Abstract
Gene expression profiles from the anterior cingulate cortex (ACC) of human, chimpanzee, gorilla, and macaque samples provide clues about genetic regulatory changes in human and other catarrhine primate brains. The ACC, a cerebral neocortical region, has human-specific histological features. Physiologically, an individual's ACC displays increased activity during that individual's performance of cognitive tasks. Of ≈45,000 probe sets on microarray chips representing transcripts of all or most human genes, ≈16,000 were commonly detected in human ACC samples and comparable numbers, 14,000–15,000, in gorilla and chimpanzee ACC samples. Phylogenetic results obtained from gene expression profiles contradict the traditional expectation that the non-human African apes (i.e., chimpanzee and gorilla) should be more like each other than either should be like humans. Instead, the chimpanzee ACC profiles are more like the human than like the gorilla; these profiles demonstrate that chimpanzees are the sister group of humans. Moreover, for those unambiguous expression changes mapping to important biological processes and molecular functions that statistically are significantly represented in the data, the chimpanzee clade shows at least as much apparent regulatory evolution as does the human clade. Among important changes in the ancestry of both humans and chimpanzees, but to a greater extent in humans, are the up-regulated expression profiles of aerobic energy metabolism genes and neuronal function-related genes, suggesting that increased neuronal activity required increased supplies of energy.
Traditionally, humans are presumed to have superior cognitive abilities and, thereby, to be very different from other animals. This presumed superiority lies in the supposed uniqueness of such human abilities as producing cultural artifacts and engaging in language and symbolic thought. Recent work, however, shows that chimpanzees, who are the sister group of humans (1–6), engage in culture (7), use tools (8–10), and display rudimentary forms of language (11–13). Moreover, with regard to DNA changes that alter proteins and are favored by natural selection, chimpanzees diverge about as much from the most recent common human–chimpanzee ancestor as do humans (1, 14). Here, by estimating the relative abundance of transcribed messages of different expressed genes, we examine in humans and several other catarrhine primates gene expression profiles in an important cerebral region involved in cognition, the anterior cingulate cortex (ACC).
The ACC is typically viewed as a bridge between paleocortex and neocortex but is actually part of the neocortex (15). Histologically, the ACC shows human-specific features. For example, clusters of spindle cell pyramidal neurons occur in the ACC of humans, lesser numbers in bonobo and common chimpanzees, lesser yet in gorillas, least in orangutans, and not at all in other primates and other mammals. Moreover, the spindle cells in humans are more than twice as large as in common and bonobo chimpanzees and three times as large as in gorillas and orangutans (16). That the human ACC's spindle cells may play some special role in cognition is supported by the finding that spindle cells in the human ACC are more vulnerable to Alzheimer's disease than are other pyramidal neurons (17, 18). Physiologically, brain imaging results show increased activity in an individual's ACC when that individual is engaged in cognitive tasks (19–21). The ACC participates in decision making when interfering choices are present, a cognitive role involved in executive function (22). In view of these histological and physiological findings, it seemed likely to us that comparative data on ACC gene expression profiles from humans and their closer living relatives could provide clues about important biological processes and molecular functions associated with the evolution of human cognition. In this context, if the data were to indicate much greater regulatory evolution in the human lineage than in the chimpanzee lineage, that would be consistent with the presumption (23) that humans in their cognitive capacities are greatly different from any other animals.
Using Affymetrix (Santa Clara, CA) microarray chips with oligonucleotides (probe sets) designed to represent the transcribed messages of all or most human genes, we have now estimated the expression profiles of the genes expressed in the ACC; also on examining human, chimpanzee, gorilla, and macaque ACC samples, we have identified profiles that are species-specific and other profiles that are shared by different species. To do so, we reconstructed the phylogenetic history of the ACC gene expression profiles by treating each probe set as a single character, e.g., analogous to a single genomic locus or a single position in a sequence alignment. The relative abundance of transcribed message recognized by each probe set was then treated as a character state in a scale or ladder with steps progressing from absent to highest abundance. Using a data matrix of the character states of each character, we reconstructed by parsimony a phylogenetic tree for the four taxa represented by the ACC samples; next we determined for each character, on each of the tree's relevant branches, any changes in character states in descent from ancestors to descendants. As our results show, the most parsimonious tree constructed from these character-state transformations demonstrates that humans and chimpanzees are closest relatives, not chimpanzees and gorillas; also, simply in terms of degrees of divergence, there are fewer character-state differences between human and chimpanzee than between chimpanzee and gorilla. To obtain clues about regulatory changes associated with important biochemical pathways in the ACC's evolution, we used, from the Gene Ontology (GO) public database (24), information on the biological process(es) and molecular function(s) associated with each ACC expressed gene. We could determine that during descent, certain important biological processes and/or molecular functions are statistically significantly represented in the data. With regard to these important and significant changes, the chimpanzee lineage shows at least as much apparent regulatory evolution as does the human lineage. Especially striking in both lineages is an up-regulation of neuronal function-related genes as well as an up-regulation of aerobic energy metabolism genes, particularly in the human lineage.
Materials and Methods
Study Samples. No primates were killed for the purpose of this research. Postmortem tissue samples were obtained from the brains of three human [Homo (Homo) sapiens], one chimpanzee [Homo (Pan) troglodytes] (1), one gorilla (Gorilla gorilla) and three macaque (Macaca mulatta) individuals (see Table 1 for age and sex characteristics of each individual). Human, chimpanzee, and gorilla specimens were subjected to routine neuropathological evaluation to ensure that only normal samples were selected. Rhesus macaque brain tissue was collected from individuals who showed no indication of neurological abnormality. To prevent problems and potential artifacts related to long autolysis time, all specimens were acquired with postmortem delays that were <12 h. ACC, Brodmann's area 24, was dissected from each fresh whole-brain specimen (or from a 2-cm-thick frozen coronal slab in chimpanzee and gorilla) and frozen immediately at -70°C or below until RNA isolation and purification. Details regarding RNA isolation procedures, as well as preparation and hybridization of RNA targets, are available as Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Table 1. Expression analyses of Affymetrix HG-U133 microarray ACC data per individual and taxon.
| Probe sets detected present*
|
|||||
|---|---|---|---|---|---|
| Sample | Sex | Age | n | Percent | Total signal value of probe sets detected present |
| Human 1 | F | 24 | 19,940 | 44.38 | 1,099, 127.80 |
| Human 2 | F | 28 | 18,577 | 41.35 | 1,023,819.30 |
| Human 3 | F | 14 | 18,414 | 40.99 | 1,090,943.50 |
| Chimpanzee† | F | 34 | 17,125 | 38.11 | 1,072,568.80 |
| Gorilla† | M | 41 | 17,383 | 38.69 | 1,203,791.95 |
| Macaque 1 | M | 11 | 12,711 | 28.29 | 977,826.90 |
| Macaque 2 | F | 11 | 12,272 | 27.31 | 963,187.90 |
| Macaque 3 | M | 4 | 12,283 | 27.34 | 967,674.80 |
| Human consensus‡ | - | - | 16,076 | 35.78 | 1,033,553.73 |
| Chimpanzee consensus‡ | - | - | 14,707 | 32.73 | 1,031,384.15 |
| Gorilla consensus‡ | - | - | 14,401 | 32.05 | 1,146,903.70 |
| Macaque consensus‡ | - | - | 9,707 | 21.61 | 907,830.27 |
| Human Data Matrix B | - | - | 8,142 | 18.12 | 804,847.53 |
| Chimpanzee data | - | - | 8,142 | 18.12 | 822,550.70 |
| Matrix B | |||||
| Gorilla Data Matrix B | - | - | 8,142 | 18.12 | 900,239.85 |
| Macaque Data Matrix B | - | - | 8,142 | 18.12 | 855,939.40 |
F, female; M, male. Age is shown in years.
Of 44,928 total on the A and B chips combined
Average of two replicates
Commonly detected among all members of the species
Oligonucleotide Arrays. Samples were hybridized to Affymetrix's HG-U133 array set, which consists of two GeneChip arrays, HG-U133A and -B, that together cover >39,000 transcript variants from the human genome. Due to built-in redundancy in the array design, multiple different probe sets sometimes map to the same gene or transcript, contributing to a final total of 44,928 probe sets on the full HG-U133 array set. Each of the three human and three rhesus ACC samples was hybridized once per individual. For the single chimpanzee and gorilla ACC specimens, duplicate hybridizations were conducted by using different aliquots of each individual's RNA preparation. Thus, all together, hybridizations were conducted on 10 samples (three human, two chimpanzee, two gorilla, and three macaque). Results for all arrays may be obtained at www.genetics.wayne.edu/lgross/primates.htm.
Data Analysis. Signal values and detection calls for all samples (n = 10) were determined by using microarray suite (Ver. 5.0, Affymetrix). Signal values were scaled by using the 100 normalization controls option (target intensity = 250), and scaling factors for all hybridizations were <0.65. Three data matrices were constructed for subsequent analyses, one using detection calls (present, absent, or marginal) for each of the 44,928 probe sets in the HG-U133 set (Data Matrix A); another using only log-transformed signal values, binned into 25 categories (e.g., refs. 25 and 26 and Table 5, which is published as supporting information on the PNAS web site) from the 8,142 probe sets commonly detected among all 10 samples (Data Matrix B); and another that combined these two data matrices (Data Matrix C). In all three data matrices, each probe set corresponded to a single character. Additional details regarding each data matrix are available in the supporting information.
For all three data matrices, all character-state transformations were unordered, reversible, and equally weighted in subsequent parsimony analyses. Phylogenetic relationships were inferred and ancestral character states reconstructed by using the maximum parsimony method in paup* (27). Searches for the most-parsimonious trees were conducted by using the branch and bound algorithm performed with 500 bootstrap replicates to assess topological support. Trees were rooted by using macaque(s) as the outgroup, and branch lengths were assigned by using the DELTRAN algorithm for choosing among alternative parsimony solutions the one that places the most changes on terminal branches and the fewest on stems.
For functional analyses, we used the phylogenetic reconstruction obtained from Data Matrix B and focused on unambiguous changes that occurred on each of the four stem lineages (human, chimpanzee, human–chimpanzee, and gorilla) and the African ape–macaque internode. Functional information on these lineage-specific changes was obtained by using ontoexpress (OE, Ver. 2.0) (28, 29). OE takes a user-identified list of probe sets (or genes), assigns functional annotations to these probe sets using GO data as listed by Locus Link (30, 31), and assesses their statistical significance by comparing them to functional annotations for a reference microarray, in this case either the HG-U133A or -B array chip. Through this comparison, OE determines the probability that the functions on the user-identified list would be observed by chance given the distribution of functions on an entire chip: in essence, the lower the probability, the more significant the function due to its overrepresentation on the user-identified list.
In this study, probe sets changing from lower to higher bins on each stem were designated as up-regulated and were analyzed for functional significance separately from those changing from higher to lower bins, which were designated as down-regulated. Results were compiled only for the GO categories of biological process and molecular function. Statistical significance was determined by using OE's binomial probability distribution and Bonferroni correction options, and annotations with P ≤ 0.05 were accepted as significant.
Results
Expression Analyses per Taxon. Hybridization to the full HG-U133 array set revealed that humans are quite similar to their closer living relatives in terms of the number of probe sets detected in the ACC (Table 1). For example, of the nearly 45,000 probe sets representing the transcribed sequences of all or most human genes, ≈16,000 were commonly detected in the human ACC and comparable numbers, 14,000–15,000, in the gorilla and chimpanzee ACC samples. However, reflecting the greater phylogenetic distance of macaques from humans, somewhat <10,000 probe sets were commonly detected in the macaque ACC samples (Table 1). An additional feature of our analyses was to total the signal values of the detected probe sets for each sample, for the commonly detected probe sets in each species, and for commonly detected probe sets in all 10 samples. These results show (Table 1) that, although the number of detected probe sets decreases with phylogenetic distance, their totaled signal values per nonhuman sample or species do not decrease with increasing phylogenetic distance but remain about the same as the human total. Furthermore, although the 8,142 probe sets commonly detected in all 10 samples are <50% of the probe sets detected in each human sample, their totaled signal values account for ≈78–87% of the total signal value calculated for the 16,076 probe sets commonly detected among all three humans. Apparently the probe sets with low-level expression profiles in the human samples are more apt to be judged as absent in phylogenetically distant (e.g., macaque) samples than are the probe sets with higher-level expression profiles. Moreover, for the genes commonly present in both human and nonhuman samples, there are a greater number of probe sets with higher signal values in the nonhuman than in the human samples (Table 2).
Table 2. Probe sets showing higher expression in pairwise comparisons among primates among commonly detected probe sets (n = 8,142).
| Human | Chimpanzee | Gorilla | Macaque | |
|---|---|---|---|---|
| Human | — | 1,678 | 1,454 | 1,915 |
| Chimpanzee | 2,481 | — | 1,642 | 2,140 |
| Gorilla | 2,853 | 2,412 | — | 2,410 |
| Macaque | 2,856 | 2,415 | 2,006 | — |
Numbers above or below diagonal represent the total number of probe sets with expression values higher in the species indicated in the row when compared to the species indicated in the column. Only those probe sets for which expression values in all representatives of a species were greater than expression values in all representatives of the compared species are shown.
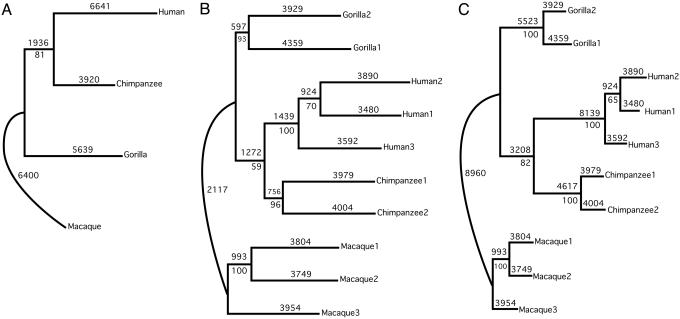
Phylogenetic Analyses. All three data matrices yielded the same most-parsimonious branching arrangement: human and chimpanzee clades are sister taxa to the exclusion of the gorilla clade, which forms a sister group to the human–chimpanzee clade (Fig. 1). In addition, results from Data Matrices A and C (Fig. 1 A and C) show reasonably good bootstrap support for the human–chimpanzee clade (81–82%). These results agree with DNA evidence on African ape phylogeny (1–6, 32) and further justify treating humans as members of an African ape clade. Results from reconstructing ancestral ACC expression patterns from all three data matrices, however, show that the lineages leading to the three humans diverge more from the ancestral African ape state than do the chimpanzee and gorilla lineages. Nevertheless, with each of the three data matrices, the chimpanzee clade diverges more from the gorilla clade than from the human clade.
Fig. 1.
The optimal maximum parsimony trees inferred by phylogenetic analysis of primate ACC microarray data. Trees are shown in phylogram format (except the lineage leading to the macaques), with branch lengths proportional to the assigned changes for that branch as determined with the DELTRAN option in parsimony analysis. Numbers above a branch indicate the assigned branch length. Number below an internode indicates the bootstrap support value for that node (500 replicates). (A) Topology obtained from Data Matrix A, the detection call analysis. Tree length, 24,266. Detection calls were generated for each taxon by assigning as present or absent only those probe sets consistently called present or absent among all three biological replicates or between duplicates. Probe sets with variable detection calls within species, or those consistently detected as marginal within a species, were assigned a call of marginal. (B) Topology obtained from Data Matrix B, the binned signal values of commonly detected probe sets. Tree length, 46,838. Continuous characters were converted into one of 25 discrete bins before analysis (see Materials and Methods for details). (C) Topology obtained from Data Matrix C, the combined data matrix. Tree length, 71,104. Bins from Data Matrix B were retained and combined into an alignment with Data Matrix A. Values assigned by the latter on a per-taxon basis were further assigned to each individual (or replicate of an individual) included in that taxon.
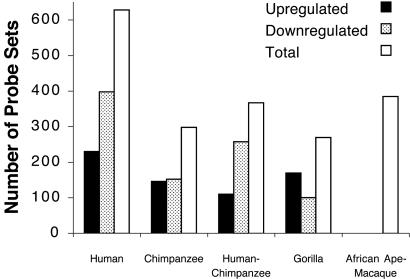
Functional Analyses. Phylogenetic analysis and ancestral character-state reconstruction using Data Matrix B indicated that the greatest number of unambiguous changes occurred on the human stem, and that there were more down-than up-regulated changes (Fig. 2). The human stem is defined as the lineage leading from the most recent common ancestor (mrca) of humans and chimpanzees to the mrca of the three humans. Similarly, the chimpanzee stem is defined as the lineage leading from the mrca of humans and chimpanzees to the joining of the two chimpanzee samples; the human–chimpanzee stem is defined as the lineage leading from the mrca of African apes to the mrca of humans and chimpanzees; the gorilla stem is defined as the lineage leading from the mrca of African apes to the joining of the two gorilla samples; and the African ape–macaque internode is defined as the lineage leading from the mrca of the three macaques to the mrca of the African apes.
Fig. 2.
Number of probe sets changing unambiguously on each stem lineage and/or internode. Shown are the number of up- and down-regulated probe sets analyzed for functional information using oe (Ver. 2.0) (28, 29). Because expression levels at the ancestral catarrhine node are unknown, the direction of expression change was not reconstructed for the African ape–macaque internode.
Among the 1,947 unambiguous changes, OE analyses of the genes showing these apparent expression changes revealed 382 biological processes and molecular functions that meet the Bonferroni criterion of statistical significance (Tables 6–10, which are published as supporting information on the PNAS web site): 69 on the human stem, mapping to a total of 71 genes (32 up-regulated, 39 down-regulated); 106 on the chimpanzee stem, mapping to a total of 66 genes (34 up-regulated, 32 down-regulated); 88 on the human–chimpanzee stem, mapping to a total of 56 genes (25 up-regulated, 31 down-regulated); 91 on the gorilla stem, mapping to a total of 68 genes (37 up-regulated, 31 down-regulated); and 82 on the African ape–macaque internode, mapping to a total of 72 genes. Furthermore, of the GO annotations judged to be significantly represented in the data, 50 were not species-specific in that OE analysis found each of these to be significant on two or more stems (Table 11, which is published as supporting information on the PNAS web site).
In terms of the percentage of significant GO annotations that were species-specific, i.e., each unique to a stem, the human stem was very similar to the chimpanzee stem: 75.3% of significant GO annotations on the human stem being human-specific, and 74.5% of significant GO annotations on the chimpanzee stem being chimpanzee-specific. This finding suggests that regulatory evolution was as extensive in the chimpanzee as in the human stem.
Tables 6–10 give a full listing of the GO annotations meeting the Bonferroni criterion of statistical significance for each of the four stems and the African ape–macaque internode, along with the genes to which they map. This full listing reveals that, on both the human–chimpanzee stem (Table 8) and descendant human stem (Table 6), there is a striking pattern of down-regulation in genes with functions relating to transcription, translation, and mRNA splicing and processing. In turn, on both the human and chimpanzee stems, there is a striking pattern of up-regulation in neuronal function-related genes and also of genes involved in aerobic energy metabolism, in particular electron transport chain (ETC) functioning genes (Tables 3, 4, 6, and 7). The striking pattern of up-regulated ETC genes is further shown on the human–chimpanzee stem (Tables 4 and 8).
Table 3. Significant neuronal function-related GO annotations and associated genes and lineages.
| Annotation | Gene | Human | Chimpanzee | Human–chimpanzee | Gorilla | African ape–macaque* |
|---|---|---|---|---|---|---|
| Axon guidance | NRXN3 | U (19–21) | ||||
| Dendrite morphogencsis | PTD009 | U (12–15) | ||||
| GABAA receptor activity | GABRB3 | U (12–20) | D (22–20) | |||
| GABA/sodium symporter activity | SLC6A1 | D (22–20) | ||||
| GABA acid transporter activity | SLC6A1 | D (22–20) | ||||
| Glia cell migration | SLIT2 | U (12–14) | ||||
| Motor axon guidance | SLIT2 | U (12–14) | ||||
| Neuromuscular junction development | CUGBP2 | D (24–22) | ||||
| Neuromuscular synaptic transmission | DTNA | D (20–17) | ||||
| Neuron differentiation | STMN2 | D (24–23) | ||||
| Neuronal cell recognition | SLIT2 | U (12–14) | ||||
| Neurotransmitter receptor activity | GABRB3 | U (12–20) | D (22–20) | |||
| Neurotransmitter receptor biosynthesis | PTD009 | U (12–15) | ||||
| Neurotrophin TRKB receptor activity | NTRK2 | U (8–9) | U (16–23) | |||
| Regulation of action potential | SRI | U (19–20) | ||||
| Regulation of neurotransmitter levels | GLUL | U (23–24) | ||||
| Regulation of synapse | NPY | U (18–19) | ||||
| Synaptic transmission | GABRB3 | U (12–20) | D (22–20) | |||
| KCNIP2 | D (18–17) | |||||
| MAPK1 | U (22–23) | |||||
| Synaptic vesicle transport | SYT4 | U (20–23) | ||||
| Total number of genes | 15 | 4 | 3 | 4 | 5 | 1 |
| Total number of GO annotations | 19 | 5 | 6 | 5 | 6 | 1 |
GABA, γ-amino butyric acid; GABAA, GABA type A. U, up-regulated; D, down-regulated. Numbers in parentheses indicate the bin categories spanned by each change.
Change on the ancestral African ape node with respect to the ancestral macaque node
Table 4. ETC genes and lineages with significant GO annotations and additional ETC genes and lineages that change unambiguously.
| Complex | Gene | Human | Chimpanzee | Human–chimpanzee | Gorilla | African ape–macaque* |
|---|---|---|---|---|---|---|
| I | GRIM19 | U (21–22) | U (11–21) | |||
| NDUFA2 | D (22–18) | |||||
| NDUFA4 | U (22–24) | |||||
| NDUFA10 | U (21–22) | U (10–21) | ||||
| NDUFA11 | U (11–22) | |||||
| NDUFAB1 | U (19–23) | |||||
| NDUFB1 | U (21–22) | |||||
| NDUFB4 | U (22–23) | |||||
| NDUFB8 | U (3–8) | U (12–23) | ||||
| NDUFC2 | U (22–23) | |||||
| NDUFS4 | U (17–18) | U (15–17) | ||||
| NDUFS6 | U (15–17) | |||||
| III | HSPC051 | U (14–23) | ||||
| UQCRC2 | U (18–20) | D (18–14) | ||||
| UQCRH | U (21–23) | |||||
| IV | COX4I1 | U (20–23) | ||||
| COX5A | U (20–22) | |||||
| COX5B | U (22–23) | U (11–23) | ||||
| COX7C | U (23–24) | |||||
| V | ATP5C1 | U (22–23) | U (21–22) | |||
| ATP5G3 | D (22–20) | |||||
| ATP5H | U (23–24) | |||||
| ATP5J2 | U (20–22) | |||||
| ATP1F1 | N | D (19–11) | ||||
| Other | CYCS | U (23–24) | ||||
| Total genes | 25 | 10 | 6 | 3 | 3 | 11 |
See Results and Tables 6–10 for a full listing of these ETC-related GO annotations. Bold, ETC genes that change unambiguously on one or more relevant branches. U, up-regulated; D, down-regulated. Numbers in parentheses indicate the bin categories spanned by each change.
Change on the ancestral African ape node with respect to the ancestral macaque node; N, indeterminate, because different probe sets for the same gene showed opposite directions of change
Because the ACC plays a key role in cognition, it probably functions at high energy levels in humans and their close relatives. To test this hypothesis, we determined how many gene expression profiles represented ETC genes. Of the 79 nuclear-encoded genes currently mapped to one of the five ETC complexes on the HG-U133 array set, 67 were detected as present in all 10 samples and represent genes from all five complexes (data not shown). The remaining 12 were not uniformly detected among all samples, largely due to a lack of detection in one or more of the macaques. OE analyses identified six GO annotations involving genes mapping exclusively to complexes I, III, and IV to be statistically significant on one or more of the four stems and/or the African ape–macaque internode (Tables 4, 6–10). These annotations are: mitochondrial electron transport, NADH to ubiquinone; NADH dehydrogenase activity; NADH dehydrogenase (ubiquinone) activity; mitochondrial electron transport, ubiquinol to cytochrome c; oxidative phosphorylation; and cytochrome c oxidase activity.
In addition to the 14 genes identified through OE analysis, 11 more ETC genes change unambiguously on one or more of the relevant branches (Table 4). Five of these 11 genes belong to Complex V, the energy-generating step of the ETC. The extended analysis of all unambiguous changes indicates that genes of the ETC are well-represented as changing on most stems and most frequently in an up-regulated direction. On two of the four stems (chimpanzee and human–chimpanzee), changes are observed in genes belonging to complexes I, III, and V and are up-regulated in nearly all cases. In addition, the human stem shows up-regulated changes in genes mapping to complex IV (cytochrome c oxidase, or COX) and the gene encoding cytochrome c (CYCS), the carrier protein that shuttles electrons between complexes III and IV, contributing to a total of 10 ETC genes that all change in an up-regulated direction on this stem.
Discussion
Analyses of the ACC gene expression profiles obtained in this study did not show an unusual amount of regulatory evolution in the human clade. Despite the bias of human-based probe sets, the chimpanzee clade shows at least as much apparent regulatory evolution as the human clade does for those unambiguous expression changes mapping to important biological processes and molecular functions that statistically are significantly represented in the data. The ACC is a specialized region of the neocortex involved in the regulation of emotional and cognitive behavior (15). Its involvement in the proper performance of executive functions, i.e., the processes involved in maintaining goals while also managing constraints on achieving those goals (22), confirms its important role in human cognition. Anatomical evidence suggests that the ACC may have acquired specialized functions during the evolution of human cognition: novel pyramidal neuronal cell types, spindle-cell projection neurons, and calretinin-expressing projection neurons are found in the ACC of humans and other large-bodied apes (common and bonobo chimpanzees, gorillas, and orangutans) but not in any other primate species or other mammal and are found in greatest numbers in humans (16, 33). Nevertheless, microarray results from the ACC show that humans and chimpanzees are more similar to each other than either is to other catarrhines, and that they share similar proportions of statistically significant unique biological processes and molecular functions, suggesting that each of these lineages has acquired important and significant regulatory changes since the divergence from their last common ancestor.
These findings contrast somewhat with previous reports of primate brain gene expression profiles investigated by using human-based microarrays (23, 34), which indicated an up-regulated pattern of gene expression in humans (34) and an accelerated rate of evolutionary change in humans vs. chimpanzee brain gene expression levels (23) [although subsequent reanalyses of these data provided contradictory views of this finding (35, 36)]. These reports present results from microarray chips representing transcripts from ≈10,000–18,000 human genes; in contrast, our report presents results from >39,000 transcript variants derived from ≈33,000 human genes recognized by Affymetrix. Moreover, our phylogenetic results revealed during descent changes in gene expression profiles that, when coupled to GO information, identified those changes associated with important and significant biological processes and molecular functions.
One criticism of using human-based chips to estimate abundance of transcripts in nonhuman samples is that sequence variation among human and nonhuman samples may confound efforts to identify true differences in gene expression. If sequence variation between a human-designed probe set and a nonhuman transcript lowered hybridization efficiency between the two, this could cause either a false negative or detection of a lower abundance than truly exists in the nonhuman primate ACC sample. This problem is expected to be more pronounced when comparing macaque to human samples than when comparing chimpanzee to human samples because of the higher sequence divergence in the former comparison (1). Because similar quantities of RNA were used per sample, an increase in sequence divergence between human and nonhuman samples should both increase the fraction of nonhuman transcripts that escape detection and decrease the nonhuman's totaled signal values compared to the human's. Contradicting this expectation, the chimpanzee's total signal value is as large as the human's, and the gorilla's total signal value is larger, but by a small amount (Table 1). Although the number of detected macaque transcripts is less than two-thirds the number of detected human transcripts (9,707–16,076), the macaque total signal value is ≈9/10ths as large as the human total signal value (907,830–1,033,554; see Table 1). Further, as shown in Table 2, among genes commonly detected among all members of the four study taxa, there is a strong tendency for the nonhuman samples to express more than the human samples. These two sets of findings (Tables 1 and 2) suggest that, for the 8,142 commonly detected transcripts, a detection of up-regulation during descent in the human stem may more likely be due to a real expression level change rather than to a transcript sequence change.
Our present results identify important changes that have occurred since the human–chimpanzee mrca. Specifically, two categories of genes show distinctive patterns of up-regulation in both the human and chimpanzee stems: genes involved in aerobic energy metabolism and genes related to neuronal functions (Tables 3 and 4). That these two categories show this up-regulated pattern may not be coincidental: studies involving macaques have shown that in neurons, COX activity (i.e., metabolic activity) is higher in dendrites than in axons (37, 38), and that this activity is tightly regulated by neuronal functional activity (39). Although these studies did not involve the ACC, this brain region, which is known to be highly active in humans during both emotional and cognitive tasks (15, 19–21, 40), could be similarly regulated. The up-regulation of ETC and neuronal function-related genes on the human and chimpanzee stems would thus indicate the relatively greater neuronal functional activity and metabolic demand of the ACC in these species relative to other species included in our study.
Our previous work on the evolution of the ETC has provided evidence for positive selection in the common ancestry of the present-day large-brained primates. We have shown upsurges of amino acid replacements relative to synonymous (amino acid maintaining) substitutions in the encoded mitochondrial-functioning proteins of the ETC (41–48). This is consistent with the hypothesis that ETC genes have coadaptively evolved under positive Darwinian selection for new or improved function in the brain (49, 50) Results from the present study indicate that several ETC genes show apparent expression differences as well (Table 4), suggesting that this pathway has been subject to both gene regulatory and protein sequence evolution during the descent of catarrhines.
To test this possibility, further research should use microarrays based on the nucleotide sequences of the actual gene transcripts for each catarrhine species tested, and these genes should be cross-referenced to their orthologous genes in other species. In addition, more rigorous quantitative measures of transcript abundance should be obtained for a variety of genes, such as those identified in the present study as associated with important significant biological processes and molecular functions. Our present phylogenetic results depict, during descent of several catarrhine primates, gene expression profile changes that group the chimpanzee clade closest to the human rather than to the gorilla clade. Future research should establish whether this is due to real evolutionary changes in levels of gene expression or simply to interspecies differences in transcript nucleotide sequences.
Supplementary Material
Acknowledgments
We thank Dr. Don R. Canfield (California National Primate Research Center at the University of California, Davis) for providing the rhesus macaque samples and Dr. Joseph M. Erwin (Great Apes Aging Project) for providing the chimpanzee and gorilla samples used in this study. We also thank Drs. Jerry Slightom and David Fitch for insightful discussion and Dr. Gerardus Tromp for statistical advice. Wayne State University's Applied Genomics Technology Facility provided invaluable technical assistance during the course of this study. P.R.H. is the Regenstreif Professor of Neuroscience. This work was supported by National Science Foundation Grants 0118696 and 0318375 and National Institutes of Health Grant GM65580.
Abbreviations: ACC, anterior cingulate cortex; ETC, electron transport chain; GO, Gene Ontology; OE, ontoexpress; mrca, most recent common ancestor.
References
- 1.Wildman, D. E., Uddin, M., Liu, G., Grossman, L. I. & Goodman, M. (2003) Proc. Natl. Acad. Sci. USA 100, 7181-7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyamoto, M. M., Slightom, J. L. & Goodman, M. (1987) Science 238, 369-373. [DOI] [PubMed] [Google Scholar]
- 3.Sibley, C. G. & Ahlquist, J. E. (1987) J. Mol. Evol. 26, 99-121. [DOI] [PubMed] [Google Scholar]
- 4.Goodman, M., Tagle, D. A., Fitch, D. H., Bailey, W., Czelusniak, J., Koop, B. F., Benson, P. & Slightom, J. L. (1990) J. Mol. Evol. 30, 260-266. [DOI] [PubMed] [Google Scholar]
- 5.Goodman, M., Porter, C. A., Czelusniak, J., Page, S. L., Schneider, H., Shoshani, J., Gunnell, G. & Groves, C. P. (1998) Mol. Phylogenet. Evol. 9, 585-598. [DOI] [PubMed] [Google Scholar]
- 6.Salem, A. H., Ray, D. A., Xing, J., Callinan, P. A., Myers, J. S., Hedges, D. J., Garber, R. K., Witherspoon, D. J., Jorde, L. B. & Batzer, M. A. (2003) Proc. Natl. Acad. Sci. USA 100, 12787-12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiten, A., Goodall, J., McGrew, W. C., Nishida, T., Reynolds, V., Sugiyama, Y., Tutin, C. E., Wrangham, R. W. & Boesch, C. (1999) Nature 399, 682-685. [DOI] [PubMed] [Google Scholar]
- 8.Goodall, J. [v. L.] (1968) Anim. Behav. Monogr. 1, 165-311. [Google Scholar]
- 9.Biro, D., Inoue-Nakamura, N., Tonooka, R., Yamakoshi, G., Sousa, C. & Matsuzawa, T. (2003) Anim. Cognit. 6, 213-223. [DOI] [PubMed] [Google Scholar]
- 10.Yamakoshi, G. & Myowa-Yamakoshi, M. (2003) Primates 45, 25-32. [DOI] [PubMed] [Google Scholar]
- 11.Savage-Rumbaugh, E. S., Rumbaugh, D. M. & Boysen, S. (1978) Science 201, 641-644. [DOI] [PubMed] [Google Scholar]
- 12.Savage-Rumbaugh, S., Rumbaugh, D. M. & McDonald, K. (1985) Neurosci. Biobehav. Rev. 9, 653-665. [DOI] [PubMed] [Google Scholar]
- 13.Bodamer, M. D. & Gardner, R. A. (2002) J. Comp. Psychol. 116, 12-26. [DOI] [PubMed] [Google Scholar]
- 14.Clark, A. G., Glanowski, S., Nielsen, R., Thomas, P. D., Kejariwal, A., Todd, M. A., Tanenbaum, D. M., Civello, D., Lu, F., Murphy, B., et al. (2003) Science 302, 1960-1963. [DOI] [PubMed] [Google Scholar]
- 15.Allman, J. M., Hakeem, A., Erwin, J. M., Nimchinsky, E. & Hof, P. (2001) Ann. NY Acad. Sci. 935, 107-117. [PubMed] [Google Scholar]
- 16.Nimchinsky, E. A., Gilissen, E., Allman, J. M., Perl, D. P., Erwin, J. M. & Hof, P. R. (1999) Proc. Natl. Acad. Sci. USA 96, 5268-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nimchinsky, E. A., Vogt, B. A., Morrison, J. H. & Hof, P. R. (1995) J. Comp. Neurol. 355, 27-37. [DOI] [PubMed] [Google Scholar]
- 18.Morrison, J. H. & Hof, P. R. (1997) Science 278, 412-419. [DOI] [PubMed] [Google Scholar]
- 19.Pardo, J. V., Pardo, P. J., Janer, K. W. & Raichle, M. E. (1990) Proc. Natl. Acad. Sci. USA 87, 256-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter, C. S., Macdonald, A. M., Botvinick, M., Ross, L. L., Stenger, V. A., Noll, D. & Cohen, J. D. (2000) Proc. Natl. Acad. Sci. USA 97, 1944-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacDonald, A. W., 3rd, Cohen, J. D., Stenger, V. A. & Carter, C. S. (2000) Science 288, 1835-1838. [DOI] [PubMed] [Google Scholar]
- 22.Hartley, A. A. & Speer, N. K. (2000) Microsc. Res. Tech. 51, 45-53. [DOI] [PubMed] [Google Scholar]
- 23.Enard, W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. (2002) Science 296, 340-343. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maddison, W. P. & Maddison, D. R. (1992) macclade (Sinauer, Sunderland, MA), Ver. 3.0.
- 26.Planet, P. J., DeSalle, R., Siddall, M., Bael, T., Sarkar, I. N. & Stanley, S. E. (2001) Genome Res. 11, 1149-1155. [DOI] [PubMed] [Google Scholar]
- 27.Swofford, D. L. (2002) paup*: Phylogenetic Analysis Using Parsimony (and Other Methods), Ver. 4.0b10 (Sinauer, Sunderland, MA).
- 28.Khatri, P., Drǎghici, S., Ostermeier, G. C. & Krawetz, S. A. (2002) Genomics 79, 266-270. [DOI] [PubMed] [Google Scholar]
- 29.Drǎghici, S., Khatri, P., Bhavsar, P., Shah, A., Krawetz, S. A. & Tainsky, M. A. (2003) Nucleic Acids Res. 31, 3775-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruitt, K. D., Katz, K. S., Sicotte, H. & Maglott, D. R. (2000) Trends Genet. 16, 44-47. [DOI] [PubMed] [Google Scholar]
- 31.Pruitt, K. D. & Maglott, D. R. (2001) Nucleic Acids Res. 29, 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodman, M. (1999) Am. J. Hum. Genet. 64, 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hof, P. R., Nimchinsky, E. A., Perl, D. P. & Erwin, J. M. (2001) Neurosci. Lett. 307, 139-142. [DOI] [PubMed] [Google Scholar]
- 34.Cáceres, M., Lachuer, J., Zapala, M. A., Redmond, J. C., Kudo, L., Geschwind, D. H., Lockhart, D. J., Preuss, T. M. & Barlow, C. (2003) Proc. Natl. Acad. Sci. USA 100, 13030-13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu, J. & Gu, X. (2003) Trends Genet. 19, 63-65. [DOI] [PubMed] [Google Scholar]
- 36.Hsieh, W. P., Chu, T. M., Wolfinger, R. D. & Gibson, G. (2003) Genetics 165, 747-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong-Riley, M. T. (1989) Trends Neurosci. 12, 94-101. [DOI] [PubMed] [Google Scholar]
- 38.Hevner, R. F. & Wong-Riley, M. T. (1991) J. Neurosci. 11, 1942-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hevner, R. F. & Wong-Riley, M. T. (1990) J. Neurosci. 10, 1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisenberger, N. I., Lieberman, M. D. & Williams, K. D. (2003) Science 302, 290-292. [DOI] [PubMed] [Google Scholar]
- 41.Wu, W., Goodman, M., Lomax, M. I. & Grossman, L. I. (1997) J. Mol. Evol. 44, 477-491. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, T. R., Jaradat, S. A., Goodman, M., Lomax, M. I. & Grossman, L. I. (1997) Mol. Biol. Evol. 14, 595-601. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt, T. R., Goodman, M. & Grossman, L. I. (1999) Mol. Biol. Evol. 16, 619-626. [DOI] [PubMed] [Google Scholar]
- 44.Wu, W., Schmidt, T. R., Goodman, M. & Grossman, L. I. (2000) Mol. Phylogenet. Evol. 17, 294-304. [DOI] [PubMed] [Google Scholar]
- 45.Baba, M. L., Darga, L. L., Goodman, M. & Czelusniak, J. (1981) J. Mol. Evol. 17, 197-213. [DOI] [PubMed] [Google Scholar]
- 46.Wildman, D. E., Wu, W., Goodman, M. & Grossman, L. I. (2002) Mol. Biol. Evol. 19, 1812-1815. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt, T. R., Goodman, M. & Grossman, L. I. (2002) Gene 286, 13-19. [DOI] [PubMed] [Google Scholar]
- 48.Goldberg, A., Wildman, D. E., Schmidt, T. R., Hüttemann, M., Goodman, M., Weiss, M. L. & Grossman, L. I. (2003) Proc. Natl. Acad. Sci. USA 100, 5873-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grossman, L. I., Schmidt, T. R., Wildman, D. E. & Goodman, M. (2001) Mol. Phylogenet. Evol. 18, 26-36. [DOI] [PubMed] [Google Scholar]
- 50.Goodman, M., McConkey, E. H. & Page, S. L. (2002) in New Perspectives in Primate Evolution and Behaviour, eds. Harcourt, C. S. & Sherwood, B. R. (Westbury Academic and Scientific, Otley, West Yorkshire, U.K.), pp. 47-70.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.