Abstract
The mechanism of an organism's adaptation to high temperatures has been investigated intensively in recent years. It was suggested that the macromolecules of thermophilic microorganisms (especially proteins) have structural features that enhance their thermostability. We compared mRNA sequences of 72 fully sequenced prokaryotic proteomes (14 thermophilic and 58 mesophilic species). Although the differences between the percentage of adenine plus guanine content of whole mRNAs of different prokaryotic species are much lower than those of guanine plus cytosine content, the thermophile purine-pyrimidine (R/Y) ratio within their mRNAs is significantly higher than that of the mesophiles. The first and third codon positions of both thermophiles and mesophiles are purine-biased, with the bias more pronounced by the thermophiles. Thermophile mRNAs that display the highest R/Y ratio (1.43–1.69) are those of the ribosomal proteins, histone-like proteins, DNA-dependent RNA polymerase subunits, and heat-shock proteins. Within mesophilic prokaryotes and five eukaryotic species, the R/Y ratio of the mRNAs of heat-shock proteins is higher than their average over coding part of the genome. Polypurine tracts (R)n (with n ≥ 5) are much more abundant within the thermophile mRNAs compared with mesophiles. Between two sequential pure-purinic codons of thermophile mRNAs, there is a rather strong tendency for the occurrence of adenine but not guanine tracts. The data suggest that mixed adenine·guanine and polyadenine tracts in mRNAs increase the thermostability beyond the contribution of amino acids encoded by purine tracts, which highlights the importance of ecological stress in the evolution of genome architecture.
Adaptive strategies of organisms to extreme environments such as exceptional salinity, high pressure, nonphysiological pH, anaerobic conditions, and high and low temperatures are of primary importance for evolutionary studies (1). Revealing and understanding the special features of the macromolecules of thermophilic prokaryotes with high to very high optimum growth temperatures (OGTs) (50–113°C), compared with much lower ranges (20–37°C) of prokaryotic mesophiles, is of particular interest. Historically, investigators first were interested in revealing the unique features of the thermophile proteins that contribute to their thermostability (2, 3). Clarifying the principles of enhanced thermostability is important theoretically and practically. Deciphering improved enzymes with higher thermostability is of significant economic value to some industries. In this study, our aim was to unravel differences between mRNAs and the proteins of thermophiles and mesophiles to identify common features of the thermophiles' molecules that might contribute to thermostability. We restricted this study to the protein-coding transcripts. Understanding how the adaptation of the transcription and translation machinery (and products) to high temperature is achieved is central to both theoretical models and in vitro experimentation. Therefore, besides the comparison of the whole proteome and mRNAs, we specifically tested RNA polymerase subunits and their coding mRNAs as well as ribosomal proteins and their coding mRNAs. Revealing the organization of the template/nascent strands and features of the thermophilic mRNAs that might contribute to thermostability have become the focus of some investigations in the last few years (4–8).
Protein Structure
Previous comparative studies of the proteins of thermophilic and extreme thermophilic prokaryotes and mesophiles revealed factors that were attributed to thermostability. These factors include (i) a higher number of salt bridges (9–14) [in this respect, it was suggested that not only a larger number of charged residues but also additional salt bridges around a particular bridge enhance the stability of the bridge (15, 16)], (ii) additional hydrogen bonds (17–21), (iii) shorter loop regions (12, 22), (iv) increasing intramolecular hydrophobic packing (11), and (v) the α-helical content of the proteins (23). These structural characteristics might result from different proportions of specific amino acids in the sequences of the thermophile proteins. Indeed, it was found that thermophiles have a high (Glu + Lys)/(Gln + His) ratio compared with mesophiles (24). This was partly because of a higher frequency of the glutamic acid and lysine in thermophile proteomes. High frequency of charged amino acids was considered to be the central contributor to thermophile protein thermostability (16, 25).
The ability of thermophile DNA polymerases and DNA-dependent RNA polymerases (RNAPs) to carry out replication and transcription under high temperature is achieved by structural features similar to those of other proteins of the thermophiles mentioned above. Likewise, additional disulfide bonds of a thermophilic species DNA polymerase compared with mesophilic orthologues and enhanced electrostatic complementarity at the DNA–protein interface have been shown (26). Similarly, a greater number of charged residues that can form ion pairs were also attributed to the high thermal stability of the archaeal RNAP subunits (27).
Translation Apparatus
Data compiled on thermophile RNA components of the protein-synthesis apparatus showed that the percentage of guanine plus cytosine (GC%) of 5S, 16S, and 23S rRNAs is positively correlated with the OGTs of the species (28). Later, the GC% content of thermophile tRNAs was also found to be positively correlated with the OGTs of the species (29). Higher GC% content of these RNAs might elevate their resistance to heat by means of the formation of more-stable intramolecular double-stranded RNA structures (with additional hydrogen bonds between the G·C base pair compared with the A·U base pair). Thermostability of the ribosomal proteins might be achieved by the aforementioned structure (with regard to other thermophile proteins).
Transcription at High Temperatures and Thermostability of the mRNAs
Some features of the coding DNA sequences and mRNAs of thermophiles might be related to the efficiency of transcription and translation under high temperature:
The DNA helix conformation of some/most of the CDSs of the thermophiles might differ from the regular B-DNA: Tracts of guanines (or cytosines) longer than four nucleotides were found to prefer the A-DNA conformation (with favorable flanking sequences) (30). In contrast, stretches of adenines (thymines) do not convert to the A-DNA form but adopt a distinct right-handed form (B′) (31). It is noteworthy that the three different Pyrococcus species of Archaea all have >5% A-DNA (≈25-fold higher than expected) (32). It is possible that DNA·RNA hybrid structures with the DNA helix in A-DNA or B′-DNA forms provide better transcription at high temperatures. Transcription might be enhanced even in an earlier stage: Yagil et al. (33, 34) proposed a contribution of purine and pyrimidine tracts to the unwinding of DNA, a central pretranscription process.
A better fit of the thermophile mRNAs to translation without interruptions under high temperature: The “politeness” hypothesis (35) assumes that purine loading in the mRNAs prevents distracting RNA–RNA interactions and excessive formation of double-stranded RNA, which might trigger various intracellular alarms (36). RNA–RNA interactions have a distinct entropy-driven component; hence, Lao and Forsdyke (4) proposed that selective pressure for the evolution of purine loading might be greater in organisms living at high temperatures.
mRNAs of thermophilic species might be more stable and less sensitive to spontaneous hydrolysis: Spontaneous hydrolysis of unstructured RNAs by intramolecular transesterification could occur ≈100,000-fold faster in thermophilic organisms than in mesophilic organisms (37). In fact, this danger could be neutralized by specific base composition, increasing the stability of RNA phosphodiester bonds. This stabilization has been attributed frequently to stacking interactions between the adjacent nucleic acid bases (37–39). Thus, the exceptionally slow cleavage within adenine-rich sequences (40) was explained in this manner (37).
The aforementioned diverse suggestions that the thermophile mRNAs (or template DNA strands) have specific structures contributing to thermostability in the early stages of template-based information processing are not mutually exclusive.
Here, we present findings supporting the general role for purine tracts (adenine tracts, especially) within the mRNAs of the thermophiles to thermoadaptation. Presumably, the contribution of these tracts to thermoadaptation may derive not only from the stabilization of proteins due to the encoded amino acids but also, to a large extent, from the earlier stages of template-based information processing.
Materials and Methods
We analyzed mRNA sequences of 72 fully sequenced genomes including 13 Archaea, 59 Bacteria, and 5 Eukarya species (Table 5, which is published as supporting information on the PNAS web site). The sequences were obtained from public databases available from the Comprehensive Microbial Resource (www.tigr.org) and GenBank (www.ncbi.nlm.nih.gov/GenBank/index.html). Because of redundancy of the prokaryotic species list (some species are represented by several strains or subspecies), the final results involved 59 prokaryotic species (14 thermophiles and 45 mesophiles; the thermophiles included 11 Archaea and 3 Bacteria, and the mesophiles included 43 Bacteria and 2 Archaea species). Calculations of codon counts, frequencies of nucleotides within the three codon positions, neighbor codon frequencies, purine and pyrimidine abundances, as well as distribution of homotract lengths of purines, pyrimidines, adenines, and guanines were conducted with our own designed programs.
Results
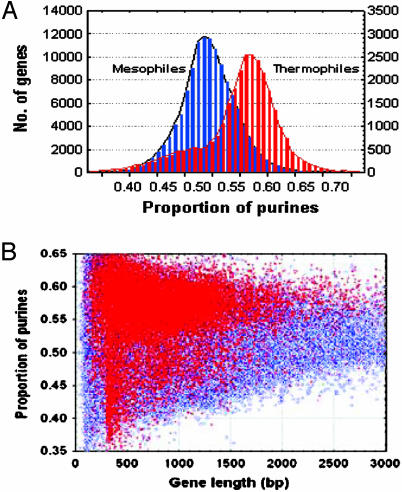
Comparing mRNAs of Thermophilic and Mesophilic Species for Purine Content and Distribution. Table 1 demonstrates that the average percentage of adenine plus guanine (AG%) content of the mRNAs is considerably higher in thermophiles as compared with mesophiles: 55.981 vs. 52.099 (P < 10-6) (for detailed data on AG% in thermophiles and mesophiles, see Tables 6 and 7, which are published as supporting information on the PNAS web site). An interesting observation was made when we compared the thermophilic and mesophilic species for the distribution of the purine abundance in their mRNA molecules (Fig. 1A). As expected, the frequency distribution of the thermophiles is shifted relative to the mesophiles in favor of higher AG proportions. However, less expected is the heterogeneity of the mRNA population of thermophiles that included, as a major fraction, the AG-enriched part and, as a minor fraction, an AG-poor part that might be related, to some extent, to horizontal gene transfer (41). Additional analysis showed that there might be some relationship between the AG% content of the mRNA molecules and their length in thermophiles but not in mesophiles (Fig. 1B). Namely, unlike mesophiles, short thermophilic mRNAs tend to avoid low-to-medium AG% content. Fig. 1B can probably be interpreted as an existence of some minimum-threshold mRNA length (≈300 bp) of thermophiles for AG% < ≈45%. Mesophiles do not seem to display such constraints.
Table 1. Prokaryote protein groups biased toward high AG% content in their mRNAs.
| No. of species | No. of genes | Average AG% | R/Y | |
|---|---|---|---|---|
| All thermophile genes | 14 | 30,374 | 55.981 | 1.272 |
| Ribosomal proteins | 14 | 849 | 61.841 | 1.620 |
| Histone-like proteins | 7 | 16 | 62.882 | 1.694 |
| RNA polymerase subunits | 14 | 165 | 58.904 | 1.433 |
| HSPs | 14 | 102 | 58.640 | 1.427 |
| All mesophile genes | 45 | 129,138 | 52.099 | 1.088 |
| Ribosomal proteins | 45 | 2,160 | 54.606 | 1.202 |
| Histone-like proteins | 15 | 39 | 54.803 | 1.212 |
| RNA polymerase subunits | 45 | 407 | 53.885 | 1.168 |
| HSPs | 45 | 795 | 54.169 | 1.174 |
Fig. 1.
Distribution of mRNA purine content in prokaryotic species. (A) Comparison of AG% between thermophilic and mesophilic species. (B) Scatter plot of AG% content and mRNA length. Thermophiles are shown in red, and mesophiles are shown in blue.
The contrasting thermal ecological groups differed not only in the relative content but also in the distribution of AG and CT tracts (Fig. 2A). Namely, polypurine tracts with five or more purines are more abundant in the thermophiles [1.7-fold compared with the mesophiles (P < 10-6) or 1.9-fold based on the total sum of purines in the tracts (P < 10-6)]. Comparison between thermophiles with the lowest (60–65°C) and highest (≥95°C) OGTs did not reveal significant differences in the average length of AG tracts (6.455 vs. 6.783; P > 0.05), but when we excluded Aeropyrum pernix (an aerobic species with high GC%), from the second subgroup the difference became highly significant (6.455 ± 0.061 vs. 6.845 ± 0.220; P < 0.01). Because of a different abundance of purines in the considered ecological groups, comparisons of the tract-length distributions will be more objective if corresponding expected distributions are taken into account. Thus, we compared the deviations of the observed distributions from the expected ones (assuming binomial distribution with parameters P = 0.56 for thermophiles and P = 0.52 for mesophiles; Fig. 2B). As can be seen from Fig. 2B, the observed distributions of polypurines and polypyrimidines in thermophiles are shifted toward higher lengths compared with the expected distributions, whereas in mesophiles this tendency is much less pronounced. In particular, the frequency of polypurine tracts with length (number of purines) within purine tracts ≥5 in thermophiles is 36% higher than expected based on the AG% content of the mRNAs (the total sum of purines in these tracts is 73% higher than expected). The frequency of pyrimidine tracts with length (number of pyrimidines) within pyrimidine tracts ≥5 in the thermophiles (albeit much lower than that of the purine tracts) is also 46% higher than expected based on the percentage of cytosine plus thymine (the total sum of pyrimidines in these tracts is 82% higher than expected). The frequencies of the pure-purinic codons within the thermophiles were 57% higher than those within the mesophiles and 43% as compared with 26% in mesophiles of these codons reside within pure-purinic tracts with length (number of purines) within purine tracts >5 (Table 2).
Fig. 2.
Frequencies of purine and pyrimidine tracts in prokaryotes. (A) Comparisons between thermophiles and mesophiles for the number of purine and pyrimidine tracts per Mb. (B) The observed vs. expected frequencies of purine tracts. thermo, thermophiles; meso, mesophiles; pur, purines; obs, observed; exp, expected.
Table 2. Prokaryote pure-purinic codon frequencies (per Mb) and codons distribution within polypurine tracts of various lengths.
| Lysine
|
Arginine
|
Glutamic acid
|
Glycine
|
|||||
|---|---|---|---|---|---|---|---|---|
| Tract size | AAA | AAG | AGA | AGG | GAA | GAG | GGA | GGG |
| Thermophiles | ||||||||
| 3–5 | 6,062 | 6,284 | 3,518 | 4,475 | 6,422 | 6,811 | 4,928 | 2,819 |
| 6–8 | 2,889 | 3,030 | 1,786 | 2,099 | 3,471 | 3,451 | 2,017 | 1,036 |
| 9–11 | 1,030 | 1,184 | 689 | 792 | 1,306 | 1,287 | 847 | 423 |
| ≥12 | 593 | 685 | 391 | 415 | 768 | 688 | 458 | 227 |
| Σ | 10,574 | 11,183 | 6,384 | 7,781 | 11,967 | 12,237 | 8,250 | 4,505 |
| Mesophiles | ||||||||
| 3–5 | 7,337 | 4,373 | 1,150 | 847 | 8,820 | 5,649 | 3,008 | 3,073 |
| 6–8 | 2,403 | 1,025 | 396 | 193 | 2,643 | 1,169 | 729 | 557 |
| 9–11 | 629 | 264 | 126 | 53 | 693 | 279 | 198 | 145 |
| ≥12 | 208 | 66 | 49 | 19 | 215 | 84 | 63 | 44 |
| Σ | 10,577 | 5,750 | 1,721 | 1,113 | 12,372 | 7,182 | 3,998 | 3,819 |
The higher abundance of purine tracts (R ≥ 5) in thermophiles compared with mesophiles corroborates well with the known higher (Glu + Lys)/(Gln + His) ratio in thermophiles (24), because the glutamic acid and lysine are encoded only by pure-purinic codons. As expected, we found higher frequencies of glutamic acid and lysine codons in thermophiles compared with mesophiles (33% and 24%, respectively; see Table 2). Likewise, the two pure-purinic codons of arginine in thermophiles are 5-fold more abundant than in mesophiles, whereas the ratio for glycine is 1.63. These findings confirmed the recent results of other authors (4, 6, 42).
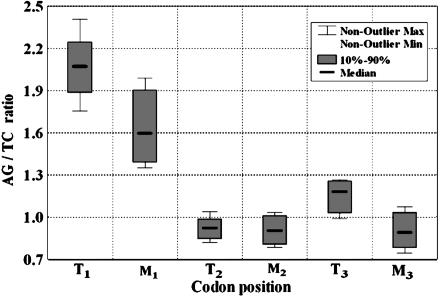
Ecological Trends in Purine vs. Pyrimidine Usage in the Three Codon Positions. Fig. 3 demonstrates a significantly higher adenine plus guanine/cytosine plus thymine (R/Y) ratio at the first position in codons used by thermophiles compared with mesophiles (R/Y ratio = 2.083 ± 0.0456 vs. 1.635 ± 0.0268; P < 10-6). For the third positions, the difference between the ecological groups in R/Y ratio was smaller but still significant (1.116 ± 0.090 in thermophiles vs. 0.897 ± 0.081 in mesophiles; P < 10-6). By contrast, for the second position, a tendency for pyrimidine preference was found for both groups (R/Y ratio = 0.919 ± 0.058 for thermophiles and 0.900 ± 0.066 for mesophiles; P > 0.1).
Fig. 3.
AG/TC in the three codon positions within the thermophiles and mesophiles [Ti and Mi; AG/CT ratio within the ith (i = 1, 2, 3) codon position in thermophiles and mesophiles].
Ecological Trends in Adenine and Guanine Usage in the Eight Pure-Purinic Codons. For the eight pure-purinic codons, mesophiles preferred adenine rather than guanine at the third position (see Table 2). In particular, for the Lys codons, the frequency of AAA is 2-fold higher than that of AAG [10,576 vs. 5,750 per megabase (Mb)]. Likewise, for the Arg codons, the ratio was AGA/AGG = 1,721:1,113; for Glu, the ratio was GAA/GAG = 12,371:7,182; and for Gly, the ratio was GGA/GGG = 3,997:3,819. This bias is not a trivial consequence of the higher adenine content of the mRNAs. Indeed, the genome-wise adenine/guanine ratio of the mRNAs was similar in mesophiles and thermophiles (1.17 and 1.21, respectively), but the thermophiles did not show this preference of adenine at the third position of pure-purinic codons (Table 2).
In cases with two sequential pure-purinic codons (SPPCs), the preferred order between two SPPCs that can form three or more runs of adenine provides longer adenine tracts. For example, between two sequential lysine codons, the order AAA–AAG appears more frequently than AAG–AAA (291 vs. 254 per Mb in thermophiles and 164 vs. 155 per Mb in mesophiles, respectively). A similar pattern was observed for the other eight combinations of SPPCs that can form (A)n with n ≥ 3 (Table 3). For thermophiles (but not mesophiles), an opposite tendency (i.e., avoidance of guanine runs) was revealed for the order of two SPPCs that might form (G)n with n ≥ 3. Thus, between the two possible sequential glycine codons, GGA–GGG was more frequent than GGG–GGA (86 and 59 per Mb, respectively). Again, a similar pattern was observed for the other eight combinations of two SPPCs with n ≥ 3 runs of guanine (Table 3).
Table 3. Sequential pure-purinic codons (frequencies per Mb) in prokaryotic genomes.
| Lysine
|
Arginine
|
Glutamic acid
|
Glycine
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Codon | AAA | AAG | AGA | AGG | GAA | GAG | GGA | GGG | Σ |
| Thermophiles | |||||||||
| AAA | 332 | 291 | 147 | 133 | 363 | 306 | 130 | 76 | 1,778 |
| AAG | 254 | 313 | 163 | 218 | 316 | 325 | 147 | 90 | 1,827 |
| AGA | 205 | 184 | 109 | 117 | 204 | 184 | 93 | 56 | 1,154 |
| AGG | 139 | 200 | 111 | 183 | 231 | 273 | 113 | 81 | 1,331 |
| GAA | 423 | 375 | 164 | 184 | 404 | 378 | 148 | 90 | 2,167 |
| GAG | 256 | 367 | 161 | 290 | 325 | 480 | 134 | 124 | 2,137 |
| GGA | 227 | 250 | 126 | 146 | 192 | 199 | 157 | 86 | 1,382 |
| GGG | 104 | 121 | 57 | 95 | 85 | 124 | 59 | 36 | 681 |
| Σ | 1,940 | 2,102 | 1,037 | 1,365 | 2,121 | 2,270 | 982 | 638 | |
| Mesophiles | |||||||||
| AAA | 436 | 164 | 59 | 25 | 421 | 207 | 97 | 89 | 1,499 |
| AAG | 155 | 106 | 19 | 12 | 158 | 93 | 45 | 50 | 639 |
| AGA | 80 | 29 | 19 | 9 | 64 | 31 | 21 | 14 | 266 |
| AGG | 25 | 16 | 8 | 6 | 32 | 21 | 12 | 11 | 130 |
| GAA | 506 | 219 | 60 | 36 | 429 | 277 | 106 | 116 | 1,749 |
| GAG | 147 | 124 | 22 | 18 | 167 | 143 | 45 | 67 | 733 |
| GGA | 148 | 57 | 29 | 15 | 114 | 65 | 49 | 31 | 508 |
| GGG | 90 | 40 | 11 | 8 | 86 | 58 | 35 | 32 | 361 |
| Σ | 1,586 | 755 | 226 | 130 | 1,471 | 895 | 410 | 410 | |
Strong Ecological Bias in Purine Abundance in mRNAs Coding for Central Elements of Transcription and Translation Machinery and Heat-Shock Proteins (HSPs). We expected that the need for more temperature-resistant structures of thermophile proteins should become mandatory to proteins involved in template-related information processing. Stronger and tighter DNA–protein and RNA–protein bonds in species living in high and very high temperatures could be achieved by the elevation of frequencies of positively charged amino acids within these proteins because of the negative charge of the phosphates within the sugar-phosphate skeleton of the nucleic acids. Elevation of the frequencies of both positively and negatively charged amino acids within protein sequences of multisubunit, large, complex machineries for transcription and translation could also contribute to thermostability of these complexes. The tests confirmed our expectations: The proteins that displayed the highest bias toward preference of purines in their mRNAs are the histone-like, ribosomal proteins and the RNAP subunits. HSP genes displayed the same pattern (Table 1). Although the purine bias within the heat-shock mRNAs is shared also by the prokaryotic mesophiles (and the eukaryotes, see below), this bias is stronger within the thermophiles.
The AG% Content Within Exons of the mRNAs of HSPs of Five Eukaryotes Examined Have Higher AG% Content than the Average for the Exons over Their Entire Genomes. The findings of higher purine content in heat-stress-related genes of both ecological groups of prokaryotes motivated us to check whether this may also be the case in eukaryotes. The results of a test conducted on five eukaryotic species basically fit this expectation (Table 4; for comparison, we also provide some results on prokaryotes from Table 1). In all tested cases, heat-stress-related genes contained a higher proportion of purines compared with the total protein-coding genome. Some differences were not high but were still highly significant (P < 10-6).
Table 4. AG% content in mRNAs of species from the three domains of life.
| All mRNAs
|
Ribosomal proteins
|
HSPs
|
||||
|---|---|---|---|---|---|---|
| AG% | Total ORFs | AG% | ORFs | AG% | ORFs | |
| Prokaryotes | ||||||
| Thermophiles | 55.981 | 29,187 | 61.84 | 849 | 58.64 | 47 |
| Mesophiles | 52.099 | 129,134 | 54.82 | 2,218 | 54.01 | 314 |
| Eukaryotes | ||||||
| Saccharomyces cerevisiae | 52.672 | 6,407 | 54.252 | 197 | 53.163 | 32 |
| Plasmodium falciparum | 58.273 | 5,408 | 59.963 | 91 | 59.080 | 18 |
| Drosophila melanogaster | 51.636 | 31,622 | 51.772 | 41 | 52.833 | 21 |
| Arabidopsis thaliana | 52.600 | 28,580 | 54.808 | 358 | 56.441 | 80 |
| Oryza sativa | 52.396 | 43,129 | 53.298 | 262 | 53.919 | 51 |
For a putative mechanism for the very high AG% in P. falciparum, see ref. 7.
Discussion
Structures and Processes That May Be Influenced by Purine Content and Distribution and Differentiate Thermophilic from Mesophilic Prokaryotes. Several potential explanations of high thermostability of some archaeal and bacterial species included specific DNA organization and processing (43–46), adaptation of the transcription apparatus (44), stability of mRNAs, efficiency of translation machinery, and protein stability. The results of our study may relate to the last four components starting with transcription. In fact, some of the revealed patterns cannot be equivocally ascribed to only one of these stages. Transcription, the DNA-directed synthesis of RNA, is the first step in the cascade of events that leads to gene expression. The elongation of the nascent mRNA strand and its processivity could be influenced by local configuration of the template DNA strand in the vicinity of, as well as at some intermediate distance from, the insertion site of the new nucleotide in the growing mRNA strand. Likewise, the elongation process may be influenced by the mRNA configuration itself and by double-stranded DNA·RNA hybrid structures. Within certain template sequences, RNAP might be prone to stall polymerization or abortive transcription (47).
Both template and nascent strand configurations might be different in medium and very high temperatures. Tracts of purines or pyrimidines and purine-pyrimidine alternations within the template strands and mRNAs could form different structures that display differential fitness under high temperatures. Variation in mRNA structure may determine its rate of translation under extreme environmental conditions (48). In addition, some structures might be less or more prone to disturbances caused by high temperatures, for example, the formation of “forbidden” RNA–RNA double strands (except the mandatory codon–anticodon contacts). Also important to the overall rate of protein synthesis is the stability of the mRNAs to spontaneous hydrolysis by intramolecular transesterification. It is noteworthy that hydrolysis is elevated dramatically at high temperature but might depend on the RNA-specific sequence organization (37–40).
Protein thermostability might be achieved by the elevation of the charged amino acid frequency within their sequences. High usage of charged amino acids could enable more inter- and intraelectrical bonds (16, 25), therefore contributing to protein stabilization. For some groups of proteins, an elevated frequency of charged amino acids within their sequences might be mandatory. Indeed, within thermophile DNA-dependent RNAP subunits, a greater number of charged residues that can form ion pairs (compared with their mesophiles' counterparts) were found (27). The affinity of thermophile ribosomal proteins to the rRNA subunits (49) might also derive from the elevation of charged amino acids within their sequences (50). For their own thermostability, HSPs might require higher levels of charged amino acids within their sequences that also could possibly enhance the ability to fulfill their role as chaperones and facilitate their contacts with other proteins and nucleic acids. HSPs are highly conserved within the three domains of life. Some of their general roles might be regarded as complementary to the cascade of events starting from transcription of mRNA resulting in the production of functioning proteins. Small HSPs have been shown to include within their sequences some “crowded” charged amino acids (51). Another example is the murine HSP86, which was found to contain internal peptide repeats of Glu-Lys-Glu within a region of highly charged amino acid residues (52).
A reasonable explanation for the high AG% within the thermophile mRNAs derives from the need for higher frequencies of Lys and Glu to stabilize their protein structures, which might also explain the relatively high AG% content in the mRNAs of HSPs in the mesophilic prokaryotes and eukaryotes compared with most of the other mRNAs. Other explanations are discussed below.
The Proposed Hypothesis. We suggest that a high AG% content contributes to thermostability already at early stages of template-based information processing starting from transcription. This ecologically associated feature seems to precede the specific stabilizing amino acids patterns determined by purine tracts. Extensive evidence supports our suggestion:
Very high frequency of purine tracts (five or more purines) within the mRNAs of thermophiles (much higher than one would expect based on the abundance of purines alone).
Preferred use of synonymous pure-purinic codons of glycine and arginine by the thermophiles.
Preferred use of two successive (neighbor) pure-purinic codons that enable the longest possible adenine tracts.
A tendency to avoid formation of guanine tracts by two successive pure-purinic codons. It might derive from the need to reduce the chances of formation of “forbidden” bonds between the nucleotides in these tracts and cytosine nucleotides in other RNA sequences.
The last tendency may possibly be due to the fact that guanine tracts are under a higher risk of forming undesired and stronger hydrogen bonds with cytosines within RNA molecules (including the rRNA), competing with the desired bonds with tRNA anticodon.
There are some possible explanations for the importance of purine tracts (and polyadenine tracts specifically) in thermophile thermostability in the early stages of template-based information processing. (i) The high transcription rate of some or most mRNAs might be facilitated by tracts of pyrimidines in the transcribed (template) DNA strand, resulting in purine tracts within the mRNAs. Specific single-stranded DNA or DNA·RNA hybrid configurations might allow a higher rate of transcription by RNAP (possibly through mediation of other proteins involved in the transcription process). (ii) The higher purine content in the mRNAs could lessen undesired RNA–RNA interactions (besides the desired codon–anticodon interaction with specific tRNAs). Because of their considerable entropy level (53), undesired RNA–RNA interactions might be favored at high temperatures. Thus, RNA sequences would possibly have adapted to avoid undesirable interactions without impairing the desirable ones (4). The biased codon usage by the thermophiles (i.e., their high bias in favor of pure-purinic codons for Arg and Gly compared with the mesophiles) supports this hypothesis. (iii) mRNAs enriched with purines (in general) and adenines (in particular) are more stable to spontaneous hydrolysis by intramolecular transesterification. Our results (see Table 3) indicate that there also might be more-subtle differences between the usages of each purine (e.g., an even higher bias to adenine rather than to guanine within two successive pure-purinic codons support the last two explanations).
It is quite possible that the primary scenario(s) of the RNA world included segments with higher-than-average GC% content, including tracts of guanine or cytosine that might have distinct helix conformations (30). Some of these islands that have relatively high GC% content may still remain in the extant genomes as remnants in the form of stable RNAs (such as rRNA and tRNA). Similarly, one may also assume that there were segments with a higher-than-average percentage of adenine plus uridine content including tracts of purines or pyrimidines that might have distinct helix conformations (31). There is a preference for mRNAs to have high purine content, and this bias is more pronounced within the thermophiles, presumably because of their higher risk for undesired RNA contacts or demands for higher mRNA stability. Universal genetic code preferences (for pure-purinic codons in three charged amino acids) that may have evolved at further steps of evolution might have resulted from the possible contribution of these charged amino acids to stabilization of proteins. It is noteworthy that each of the three stop codons, UAA, UAG, and UGA, needs only one point mutation in the first position to become pure-purine codons. This “coincidence” might have accelerated new “evolutionary trials” for building proteins (in the presumably hot world), because some of these mutations could form not only larger proteins but also proteins with higher thermostability and/or ability to participate in large protein complexes or in contacts with nucleic acids and ions.
The aforementioned evidence and analysis suggest that ecological pressures seem to shape genomic architecture and evolution, leading to improved adaptation to stressful conditions of their information-processing molecules and structures including transcription and translation machinery. Direct experimental support by protein and DNA engineering is still needed to validate the hypothesized ecological-genetic selective pressures involved in genome structural-functional evolution.
Note. As of the last correction of this article, we noticed that Lambros et al. (8) recently published their findings on the topic. In the overlap of their article and ours, there is very good agreement. Our study extends the ecological perspective by adding important data on (i) differences between thermophiles and mesophiles in relation to pure-purine tract lengths and distribution, (ii) thermophile preference of pure-purinic neighbor codons and adenine/guanine order within the pure-purinic tracts, and (iii) purine bias within heat-shock mRNAs in all three domains of life.
Supplementary Material
Acknowledgments
We thank R. Nussinov, D. R. Forsdyke, and E. Trifonov for helpful comments and suggestions. This work was supported by the Israeli Ministry of Absorption and the Ancell-Teicher Research Foundation for Molecular Genetics and Evolution.
Abbreviations: OGT, optimum growth temperature; RNAP, DNA-dependent RNA polymerase; GC%, percentage of guanine plus cytosine; AG%, percentage of adenine plus guanine; R/Y, adenine plus guanine/cytosine plus thymine; Mb, megabase; SPPC, sequential pure-purinic codon; HSP, heat-shock protein.
References
- 1.Nevo, E., Oren, A. & Wasser, S. P., eds. (2003) Fungal Life in the Dead Sea (AEG Gantner, Ruggel, Lichtenstein).
- 2.Matthews, B. W., Weaver, L. H. & Kester, W. R. (1974) J. Biol. Chem. 249, 8030-8044. [PubMed] [Google Scholar]
- 3.Perutz, M. & Raidt, H. (1975) Nature 255, 256-259. [DOI] [PubMed] [Google Scholar]
- 4.Lao, P. J. & Forsdyke, D. R. (2000) Genome Res. 10, 228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seffens, W. & Digby, D. (1999) Nucleic Acids Res. 27, 1578-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lobry, J. R. & Chessel, D. (2003) J. Appl. Genet. 44, 235-261. [PubMed] [Google Scholar]
- 7.Xue, H. Y. & Forsdyke, D. R. (2003) Mol. Biochem. Parasitol. 128, 21-32. [DOI] [PubMed] [Google Scholar]
- 8.Lambros, R. J., Mortimer, J. R. & Forsdyke, D. R. (2003) Extremophiles 7, 443-450. [DOI] [PubMed] [Google Scholar]
- 9.Yip, K. S. P., Stillman, T. J., Britton, K. L., Artymiuk, P. J., Baker, P. J., Sedelnikova, S. E., Engel, P. C., Pasquo, A., Chiaraluce, R., Consalvi, V., et al. (1995) Structure (London) 3, 1147-1158. [DOI] [PubMed] [Google Scholar]
- 10.Yip, K. S. P., Briton, K. L., Stillman, T. J., Lebbink, J., De Vos, W. M., Robb, F. T., Vetriani, C., Maeder, D. & Rice, D. W. (1998) Eur. J. Biochem. 255, 336-346. [DOI] [PubMed] [Google Scholar]
- 11.Haney, P., Konisky, J., Koretke, K. K., Luthey-Schulten, Z. & Wolynes, P. G. (1997) Proteins 28, 117-130. [DOI] [PubMed] [Google Scholar]
- 12.Russell, R. J. M., Ferguson, J. M. C., Haugh, D. W., Danson, M. J., Taylor, G. L. (1997) Biochemistry 36, 9983-9994. [DOI] [PubMed] [Google Scholar]
- 13.Russell, R. J. M., Gerike, U., Danson, M. J., Hough, D. W. & Taylor, G. L. (1998) Structure (London) 6, 351-361. [DOI] [PubMed] [Google Scholar]
- 14.Elcock, A. H. (1998) J. Mol. Biol. 284, 489-502. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, S., Ma, B., Tsai, C. J. & Nussinov, R. (2000) Proteins 38, 368-383. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, S. & Nussinov, R. (2002) Chembiochem 3, 604-617. [DOI] [PubMed] [Google Scholar]
- 17.Querol, E., Perez-Pons, J. A. & Mozo-Villarias, A. (1996) Protein Eng. 9, 256-271. [DOI] [PubMed] [Google Scholar]
- 18.Vogt, G., Woell, S. & Argos, P. (1997) J. Mol. Biol. 269, 631-643. [DOI] [PubMed] [Google Scholar]
- 19.Vogt, G. & Argos, P. (1997) Folding Des. 2, S40-S46. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., Tsai, C. J. & Nussinov, R. (2000) Protein Eng. 3, 179-191. [DOI] [PubMed] [Google Scholar]
- 21.Szilagyi, A. & Zavodszky, P. (2000) Structure (London) 8, 493-504. [DOI] [PubMed] [Google Scholar]
- 22.Thompson, M. J. & Eisenberg, D. (1999) J. Mol. Biol. 290, 595-604. [DOI] [PubMed] [Google Scholar]
- 23.Warren, G. L. & Petsko, G. A. (1995) Protein Eng. 8, 905-913. [DOI] [PubMed] [Google Scholar]
- 24.de Farias, S. T. & Bonato, M. C. M. (2002) Genome Biol. 3, preprint 0006.
- 25.Sterner, R. & Liebl, W. (2001) Crit. Rev. Biochem. Mol. Biol. 36, 39-106. [DOI] [PubMed] [Google Scholar]
- 26.Hopfner, K. P., Eichinger, A, Engh, R. A., Laue, F., Ankenbauer, W., Huber, R. & Angerer, B. (1999) Proc. Natl. Acad. Sci. USA 96, 3600-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yee, A., Booth, V., Dharamsi, A., Engel, A., Edwards, A. M. & Arrowsmith, C. H. (2000) Proc. Natl. Acad. Sci. USA 97, 6311-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galtier, N. & Lobry, J. R. (1997) J. Mol. Evol. 44, 632-636. [DOI] [PubMed] [Google Scholar]
- 29.Nakashima, H., Fukuchi, S. & Nishikawa, K. (2003) J. Biochem. 133, 507-513. [DOI] [PubMed] [Google Scholar]
- 30.Ng, H., Kopka, M. & Dickerson, R. (2000) Proc. Natl. Acad. Sci. USA 97, 2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopka, M., Fratini, A., Drew, H. & Dickerson, R. (1983) J. Mol. Biol. 163, 129-146. [DOI] [PubMed] [Google Scholar]
- 32.Ussery, D., Soumpasis, D. M., Brunak, S., Staerfeldt, H. H., Worning, P. & Krogh, A. (2002) Comput. Chem. 26, 531-541. [DOI] [PubMed] [Google Scholar]
- 33.Yagil, G. (1993) J Mol Evol. 37, 123-130. [DOI] [PubMed] [Google Scholar]
- 34.Yagil, G., Shimron, F. & Tal, M. (1998) Gene 225, 152-163. [DOI] [PubMed] [Google Scholar]
- 35.Zuckerkandle, E. (1986) J. Mol. Evol. 24, 12-27. [DOI] [PubMed] [Google Scholar]
- 36.Forsdyke, D. R. (1999) Cell Stress Chaperones 4, 205-210. [PMC free article] [PubMed] [Google Scholar]
- 37.Li, Y. & Breaker, R. R. (1999) J. Am. Chem. Soc. 121, 5364-5372. [Google Scholar]
- 38.Bibillo, A., Figlerowicz, M. & Kierzek, R. (1999) Nucleic Acids Res. 27, 3931-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kierzek, R. (1992) Nucleic Acids Res. 20, 5079-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lane, B. G. & Butler, G. C. (1959) Biochim. Biophys. Acta 33, 281-283. [DOI] [PubMed] [Google Scholar]
- 41.Koonin, E. V., Makarova, K. S. & Aravind, L. (2001) Annu. Rev. Microbiol. 55, 709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynn, D. J., Singer, G. A. C. & Hickey, D. A. (2002) Nucleic Acids Res. 30, 4272-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez-Garcia, P., Knapp, S., Ladenstein, R. & Forterre, P. (1998) Nucleic Acids Res. 26, 2322-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell, S. D., Jaxel, C., Nadal, M., Kosa, P. F. & Jackson, S. P. (1998) Proc. Natl. Acad. Sci. USA. 95, 15218-15222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marguet, E. & Forterre, P. (1998) Extremophiles 2, 115-122. [DOI] [PubMed] [Google Scholar]
- 46.Makarova, K. S., Aravind, L., Grishin, N. V., Rogozin, I. B. & Koonin, E. V. (2002) Nucleic Acids Res. 30, 482-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Droge, P. & Pohl, F. M. (1991) Nucleic Acids Res. 19, 5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storz, G. (1999) Genes Dev. 13, 633-636. [DOI] [PubMed] [Google Scholar]
- 49.Gruber, T., Kohrer, C., Lung, B., Shcherbakov, D. & Piendl, W. (2003) FEBS Lett. 549, 123-128. [DOI] [PubMed] [Google Scholar]
- 50.Härd, T., Rak, A., Allard, P., Kloo, L. & Garber, M. (2000) J. Mol. Biol. 296, 169-180. [DOI] [PubMed] [Google Scholar]
- 51.Farnsworth, P. N. & Singh, K. (2000) FEBS Lett. 482, 175-179. [DOI] [PubMed] [Google Scholar]
- 52.Moore, S. K, Kozak, C., Robinson, E. A., Ullrich, S. J. & Appella, E. (1989) J. Biol. Chem. 264, 5343-5351. [PubMed] [Google Scholar]
- 53.Cantor, C. R. & Schimmel, P. R. (1980) Biophysical Chemistry, (Freeman, San Francisco), pp. 1183-1264.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.