frame in the MD trajectory between 50 and 350 ps (i.e., corresponding to a 300-ps trajectory). The root-mean-square deviation (rmsd) for each residue's backbone around the average structure during the MD trajectory, was calculated to quantify the structural flexibilities of the two CYP6B proteins.
Reagents and Chemicals. Analytical grade plant allelochemicals (xanthotoxin, flavone, chlorogenic acid, indole-3-carbinol, coumarin, quercetin, and rutin) were obtained from Sigma. NADPH, hemin, and heat-inactivated fetal bovine serum (FBS) were also from Sigma. Penicillin and streptomycin were from BioWhittaker. The Sf9 insect cell line, SF-900 serum-free medium, Bac-to-Bac baculovirus expression system, and restriction enzymes were from GIBCO/BRL/Life Technologies.
CYP6B and House Fly P450 Reductase Recombinant Virus Preparations. CYP6B1, CYP6B8, and house fly P450 reductase recombinant baculoviruses were generated by using the Bac-to-Bac baculovirus expression system after subcloning the cDNA sequences for the three genes into the pFastBac shuttle vector. The restriction sites used for subcloning the CYP6B8 cDNA were EcoRI and KpnI. Constructions of CYP6B1 and P450 reductase cDNA clones in pFastBac were conducted as described in Chen et al. (8). Positive pFastBac clones were transformed into competent DH10Bac cells, and recombinant bacmids were confirmed by restriction digestion and PCR amplification, and recombinant virus were produced in Sf9 insect cells. Sf9 cells were infected with these initial viruses at a multiplicity of infection of 0.05, and the final viral stocks were harvested 48 h after infection and stored at 4°C for up to 1 year.
Coexpression of CYP6B and House Fly P450 Reductase in Sf9 Cells. To produce CYP6B proteins for metabolism assays, Sf9 cells at a density of 0.9 × 106 cells per ml were coinfected with recombinant CYP6B1 (or CYP6B8) virus and house fly P450 reductase virus at multiplicities of infection of 2 (P450 virus) and 0.1 (house fly P450 reductase virus), corresponding to a multiplicity of infection ratio of 20. Sf9 cells were cultured in SF-900 serum-free medium supplemented with 9.1% FBS, 50 μg/ml streptomycin, and 50 units/ml penicillin. Hemin was added to the medium at a final concentration of 4 μg/ml 20 h after infection. The infected cells were harvested 72 h after infection by a brief spin at 1,000 × g for 2 min and washed three times with 10 vol of 0.1 M sodium phosphate buffer (pH 7.8). The final cell pellets were flash-frozen in liquid nitrogen and stored in -80°C until use.
Metabolism Kinetics Assay. Sf9 cells coinfected with CYP6B1 (or CYP6B8) and house fly P450 reductase viruses were thawed and lysed in grinding buffer [0.1 M sodium phosphate buffer (pH 7.8), with 0.5 mM PMSF, 20% (vol/vol) glycerol, 0.1 mM DTT, 1.1 mM EDTA, 5 μg/ml leupeptin] by ultrasonication for 1 min in ice. The P450 content of cell lysates was determined by carbon monoxide-difference spectra analysis (22) using an extinction coefficient of 91 mM-1·cm-1. Methods for separating and characterizing substrates are summarized in Table 2, which is published as supporting information on the PNAS web site. All metabolism assays were based on disappearance of substrates rather than product quantification because of the fact that products have not yet been characterized for many of the substrates tested. All substrates were dissolved in methanol at a concentration of 1–2 mg/ml, which was further diluted to five different concentrations ranging from 2.5 to 30.0 μg/ml in sodium phosphate buffer (PB, pH 7.8) for kinetics assay. Each reaction mixture consisted of 485 μl of diluted substrate solution, 100 μl of NADPH solution (3 ng/ml in 0.1 M PB), and 100 pmol of P450 (1 pmol/μl). Zero-time controls (terminated with 50 μl 2 M HCl before addition of cell lysates) and no-NADPH controls (incubated with 100 μl of PB instead of NADPH solution) were conducted at the same time to correct for non-P450-mediated reductions in substrates. After incubation of the reactions at 30°C for 15 min, 50 μl of 2 M HCl was added to stop each reaction and unmetabolized substrates were extracted from the reaction system and quantified (Table 2). For every compound analyzed, five concentrations, three treatments (15 min with NADPH, zero-time control, no-NADPH control), and three replicates for each were conducted to calculate the Michaelis constant, Km, and the maximum velocity, Vmax. Metabolic clearance, defined as Vmax/Km (23), was also calculated as an indicator of metabolic efficiency.
Results
CYP6B1 and CYP6B8 3D Models. Comparison of the CYP6B1 and CYP6B8 3D models (Fig. 1) shows that the secondary structure elements in the two proteins are similar, with some minute variations in the composition of individual elements, mainly in β-sheet strands. The CYP6B1 (18) model has β-sheet strands 1-1, 1-2, 1-3, 1-4, 1-5, 2-1, 2-2, 3-2, 3-3, 5-1, and 5-2 and two short β-sheet strands designated as 6-1 and 6-2 surrounding the K″ helix but not β-sheet strands 3-1, 4-1, and 4-2 (Fig. 1 A) [numbered according to secondary structure elements in the CYP102 template (24)]. The CYP6B8 model has β-sheet strands 1-1, 1-2, 1-3, 1-4, 1-5, 3-2, and 3-3 but not the other β-sheet strands listed above (Fig. 1B). Less pronounced differences also exist in the composition of α-helices, with the CYP6B1 model maintaining all α-helices found in the CYP102 template and adding a short F′ helix in the F–G loop (Fig. 1 A). In contrast, the CYP6B8 model lacks the H and K″ helices, has an additional short K° helix in the β1-4/β1-3 loop, and an A helix that is destabilized by a loop (Fig. 1B).
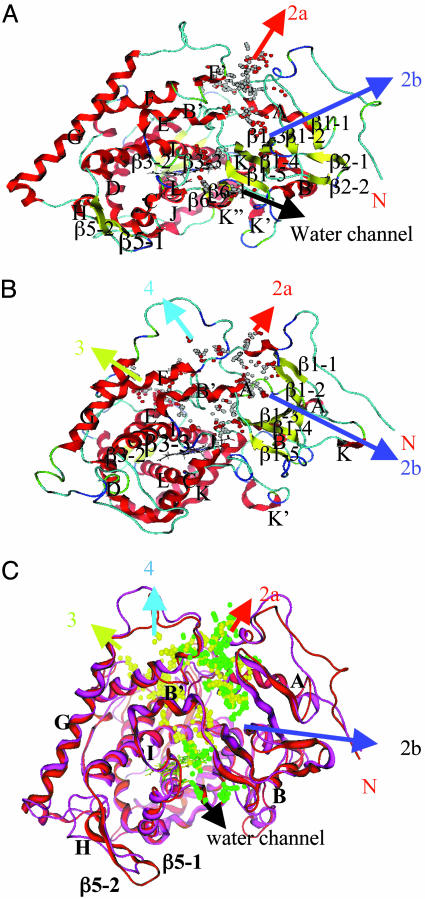
Fig. 1.
Apoprotein models and putative substrate access channels for the CYP6B1 and CYP6B8 proteins. The protein backbones of CYP6B1 (A) and CYP6B8 (B) are represented by flat ribbon drawings, and the heme structure is represented with lines. Proteins are viewed along the I helix with their α-helices (red), β-sheet strands (yellow), and N termini (red-labeled N) labeled according to the nomenclature of secondary structure elements in the CYP102 template (24). The loops (light blue) and turns (dark blue for β-turn and green for ribbon turn) are not labeled. Putative substrate access channels are indicated with different colored arrows and labeled according to the P450 substrate channel classification system (26, 27). The hydrophobic and hydrophilic sites are displayed as white and red spheres, respectively. The superposition of the CYP6B1 and CYP6B8 models (C) uses a different color scheme, with the CYP6B1 backbone in red, the CYP6B1 channel spheres in green, the CYP6B8 backbone in purple, and the CYP6B8 channel spheres in yellow.
Superimposition of the two protein models suggests that CYP6B1 and CYP6B8 differ with respect to the length, orientation, and topological positioning of some of these secondary structure elements (Fig. 1C). Specifically, the B and G helices are longer in the CYP6B1 model, and the G helix in the CYP6B1 model is sharply turned at residue Met-259 near its C terminus. In contrast, most loop structures are longer in the CYP6B8 model, with the most striking positional variations between these two models found in the F–G, B′–C, G–H, and H–I loops (Fig. 1C). While some of these differences between the overlays of the CYP6B1 and CYP6B8 structures may originate from the fact that static energy-minimized structures without dynamic perturbation are considered in this description, other differences are significant, with the MD simulations described below supporting the existence of these structural differences.
Substrate Access Channel. To determine whether these overall structural differences may contribute to divergence in the substrate access channels of these two proteins, all hydrophobic and hydrophilic cavities in these CYP6B models were searched by using the Alpha Site Finder function of the moe program. Potential substrate access channels were then identified by finding pathways with cavity sites extending from the catalytic pocket immediately above the heme ring to the exterior surface of the model.
In the CYP6B1 model, three channels were identified: one vertical channel above the heme plane (2a in Fig. 1 A and C), one vertical channel below the heme plane (labeled water channel), and one side channel above the heme plane (2b in Fig. 1 A and C). The vertical channel above the heme is located at the boundary between the α-helix and β-sheet domains, with its opening positioned between the F′–G loop, B′ helix, and A helix. The lower vertical channel is connected to the upper vertical channel at the right side of the heme ring and opens between the K″ helix and β-sheet strand 1-5. The side channel branches out from the upper vertical channel at the B helix, extends into the β-sheet domain, and opens between B–B′ loop and the β-sheet strand 1-3. The upper and lower vertical channels correspond to those proposed as the substrate access and product exit channels in the P450cam crystal structure (25). According to the nomenclature for P450 substrate access channels introduced by Lüdemann et al. (26) and Winn et al. (27), the three CYP6B1 channels are designated as the 2a, 2b, and water channels (designated as the water channel because it is filled with ordered water molecules) in Fig. 1 A.
In the CYP6B8 model, two of the three channels are also present (Fig. 1B) but they do not completely correspond to those described in the CYP6B1 model (Fig. 1C). The CYP6B8 model includes an additional vertical channel and side channel (channels 3 and 4 in Fig. 1B) not identified in the CYP6B1 model. Both of these channels initiate at the heme ring, extend upward, and separate just above the B′ helix with the vertical channel running through the F helix and opening at the F–G loop and the side channel crossing the F and G helices and opening at the E–F loop (Fig. 1 B and C). The additional side channel is equivalent to pathway 3 predicted from the P450cam and P450eryF structures (26, 27). The additional vertical channel, which is not represented in the current substrate access channel classification system (26, 27), is designated as pathway 4.
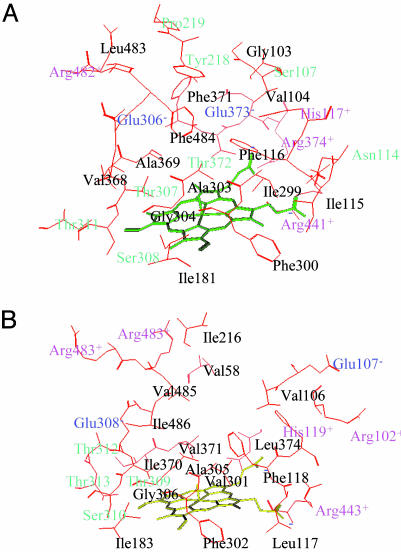
Residues in the Catalytic Pocket. Amino acids proximal to the heme ring in each of the proteins are presented in Fig. 2. The first similarity in these proteins is that both have a pair of positively charged arginines [Arg-374 of substrate recognition site (SRS)5 and Arg-441 of the FXXGXRXCXG P450 signature motif in CYP6B1 and Arg-102 of SRS1 and Arg-443 of P450 signature motif in CYP6B8] in proximity to the two negatively charged carboxyl groups of the heme ring (Fig. 2). This arginine pair contributes to stabilization of the heme ring and the protein by forming a salt bridge (by electrostatic interactions). It also may serve as a terminal electron- or proton-transferring amino acid. The second similarity is that both contain highly conserved hydrophilic microenvironments on the other side of the heme ring, consisting of Glu-306, Thr-307, Ser-308, and Thr-311 in the I helix of the CYP6B1 model and Glu-308, Thr-309, Ser-310, Thr-312, and Thr-313 in the I helix of the CYP6B8 model. The third similarity is that both conserve a number of amino acids in the hydrophobic patch near the heme ring, including Ile-299, Phe-300, Ala-303, and Gly-304 of SRS4 and Phe-116 and His-117 of SRS1 in the CYP6B1 model and Val-301, Phe-302, Ala-305, and Gly-306 of SRS4 and Phe-118 and His-119 of SRS1 in the CYP6B8 model (Fig. 2).
Fig. 2.
Catalytic pockets in the CYP6B1 (A) and CYP6B8 (B) proteins. The pockets are shown with amino acids represented by red lines and the heme represented by green or yellow lines. The biochemical properties of amino acids in the catalytic site are designated in different colors, with black representing hydrophobic amino acids, pink representing positively charged amino acids, blue representing negatively charged amino acids, and green representing polar amino acids.
The first notable divergence in these proteins is that the CYP6B1 model contains an aromatic network consisting of Phe-116 and His-117 of SRS1, Phe-484 of SRS6, Phe-371 of SRS5, and Tyr-218 of SRS2 (Fig. 2 A), which has been suggested to be involved in the stabilization of resonant furanocoumarin substrates (18). The CYP6B8 model lacks this aromatic network owing to the substitution of Val-485 in CYP6B8 for Phe-484 in CYP6B1, Ile-216 for Tyr-218, and Val-58 for Phe-371 (Fig. 2B). The second divergence is that all of the amino acids surrounding the CYP6B1 aromatic network, including Arg-482 of SRS6, Tyr-218 and Pro-219 of SRS2, Ser-107, Asn-114, and His-117 of SRS1, Thr-372, Gln-373, and Arg-374 of SRS5, and Arg-441 of the P450 signature motif, are polar amino acids (Fig. 2 A) capable of forming a polar or hydrophilic cage by means of hydrogen bond and electrostatic interactions. The CYP6B8 model does not contain this polar cage because of the substitution of nonpolar Ile-216 and Leu-374 for the polar Tyr-218 and Thr-372 found in the CYP6B1 pocket. In discussing this substitution of Ile-216 for Tyr-218, it is important to note that this difference occurs in a region that has been modeled in two different orientations in CYP6B1 (18). In one orientation, this residue projects toward the active site and participates in the resonant aromatic network of the CYP6B1 active site and, in the other orientation, this residue projects outside of the active site in a position closer to that of Tyr-222 in the CYP6B8 model. Because the orientation projecting Tyr-218 into the active site was energetically slightly more favored than the orientation projecting it outside the active site and because protein/substrate interaction geometry and energetics were correctly reproduced with Tyr-218 localized in the active site (18), this CYP6B1 model is used for comparison with the CYP6B8 model. However, because of these potential degeneracies in the folding solutions for this region, we cannot entirely eliminate the possibility that Tyr-218 in the CYP6B1 model projects outside the binding site as Tyr-222 does in the CYP6B8 model. Despite this uncertainty, it is clear that the polar cage serves as a stabilizing force in the CYP6B1 pocket. Together, these variations contribute to making the CYP6B1 catalytic pocket smaller and more rigid and the CYP6B8 hydrophobic pocket larger and more flexible.
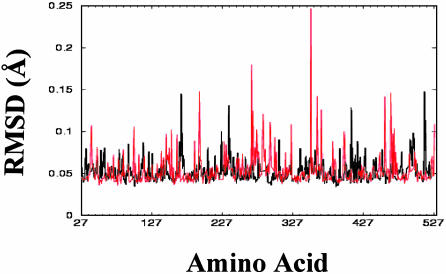
rmsd Fluctuation During the MD Trajectory. To identify and quantify thermodynamically flexible residues, rmsd values for each residue's backbone around the average structure during the 300-ps MD trajectory were calculated for both proteins. The rmsd values identify 7 amino acids with high rmsd values in the CYP6B1 model and 19 amino acids with high rmsd values in the CYP6B8 model (Fig. 3). The range of rmsd fluctuations in the CYP6B8 model is greater than in the CYP6B1 model (Fig. 3). The seven most thermodynamically flexible residues in the CYP6B1 model are Ser-161 and Gln-162 in D–E loop, Tyr-218 in F helix, Leu-229 in F–G loop, Gly-392 in β1-3 sheet, and Gly-489 and Gly-490 in the C-terminal loop. Of these, Tyr-218 and Leu-229 are located in the SRS2 domain and theoretically involved in formation of substrate access channel 2a. Tyr-218 is also the amino acid previously mentioned as difficult to model, with one possible orientation projecting into the CYP6B1 aromatic network and another projecting outside the catalytic site. The 19 thermodynamically flexible amino acids in CYP6B8 model include Phe-42 in the N-terminal loop, Phe-98 the B–B′ loop, Glu-150 in the D helix, Gly-189 in the E–F loop, Pro-257, Gly 259, and Met 264 in the G helix, Met-273, Asp-283, and Gly-284 in the G–I loop, Ser-310 in the I helix and catalytic pocket, Ala-338, Thr-347, and Glu-353 in the J–K loop, Phe-438, Ile-446, Gly-447, and Gly-451 in the K′–L loop and heme-binding region, and Ser-504 in the C-terminal loop. The relative conformational freedom found in the heme-binding region and the J–K loop immediately below the heme ring may allow the heme and its catalytic iron–oxygen to accommodate an array of structurally diverse substrates. Ser-310 movement in the I helix has potential to increase the flexibility of CYP6B8 catalytic pocket. The conformational freedom of Phe-42, Phe-98, Gly-189, Pro-257, Gly-259, Met-264, Met-273, Asp-283, and Gly-284, which are predicted to be involved in the formation of substrate channels 2a, 2b, 3, and 4 (Fig. 1B), may enhance the flexibility of these substrate access channels.
Fig. 3.
rmsd fluctuations of CYP6B1 and CYP6B8 during MD trajectory. rmsd values of each residue's backbone around the average structure (on the vertical axis) are plotted against residue number (on the horizontal axis), with CYP6B1 in black and CYP6B8 in red.
Metabolism Profiles of CYP6B1 and CYP6B8. The differences in structural folding, substrate access channel, catalytic pocket, and structural flexibility between the two CYP6B proteins suggest that CYP6B1 of the specialist might metabolize a narrow range of substrates at efficiencies that are optimized because of the positioning of preferred substrates in its rigid catalytic pocket. CYP6B8 of the generalist, on the other hand, may metabolize a wider range of substrates at efficiencies that are lower because of nonoptimal positioning of substrates in its more flexible catalytic site. To confirm that these two CYP6B proteins function differently in accordance with their structural differences, the metabolic profiles of the CYP6B1 and CYP6B8 proteins were determined. Seven naturally occurring plant allelochemicals, representing a range of ecological encounter rates by the generalist H. zea and the specialist P. polyxenes, and five synthetic insecticides, representing five classes of insecticides used for control of H. zea, were selected for testing as possible substrates. The flavonoids flavone (a flavone aglycone), quercetin (a flavonol aglycone), and rutin (a flavonol glycoside) occur in a wide range of host plant families of H. zea, as well as in the host plants of P. polyxenes (ref. 17; www.ars-grin.gov/duke/index.html). Chlorogenic acid, an intermediate in the shikimate and lignin synthetic pathways, occurs in many host families of H. zea and in Apiaceae, a preferred host plant family of P. polyxenes (www.ars-grin.gov/cgi-bin/duke/highchem.pl). Coumarin is found in legumes (Fabaceae), a preferred host plant family of H. zea, and in carrot (Daucus), a host plant of P. polyxenes (www.ars-grin.gov/cgi-bn/duke/highchem.pl; ref. 28). Indole-3-carbinol, a breakdown product of glucobrassicin, occurs primarily in Brassicaceae, a family occasionally used by H. zea but not P. polyxenes (www.dallasbutterflies.com/Butterflies/html/black.html; http://creatures.ifas.ufl.edu/veg/corn_earworm.htm#host). Xanthotoxin, a furanocoumarin, is present in Apiaceae and Rutaceae, preferred host plant families of P. polyxenes and occasional host plant families of H. zea (17; http://www.ars-grin.gov/cgi-bin/duke/highchem.pl).
The metabolic analyses, using relatively uniform reaction conditions (Table 2), demonstrate that CYP6B8 from the generalist metabolizes six of the seven plant toxins and three of five insecticides tested, with the best substrates being the insecticides diazinon and aldrin and the allelochemical quercetin (Table 1). In contrast, CYP6B1 from the specialist metabolizes only two plant allelochemicals, xanthotoxin and flavone, and one insecticide, diazinon, with the best substrate being xanthotoxin (Table 1). In contrast to the high efficiency with which CYP6B1 metabolizes xanthotoxin, CYP6B8 metabolizes the six plant allelochemicals and three insecticides at efficiencies that are substantially lower than CYP6B1 achieves in metabolizing xanthotoxin (Table 1). In terms of the Km values for xanthotoxin, there is a 5.5-fold difference between the CYP6B1 (0.012 mM) and CYP6B8 (0.0659 mM) proteins. In terms of Vmax, there is a 6.1-fold difference between the CYP6B1 (22.24 μmol/μmol of P450 per min) and CYP6B8 (3.67 μmol/μmol of P450 per min) proteins. These differences translate to a 33.4-fold difference in metabolic clearance (Vmax/Km) for these proteins. The other two common substrates, flavone and diazinon, which are rarely encountered by the specialist P. polyxenes but are frequently encountered by the generalist H. zea, are more actively metabolized by CYP6B8 than by CYP6B1 (Table 1).
Table 1. Metabolic kinetics of the CYP6B1 and CYP6B8 proteins against plant allelochemicals and insecticides.
| Kinetic parameters
|
|||
|---|---|---|---|
| Xenobiotic | Vmax, μmol/μmol of P450 per min | Km, mM | Clearance, ml/μmol of P450 per min |
| CYP6B8 | |||
| Allelochemicals | |||
| Coumarin | NDA | ||
| Flavone | 6.95 | 0.158 | 44.1 |
| Rutin | 0.89 | 0.020 | 44.3 |
| Xanthotoxin | 3.67 | 0.066 | 55.6 |
| Chlorogenic acid | 13.30 | 0.235 | 55.7 |
| Indole-3-carbinol | 1.56 | 0.013 | 121.8 |
| Quercetin | 5.30 | 0.020 | 260.5 |
| Insecticides | |||
| Carbaryl | NDA | ||
| Diflubenzuron | NDA | ||
| α-Cypermethrin | 12.72 | 0.085 | 149.3 |
| Aldrin | 22.42 | 0.084 | 267.8 |
| Diazinon | 38.92 | 0.088 | 442.9 |
| CYP6B1 | |||
| Allelochemicals | |||
| Coumarin | NDA | ||
| Flavone | 2.35 | 0.059 | 39.8 |
| Rutin | NDA | ||
| Xanthotoxin | 22.24 | 0.012 | 1,859.9 |
| Chlorogenic acid | NDA | ||
| Indole-3-carbinol | NDA | ||
| Quercetin | NDA | ||
| Insecticides | |||
| Carbaryl | NDA | ||
| Diflubenzuron | NDA | ||
| α-Cypermethrin | NDA | ||
| Aldrin | NDA | ||
| Diazinon | 50.52 | 0.197 | 256.5 |
NDA, no detectable activity.
Discussion
Molecular modeling suggested many commonalities and several potentially interesting differences between the specialist and generalist CYP6B proteins. Relative to the overall folding of the CYP6B1 model, the CYP6B8 model has reduced the number of β-sheet strands and α-helices as well as the lengths of some α-helices (Fig. 1). As a result, several of the loop structures are predicted to be longer in CYP6B8 than in CYP6B1. MD simulations suggest that the CYP6B8 protein has high rmsd fluctuations in more residues associated with the formation of the substrate access channel, catalytic pocket, and heme-binding region.
These overall folding differences lead to divergence in the substrate access channel with both the CYP6B1 and CYP6B8 models predicted to contain channels 2a and 2b but with variations in their topology that result in different channel shapes, sizes, and opening mechanisms as described for the 2a channels of P450cam, P450BM-3, and P450eryF (27). More significant substrate channel differences are associated with the existence of two putative channels (3 and 4) in the CYP6B8 model that are not predicted in the CYP6B1 model. These differences suggest that, if all of these predicted channels exist and are used for substrate and/or product movement, more substrates have potential to access the CYP6B8 catalytic pocket compared with the CYP6B1 catalytic pocket, consistent with the idea that P450s from generalists are more versatile than those from specialists.
The overall folding differences also lead to differences in the catalytic pockets of these two proteins. The two stabilizing networks in the CYP6B1 catalytic pocket (aromatic resonant and hydrophilic hydrogen bonding) are nearly nonexistent in the CYP6B8 catalytic pocket. Their presence in the CYP6B1 catalytic site appears to hold the involved amino acids in more rigid topology, making this catalytic site more structurally constrained in its substrate contacts than the CYP6B8 catalytic site. The structural differences between these two proteins proposed to contribute to CYP6B8's metabolism of a wider range of substrates (additional substrate access channels, catalytic site flexibility) also potentially account for CYP6B8's less efficient metabolism of furanocoumarins and other planar aromatic compounds. Balancing its need to metabolize many plant allelochemicals with only sporadic exposure to some, CYP6B8 appears to possess a catalytic site capable of accepting and metabolizing many types of allelochemicals and insecticides at turnover rates lower than those of comparable specialist proteins such as CYP6B1.
Evolution of host range has been a central interest in the field of plant–insect interactions (29–31), with many theories proposed to explain macroevolutionary patterns of host use and dietary breadth (32, 33). The results of our study demonstrate that functional versatility of counterdefense genes may facilitate polyphagy, whereas functional specialization of host use-related genes may promote oligophagy. Other genetic and molecular factors may contribute to polyphagy; comparative genomewide surveys of host use-related genes between pairs of closely related species that diverge in their degree of feeding specialization may resolve the question of whether multiple gene duplication events also aid generalists in coping with their diverse and unpredictable plant defense challenges (34).
Supplementary Material
Acknowledgments
We thank Xiangxia Luo for performing metabolism assays, Raelene Lawrence (Chemical Computing Group, Montreal) for help with SVL programming in moe for rmsd calculations, Sanjeewa Rupasinghe for assistance in running the moe program, Drs. James Nitao and Arthur Zangerl for technical assistance in developing HPLC/GC methods, and Liz Yang for providing CYP6B1 proteins. This research was supported by U.S. Department of Agriculture Grant 01-35302-10884 to M.A.S. and M.R.B., by National Institutes of Health Grant RO1-GM50007 to M.A.S., and by China Natural Science Foundation Grant 30325028 to X.L.
Abbreviations: SRS, substrate recognition site; rmsd, root mean square deviation; MD, molecular dynamics.
References
- 1.Gatehouse, J. A. (2002) New Phytol. 156, 145-169. [DOI] [PubMed] [Google Scholar]
- 2.Bernays, E. A. & Chapman, R. F. (1994) Host-Plant Selection by Phytophagous Insects (Chapman & Hall, New York).
- 3.Bernays, E. A. (2001) Annu. Rev. Entomol. 46, 703-727. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, M. B., Schuler, M. A. & Berenbaum, M. R. (1992) Proc. Natl. Acad. Sci. USA 89, 10920-10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma, R., Cohen, M. B., Berenbaum, M. R. & Schuler, M. A. (1994) Arch. Biochem. Biophys. 310, 332-340. [DOI] [PubMed] [Google Scholar]
- 6.Hung, C.-F., Prapaipong, H., Berenbaum, M. R. & Schuler, M. A. (1995) Insect Biochem. Mol. Biol. 25, 89-99. [Google Scholar]
- 7.Hung, C.-F., Harrison, T. L., Berenbaum, M. R. & Schuler, M. A. (1995) Insect Mol. Biol. 4, 149-160. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J.-S., Berenbaum, M. R. & Schuler, M. A. (2002) Insect Mol. Biol. 11, 175-186. [DOI] [PubMed] [Google Scholar]
- 9.Li, X.-C., Berenbaum, M. R & Schuler, M. A. (2000) Insect Biochem. Mol. Biol. 30, 75-84. [DOI] [PubMed] [Google Scholar]
- 10.Li, X., Berenbaum, M. R. & Schuler, M. A. (2002) Insect Biochem. Mol. Biol. 32, 311-320. [DOI] [PubMed] [Google Scholar]
- 11.Ranasinghe, C. & Hobbs, A. A. (1998) Insect Biochem. Mol. Biol. 28, 571-580. [DOI] [PubMed] [Google Scholar]
- 12.Fitt, D. P. (1989) Annu. Rev. Entomol. 34, 17-52. [Google Scholar]
- 13.Mitter, C., Poole, R. W. & Matthews, M. (1993) Annu. Rev. Entomol. 38, 207-225. [Google Scholar]
- 14.Li, X., Berenbaum, M. R. & Schuler, M. A. (2002) Insect Mol. Biol. 11, 343-352. [DOI] [PubMed] [Google Scholar]
- 15.Li, X., Schuler, M. A. & Berenbaum, M. R. (2002) Nature 419, 712-715. [DOI] [PubMed] [Google Scholar]
- 16.Petersen R. A., Zangerl, A. R., Berenbaum, M. R. & Schuler, M. A. (2001) Insect Biochem. Mol. Biol. 31, 679-690. [DOI] [PubMed] [Google Scholar]
- 17.Tietz, H. M. (1972) An Index to the Described Life Histories, Early Stages, and Hosts of Macrolepidoptera of the Continental United States and Canada (Allyn Museum of Entomology, Sarasota, FL), Vols. 1 and 2.
- 18.Baudry, J., Li, W., Pan, L., Berenbaum, M. R. & Schuler, M. A. (2003) Protein Eng. 16, 577-587. [DOI] [PubMed] [Google Scholar]
- 19.Rupasinghe, S., Baudry, J. & Schuler, M. A. (2003) Protein Eng. 16, 721-731. [DOI] [PubMed] [Google Scholar]
- 20.Kirton, S. B., Kemp, C. A., Tomkinson, N. P., St-Gallay, S. & Sutcliffe, M. J. (2002) Proteins Struct. Funct. Genet. 59, 216-231. [DOI] [PubMed] [Google Scholar]
- 21.MacKerell, A. D., Jr., Bashford, D., Bellott, M., Dunbrack, R. L., Jr., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., et al. (1998) J. Physical Chem. B 102, 3586-3616. [DOI] [PubMed] [Google Scholar]
- 22.Omura, T. & Sato, R. (1964) J. Biol. Chem. 239, 2379-2385. [PubMed] [Google Scholar]
- 23.Smith, D. A. (1991) Drug Metab. Rev. 23, 353-373. [DOI] [PubMed] [Google Scholar]
- 24.Graham, S. E. & Peterson, J. A. (1999) Arch. Biochem. Biophys. 369, 24-29. [DOI] [PubMed] [Google Scholar]
- 25.Poulos, T., Finzel, B. C. & Howard, A. J. (1987) J. Mol. Biol. 195, 687-700. [DOI] [PubMed] [Google Scholar]
- 26.Lüdemann, S. K., Lounnas, V. & Wade, R. C. (2000) J. Mol. Biol. 303, 797-811. [DOI] [PubMed] [Google Scholar]
- 27.Winn, P. J., Lüdemann, S. K, Gauges, R., Lounnas, V. & Wade, R. C. (2002) Proc. Natl. Acad. Sci. USA 99, 5361-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaliwal, B. B. & Rao, M. A. (1981) Indian J. Exp. Biol. 19, 1058-1060. [Google Scholar]
- 29.Dobler, S. & Farrell, B. D. (1999) Mol. Ecol. 8, 1297-1307. [DOI] [PubMed] [Google Scholar]
- 30.Agrawal, A. A. (2000) Ecology 81, 500-508. [Google Scholar]
- 31.Ueno, H., Fujiyama, N., Irie, K., Sato, Y. & Katakura, H. (1999) Entomol. Exp. Appl. 91, 245-250. [Google Scholar]
- 32.Becerra, J. X. & Venable, D. L. (1999) Proc. Natl. Acad. Sci. USA 96, 1262-12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Klinken, R. D. (2000) Ecol. Entomol. 25, 413-422. [Google Scholar]
- 34.Li, W., Schuler, M. A. & Berenbaum, M. R. (2003) Proc. Natl. Acad. Sci. USA 100, Suppl. 2, 14593-14598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.