Abstract
To date, there has been no way to examine induced human p53 gene mutations in cell cultures exposed to mutagenic factors, other than by restriction site analysis. Here, we used embryonic cells from our Hupki (human p53 knock-in) mouse strain to generate human p53 DNA-binding domain (DBD) mutations experimentally. Twenty cultures of untreated primary mouse Hupki fibroblasts and 20 short-wavelength UV light (UVC)-treated cultures (20J/m2) were passaged >20 times. Established Hupki embryonic fibroblast cell lines (HUFs) were genotyped by dideoxy DNA sequencing of p53 exons 4-9. Seven of the HUFs harbored point mutations in the humanized p53 DBD. Of the 9 mutations (6 single- and 1 triple-site mutation), 2 were at the most frequently mutated codons in human cancers (c.248 and c.273). The Affymetrix p53 GeneChip assay also readily identified the 6 single-base substitutions. All mutations in HUFs from UV-treated cultures were at dipyrimidine sites, including 3 nontranscribed strand C →T transitions. The mutant HUFs were deficient in p53 transactivation function, and missense mutants had high levels of nuclear p53 protein. In a second experiment, primary Hupki cells were exposed to the carcinogen aristolochic acid I (AAI). Five of 10 cultures that became established within 2 months harbored p53 DBD mutations. All were transversions, including 4 A → T substitutions on the nontranscribed strand, a hallmark of DNA mutation by AAI. We conclude that establishment of Hupki mouse fibroblasts in culture readily selects for p53 DBD mutations found in human tumors, providing a basis for generating experimental mutation patterns in human p53.
Keywords: tumor suppressor, mutation spectrum, gene targeting, carcinogen
The most common human tumor DNA sequence change is a p53 gene missense mutation in the segment encoding the DNA-binding domain (DBD) of the protein. This event leads to nuclear accumulation of p53 protein and loss of its normal functions, such as transcriptional transactivation of target genes that regulate the cell cycle and apoptosis (1). Over 18,000 human tumor mutations in the p53 gene have been registered in the International Agency for Research on Cancer TP53 database R8 [www.iarc.fr/p53 (2, 3)]. Scrutiny of these data has provided clues to cancer-causing agents and to endogenous promutagenic processes in humans (4-7). Furthermore, detailed analysis of this large set of mutations has substantiated inferences on mechanisms of mutagenesis originally derived from experiments with lower organisms and experimental mutagenesis assays (8-15).
Although the nature of a mutagenic agent clearly is a key factor in shaping a mutation spectrum (8), base context and biological selection are also crucial determinants. A powerful approach to testing hypotheses on the origins and modulators of p53 mutation patterns in human tumors would be a mutagenesis assay in which the exogenous or endogenous mutagen targets human p53 gene sequences in the living cell, and in which the resultant mutant p53 protein then confers a selectable phenotype, preferably one that corresponds to the aberrant functions characteristic of human tumor p53 mutants. There has been no way to accomplish this goal with human cells. The most widely used systems to study mechanisms of mutation or mutation spectra exploit the elegance and efficiency of working with either lower organisms or their genes [for example, the Ames Salmonella typhimurium test, the BigBlue (LacI) and MutaMouse (LacZ) rodent systems, and the yeast-based Functional Analysis of Separated Alleles in Yeast (FASAY)] or are based on mutation of non-cancer-related genes [e.g., hypoxanthine phosphoribosyltransferase (HPRT)] that confer drug resistance and thereby a selection method to recover mutant cells among a large population of unmutated human or rodent cells (16-22).
Here, we explore an approach for generating and selecting dysfunctional human p53 mutations experimentally. We have made use of gene-targeting in mice to humanize the endogenous murine p53 gene and have exploited the metabolic competence of primary embryonic fibroblasts, as well as the fact that mouse cells, in contrast to human cells, readily undergo spontaneous disruption of the p53/p19ARF pathways during establishment in culture (23). We reported previously the construction of the human p53 knock-in (Hupki) mouse (The Jackson Laboratory Repository designation: 129Trp53tm/Holl), which involved gene-targeted replacement of the endogenous murine p53 coding sequences encompassing exons 4-9 with the homologous WT human sequences (24, 25). Homozygous Hupki mice are phenotypically normal, not tumor-prone, and retain a variety of normal (WT) p53 functions and characteristics, including nuclear accumulation of p53 protein after exposure to DNA-damaging agents, transcriptional activation of known p53 targets, and induction of apoptosis in Hupki thymocytes after γ-irradiation, a process that depends on p53 function (24, 26, 27). We reasoned that p53 DBD mutations would arise spontaneously and after mutagen exposure in Hupki fibroblast cultures, and would be selected for during subsequent growth in vitro. We isolated Hupki fibroblast cell lines that were found to harbor functionally deficient human p53 DBD mutations typical of human tumors.
Methods
Cell Culture. Untreated and UV-treated cells. Primary Hupki (Trp-53tm/Holl; homozygous for the humanized knock-in p53 allele) mouse fibroblasts were harvested from 13.5-day-old embryos according to standard procedures (28). Cells trypsinized and replated from dishes receiving the embryonic cell suspensions were designated as passage 1 (p.1). Forty cultures of ≈2 × 105 primary embryonic fibroblasts were subcultured at a 1:4 dilution (1:2 during crisis) for >24 passages; 20 of the cultures were irradiated at p.2 with short-wavelength UV light (UVC) (20 J/m2; cell survival 72 h posttreatment, 30%). Cells were grown in DMEM supplemented with 10% FCS (15% at p.5-8), and aliquots were frozen at p.12-15 (set I) and at p.23-33 (set II). Initial p53 mutation screening of all cultures was performed with cell populations at the later passage (set II), which we designate hereafter as Hupki embryonic fibroblast cell lines (HUFs). DNA from set I frozen cell aliquots corresponding to HUFs found to have p53 mutations was reanalyzed to determine whether the mutation could be detected at this earlier passage. Cell lines with multiple mutations or with putative heterozygous mutations were subcloned by dilution cloning and resequenced to verify that mutant cells were heterozygous for the p53 mutation.
Aristolochic acid I (AAI)-treated cells. Twenty-four cultures of Hupki primary cells were exposed to 100 μM AAI (10-mM stock solution as sodium salt in sterile water; cell survival at 24 h after end of treatment, 15%) for 48 h and then passaged for 8-10 weeks (9-13 passages). The 10 cultures that seemed to be established within this time period, i.e., that had acquired a uniform morphology and population-doubling time of 72 h or less, were analyzed for the presence of p53 mutations and for nuclear p53 accumulation.
Mutation Analysis. DBD sequence changes in the p53 gene of HUFs were examined by PCR amplification of p53 exons 4-9 and fluorescent dideoxynucleotide cycle sequencing using reagents, primers, standard protocols, and equipment (Model 310 Genetic Analyzer) from Applied Biosciences International (ABI, Weiterstadt, Germany) essentially as described previously for p53 gene mutation analysis of human tumors (29). Mutations were confirmed by repeat DNA extraction, PCR, and sequencing of the opposite strand. Additionally, all p53 mutant cell lines, and 15 of the non-mutant cell lines (as determined by direct sequencing results) were coded and then retested with the Affymetrix P53 GeneChip assay (Affymetrix, Santa Clara, CA), which, in the case of Hupki DNA, interrogates the humanized p53 sequences from exon 4-9 and splice sites. The array is gene-specific. Protocols, reagents, equipment, and software were used as recommended by Affymetrix, except that the multiplex primer mix for exons 4-9 was prepared from oligodeoxynucleotides synthesized by ABI. A value of 12 for mutant signals was chosen as a cut-off score for mutations. Reanalysis of HUF-UV5, which harbored 3 substitutions in exon 7, was performed by ligating PCR product into the pGEM T-Easy vector (Promega) and by sequencing exon 7 from 8 individual clones. Classes of base substitutions in p53 mutant clones were compared in the different experiments with the Fisher's exact test and were considered significant at P < 0.05.
Immunohistochemistry. Basal immunostaining levels of p53 protein were determined by standard immunocytochemistry procedures by using anti-human p53 antiserum CM-1 (Novacastra, Newcastle upon Tyne, U.K.) at a dilution of 1:1,500. To investigate stabilization and accumulation of nuclear p53 protein after exposure to a DNA damaging agent, cells were pretreated with 1 μM adriamycin 12-16 h before fixation and immunostaining.
Expression Analysis. Induction of p53 downstream transcriptional targets of activated p53. Cells were exposed to 20 Gy γ-irradiation 4 h before extraction of total RNA with a Qiagen RNeasy Extraction Kit, cDNA synthesis with AMV reverse transcriptase (Promega), and quantitative real-time RT-PCR, performed in a LightCycler (Roche Diagnostics, Mannheim, Germany), by using the LC-FastStart DNA Master SYBR Green I kit (Roche Diagnostics). Primer sequences for amplification, synthesized by ABI are as follows: CIP1/p21/WAF1 forward, 5′-CGGTCCCGTGGACAGTGAGC-3′; reverse, 5′-AAATCTGTCAGGCTGGTCTGCC-3′; Mdm2 forward, 5′-TAGCAGCCAAGAAAGCGTGAAAG-3′; reverse, 5′-TGGCAGATCACACATGGTTCG-3′. Expression levels of GAPDH were used for standardization in calculation of fold-induction of CIP1/p21/WAF1 and Mdm2 messages. Each experiment was performed in triplicate.
RNA expression of cytochrome P450 gene family members Cyp 1a1, 1a2, 1b1, and 2e1, NAD(P)H:quinone oxidoreductase, and microsomal epoxide hydrolase genes. Primer sequences and RT-PCR conditions were used essentially as established by previous investigators (30-32). Primers were synthesized from Pro-Oligo (Paris). RNA for RT-PCR reactions was isolated from p.1-2 untreated Hupki embryonic fibroblasts during log-phase growth in normal, complete DMEM.
DNA Adducts Analysis. We have described the 32P-postlabeling method for detection of AAI-DNA base adducts (33). DNA samples from primary Hupki embryonic fibroblasts exposed in vitro to 50 or 100 μM AAI for 1-4 days were digested enzymatically to nucleoside 3′-monophosphates and enriched for adducts by nuclease P1 digestion. The digests were then analyzed by 32P-postlabeling. Nuclease P1-resistant adducts were 5′-labeled with [32P]orthophosphate, separated on polyethyleneimine-cellulose thin-layer plates, and detected by autoradiography by using a Packard Instant Imager (Canberra, Dowers Grove, IL). Adducts were measured by assay of their 32P content. Results were expressed as mean adduct levels per 106 normal nucleotides [relative adduct labeling (RAL)]. Comparison of different chromatographic mobilities on thin-layer plates of single 32P-postlabeled adducts obtained in the Hupki cells with those of in vitro-synthesized reference compounds was used for identification purposes.
Results
p53 Status in HUF Cell Lines Established from Untreated and from UV-Treated Primary Hupki Embryonic Fibroblasts. Forty HUFs were established by culturing 20 untreated and 20 short-wavelength UV light (UVC)-treated primary cultures for >24 passages and then screened by direct PCR amplification of p53 sequences and dideoxynucleotide cycle sequencing for the presence of homozygous or heterozygous mutations in the DNA-binding domain of the humanized p53 gene. Seven of the 40 HUFs established in this way (5/20 cell lines from UV-treated primary cultures, and 2/20 cell lines from untreated cells) harbored a missense or nonsense mutation in the p53 gene (Table 1 and Fig. 1A). The same mutations (i.e., identical base change at the same codon and position) have been found in human tumors [R8, IARC TP53 Database: www.iarc.fr/P53 (2)]. The base changes derived from UV-treated cultures were all at dipyrimidine sites of the p53 gene, including 3 that were C to T transitions on the nontranscribed strand, a hallmark mutation in the well studied UV signature on DNA (34). One UV-derived HUF (UV5) harbored a hat trick involving 3 single-base substitutions at codons (c.) 248, 249, and 250. P53 sequence changes identical to those found in the HUFs from UV-exposed cultures have been found in human nonmelanoma skin cancers, including 2 that are human skin cancer hotspot mutations (CAT to TAT at c.179, and CGG to CAG at c.248, where the rank order is 7th and 15th, respectively, of 113 multiple entry mutations in the human skin cancer p53 mutation database). Of the 2 mutant HUFs derived from untreated (control) primary cultures, one harbors a transition at c.273, which is the second most commonly mutated codon in human cancers (all types combined). DNA from the 7 mutant HUFs and 15 of the HUFs determined to have a WT DBD sequence were coded and retested with the semiautomated p53GeneChip oligonucleotide microarray assay, which confirmed ABI sequencing data in all 22 retested cell lines, with the exception of the mutation cluster at c.248-250 in HUF-UV5 (data not shown). Cloning of PCR product spanning the mutation, and resequencing confirmed that the 3 substitutions had occurred on the same allele. Curiously, this cell line also harbors a C to G missense mutation on the other allele identical to that in UV17. This mutation may have arisen spontaneously during initial in vitro culturing of primary cells (from p.0-2) before seeding into separate wells before UV exposure and subsequent passaging.
Table 1. P53 mutations identified in the human DBD of HUF cell lines.
| Cell line | Codon | Mutation | Description |
|---|---|---|---|
| UV-treated HUFs | |||
| UV 5 | 248 | CGG to CAG* | Missense (R/Q), DP† |
| 249 | AGG to AGA | (Silent), DP | |
| 250 | CCC to CAC | Missense (P/H), DP/NT | |
| UV 17 | 135 | TGC to TGG | Missense (C/W), DP/NT |
| UV 20 | 179 | CAT to TAT* | Missense (H/Y), DP/NT |
| UV 21 | 151 | CCC to TCC* | Missense (P/S), DP/NT |
| UV 24 | 317 | CAG to TAG* | Nonsense (Q/stop), DP/NT |
| Untreated (control) HUFs | |||
| CO 15 | 273 | CGT to TGT | Missense (R/C)‡ |
| CO 17 | 194 | CTT to TTT | Missense (L/F)§ |
The identical mutation (nucleotide position and base change) has been detected in two or more human skin tumors (H179Y, 12 ×; R248Q 8 ×; Q317Stop 3 ×; P151S 2 ×).
DP, dipyrimidine site; NT, nontranscribed strand.
7.5% of all human tumor p53 base substitutions are at c.273.
0.6% of human tumor mutations are at c. 194 (IARC TP53 DB, R8).
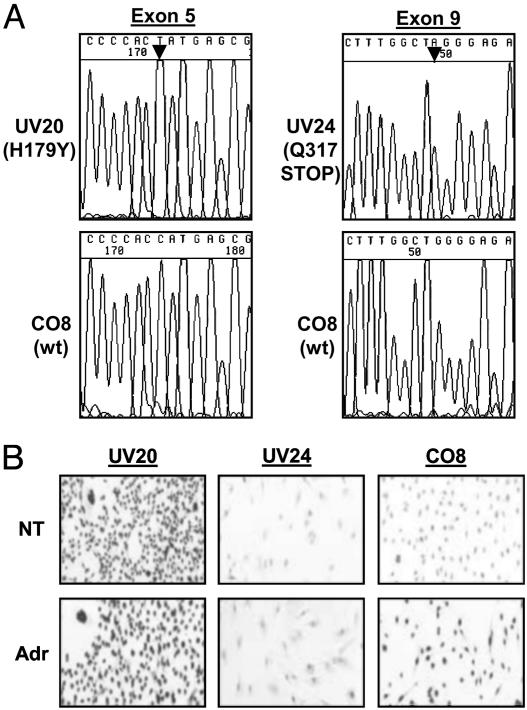
Fig. 1.
(A) Identification of p53 DBD mutations in HUFs by Sanger sequencing with fluorescent dye-labeled dideoxynucleotides. A homozygous base substitution was present in UV20 at p.24 (electropherogram shows 5′ to 3′ sequencing) and in UV24 (3′ to 5′ sequence shown) at p.25. (B) Correlation of p53 mutation status in HUF cell lines with elevated basal levels of nuclear p53 protein (HUF with missense mutation: UV20), and with accumulation of p53 protein in a HUF (CO8) with WT p53, 12 h after exposure to the DNA-damaging agent adriamycin (1 μM). UV24, which harbors a homozygous stop mutation in exon 9, shows background immunostaining and no accumulation response after exposure to adriamycin, as expected. Immunostaining was performed with CM-1 anti-human p53 polyclonal antiserum at 1:1,500 dilution. NT, untreated; Adr, adriamycin-treated.
In 5 mutant HUFs, both WT and mutant signals were present at the mutation site in sequencing electropherograms (UV5, UV17, UV21, CO15, and CO17). To examine the heterozygosity of the single-base substitutions, we subcloned these HUFs and sequenced DNA from clones derived from single cells. Again, both the mutant and WT sequence were present in UV17 and CO17 whereas, in the UV21 and CO15 subclones, only the mutant signal was present, indicating either that loss of the WT allele occurred during outgrowth of these subclones or that a subpopulation of WT cells contributed the WT signal in the cell population initially analyzed. We have maintained the 7 mutant HUFs and several that were p53 DBD WT in culture for >65 passages.
We next asked whether we could detect the HUF mutations that were found at >p.24 in aliquots of cells frozen earlier, at p.13-15. Surprisingly, the homozygous mutations we observed at >p.24 in UV20 and UV24 already were present and were hemi-/homozygous at these earlier passages (data not shown). The heterozygous exon 5 missense mutations in UV21 and UV17 were also detectable by fluorescent dideoxy sequencing at the earlier passage whereas mutant signals from the mutations in the 2 HUFs derived from untreated cultures (CO15, CO17) were not yet detectable/present at p.15.
Nuclear p53 Staining. A characteristic feature of human tumors with missense p53 mutations is an abnormally high amount of nuclear p53 protein resulting from deficiency in transcriptional transactivation by p53 of downstream targets such as Mdm2 involved in feedback control of p53 protein levels and cellular localization. Accumulation is readily detectable by immunohistochemical staining of tumors or cell lines with certain anti-p53 antibodies. We examined the basal intensity of p53 protein staining by anti-p53 antiserum CM-1 in the 7 mutant HUFs and in 7 HUFs without p53 DBD mutations. As shown in Fig. 1B for 3 HUFS, the presence of a missense mutation (UV20) correlated with intense nuclear staining in nontreated cells as anticipated whereas, in HUFs with no mutation (CO8) or harboring a homozygous stop mutation (UV24), the reactivity was at background levels. Strong nuclear staining indicating accumulation of nuclear p53 protein was induced after treatment of the p53 WT CO8 cells with adriamycin, but, as expected, not in p53 null UV24 HUFs (Fig. 1B).
P53 DBD Mutation Status in HUFs and Transactivation Function. To determine whether mutations in HUFs cause defects in p53 transactivation function similar to the defects seen in p53 mutant human cell lines, we performed real-time RT-PCR of the p53 downstream target genes CIP1/p21/WAF1 and Mdm2 in the 7 mutant HUFs and in 7 of the HUFs without DBD mutations after exposure to adriamycin and γ-irradiation. These agents are known to induce transcriptional transactivation activity of WT functional p53. As shown for p53 WT and mutant human tumor cell lines in the NCI anti-cancer drug screen panel (35), there is an overall correlation between degree of p53 target gene inducibility and HUF p53 mutation status (Table 2). After 4 h exposure to 20 Gy γ-irradiation, p53-mutant HUFs generally showed no or attenuated induction of the two downstream genes in comparison with HUFs with a WT Hupki p53 DBD. A similar trend was seen after exposure to adriamycin (data not shown).
Table 2. γ-induced expression of Mdm2 and p21/WAF1/CIP1 mRNAs.
| HUF | p53 status* | Mdm2† | p21† |
|---|---|---|---|
| UV19 | WT | 5.6 ± 1.2 | 4.2 ± 0.8 |
| CO23 | WT | 5.1 ± 0.3 | 4.6 ± 1.0 |
| UV1 | WT | 4.5 ± 0.4 | 3.1 ± 0.4 |
| UV3 | WT | 3.9 ± 1.1 | 2.5 ± 0.7 |
| CO8 | WT | 3.7 ± 0.8 | 5.0 ± 0.3 |
| UV21 | Mu | 2.6 ± 0.5 | 2.6 ± 0.2 |
| UV13 | WT | 2.4 ± 0.7 | 2.1 ± 0.2 |
| UV18 | WT | 2.3 ± 0.4 | 1.9 ± 0.1 |
| CO15 | Mu | 2.2 ± 0.5 | 2.4 ± 0.3 |
| CO17 | Mu | 1.1 ± 0.1 | 1.7 ± 0.3 |
| UV5 | Mu | 1.1 ± 0.2 | 1.7 ± 0.3 |
| UV24 | Mu | 1.1 ± 0.2 | 1.3 ± 0.5 |
| UV20 | Mu | 1.0 ± 0.1 | 1.5 ± 0.1 |
| UV17 | Mu | 0.9 ± 0.1 | 1.4 ± 0.2 |
P53 status: WT, wild-type, no p53 DBD mutation; Mu, p53 DBD mutation (see Table 1).
Level of induction with SD is given as fold-increase over nonirradiated cells and normalized against GAPDH message level. Cells were collected 4 h after 20-Gy irradiation.
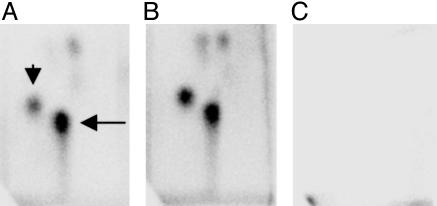
Mutation Analysis in Early Passage HUFs Derived from AAI-Treated Primary Hupki Embryonic Fibroblasts. The experiments described above with UV-treated HUFs suggested that p53 mutations could be induced and detected in HUFs at relatively early passage (<p.15), within 8-10 weeks of treatment. This simple procedure would offer potential as a mutation assay for human carcinogens, particularly if murine primary embryonic fibroblasts are efficient in metabolic activation of xenobiotics. To explore this possibility, we chose to examine in Hupki cells the mutagenicity of AAI, the major component of the plant extract aristolochic acid, a potent mutagen and human carcinogen associated with Chinese herbs nephropathy (CHN) and with development of urothelial cancer in CHN patients (36). First, we asked whether the major and most persistent mutagenic adduct, the adenosine adduct of aristolochic acid I 7-(deoxyadenosin-N6-yl) aristolactam I (dA-AAI) found in CHN patients could be detected in Hupki primary embryonic fibroblasts treated with AAI. As shown in Fig. 2 and Table 3, dA-AAI is formed and the pattern of adducts is similar to that found in AAI-treated animals (37), indicating the metabolic competence of the Hupki primary embryonic fibroblasts. Retention in early passage Hupki embryonic fibroblasts of metabolic competency described for mouse embryos (30) was indicated by presence of mRNA from four of the six genes we screened by RT-PCR (Cyp 1a2, Cyp 1b1, microsomal epoxide hydrolase, and NAD(P)H:quinone oxidoreductase), suggesting that these cells may be proficient in enzymatic activation of a variety of procarcinogens (data not shown).
Fig. 2.
Autoradiogram of 32P-postlabeling of DNA from primary (p.2) Hupki embryonic fibroblasts treated for 48 h (A) or 96 h (B) with 50 μM AAI before harvesting and DNA extraction. The arrows in A indicate the signals from the two major DNA adducts dA-AAI (long arrow) and dG-AAI [7-(deoxyguanosin-N-yl)-aristolactam I; short arrow]. (C) An autoradiogram from an experiment with DNA from solvent control Hupki cells.
Table 3. Total RAL in HUFs exposed to AAI at p.2.
| AAI, 50 μM | AAI, 50 μM | AAI, 100 μM | |
|---|---|---|---|
| Time, hr | 48 | 96 | 72 |
| RAL, × 10−6* | 0.49 | 1.1 | 5.3 |
Mean RAL of two separate incubations.
We then exposed 24 Hupki primary embryonic fibroblast cultures at p.2 to 100 μM AAI for 48 h and analyzed the p53 status of the first 10 cultures that had survived crisis, showing uniform morphology and accelerated growth, and were at p.9-13 by week 10 after exposure. P53 GeneChip screening identified 6 base substitution mutations (4 missense, 1 nonsense, and 1 splice site mutation) in 5 of the 10 established cultures, and 5 were verified by independent DNA cycle sequencing analysis (Table 4). Four of the substitutions were A to T transversions on the nontranscribed strand, a finding significantly different from the mutation data from UV-treated/untreated cultures, which were comprised exclusively of base changes at G:C pairs, primarily transitions. A to T transversion is the predominant mutation class anticipated from AAI exposure (38, 39), yet is a relatively rare class of base substitution in spontaneous or in UV mutation spectra, making it likely that these HUF mutations were induced by the AAI treatment. Consistent with the immunohistochemistry results reported above for UV-treated and control HUFs, the missense mutant HUF cell strains AA7 and AA13 derived from the AAI-exposed cultures showed intense nuclear p53 staining whereas AA6, which harbors a homo-/hemizygous nonsense mutation in exon 6, did not stain with anti-p53 antibody and failed to show induction of p53 after exposure to adriamycin, as expected.
Table 4.
P53 mutations detected* in AAI-treated HUFs at p.9-13
| Cell line | Codon | Mutation | Description |
|---|---|---|---|
| AA6 | 209 | AGA to TGA | Nonsense (R/Stop) A to T† |
| AA7 | 176 | TGC to TGG | Missense (C/W) C to G‡ |
| AA13 | 280 | AGA to AGT | Missense (R/S) A to T† |
| AA18 | 158 | CGC to GGC | Missense (R/G) C to G‡ |
| int 8 | AG to TG | Splice site A to T† | |
| AA22 | 281 | GAC to GTC§ | Missense (D/V) A to T† |
Mutation identified by p53 GeneChip microarray analysis and by DNA sequencing.
Detected only by the P53 GeneChip at p.10; detected by both methods at p.20.
A:T to T:A transversion.
C:G to G:C transversion.
Discussion
Mouse embryonic fibroblasts (MEFs) undergo spontaneous immortalization in vitro readily, and this acquired capacity to thrive in vitro typically requires a genetic alteration in the p19ARF or the p53 locus, for example, a missense mutation in the p53 gene (23). In principle, this interesting observation offers an opportunity to gain information on spontaneous mutation sites, or to produce carcinogen-derived p53 mutation patterns in MEFs, if it can be shown that p53 mutations are inducible by experimental treatment of primary MEFs and are selected for during establishment in culture. Due to the 15% discrepancy in base sequence and to amino acid differences between the human and murine DBDs, however, mouse p53 mutations are of limited use in interpreting p53 mutation patterns in human tumors. The most notable example of divergence is the 9-bp sequence at c.248-250, the most heavily mutated sequence in human tumors. Four of the 9 base residues differ between the two species. In the present study, we have made use of the inherent characteristic of murine cells to adapt to in vitro growth by p53 pathway disruption and have bypassed the drawback of examining mutation patterns in murine p53 sequences by replacing them with the human sequence in the Hupki model. P53 mutation detection in Hupki embryonic fibroblasts also offers the practical advantage of identifying p53 DBD mutations with a commercial oligonucleotide microarray specific for the human p53 sequence, and immunohistochemical prescreening of HUFs can improve detection of out-of-frame insertions/deletions, which are not readily detected by the GeneChip microarray (40).
For the development of a mutation system employing MEFs, detailed information on expressed cytochrome P450 species and DNA repair genes would be desirable. Previous studies by others have shown that murine fibroblasts (C3H10T1/2; primary MEFs) express cytochrome P450 reductase and Cyp 1b1, an enzyme involved in the metabolic activation of the carcinogens DMBA and B[a]P whereas Cyp 1a2 and Cyp 1a1 levels are typically low to absent but may be inducible (41-44). Choudhary et al. investigated the presence of 40 murine cytochrome P450 gene message species by RT-PCR in mouse embryos and in adult tissues and found that most, including Cyp 1b1, were constitutively expressed in the embryonic RNA pool at various developmental stages (30). In our initial experiments exploring the potential of primary HUFs to reveal the mutagenic activity of environmental carcinogens requiring cytochrome P450s for activation to electrophilic intermediates, we detected the expression of several species important in xenobiotic metabolism of polycyclic aromatic hydrocarbons and other classes of environmental carcinogens. In addition, we detected the major premutagenic deoxyguanosine B[a]P adduct, benzo[a]pyrene diol epoxide-N2-deoxyguanosine in 32P postlabeling experiments with DNA from primary Hupki embryonic fibroblasts exposed to 1 μM B[a]P for 24 h (H.S. and M.H., unpublished results). Given the complexity, size, and redundancies of the cytochrome P450 gene superfamily in mice and humans (45, 46) and the role of phase II enzymes in xenobiotic metabolism, we expect that DNA adduct measurement will be the more efficient and predictive means to sound out the capability of Hupki cells to process specific chemical carcinogens to reactive intermediates.
We also have examined the expression of various cytochrome P450 mRNAs in a Hupki-derived, p53 DBD WT HUF cell line (42B1) by using Affymetrix oligonucleotide expression microarray technology, and confirmed that Cyp 1b1 message was the single most prominent P450 species interrogated by the U74Av2 array, followed by substantial expression of cytochrome P450 reductase and Cyp 51a1 mRNA (Z.L. and M.H., unpublished observations). Array profiling also confirmed that expression of a large number of DNA repair genes was retained in the HUF cell line, and primary MEFs are known to have an extensive DNA repair repertoire, and thus have served as an efficient system to investigate the biological consequences of specific repair deficiencies in MEFs isolated from knock-out transgenic mice with disrupted components of the base excision repair and nucleotide excision repair pathways (47-49). Despite the overall conservation in evolution of DNA repair mechanisms, differences between humans and mice have been found, such as the efficiency of the global genomic repair subpathway of nucleotide excision repair (reviewed in ref. 50).
At the present stage, the generation of a representative mutation pattern with HUFs is labor-intensive. The number of mutants required to generate representative mutation patterns with any assay is mutagen-dependent, essentially unpredictable, and typically large. It will be necessary to explore ways to select and analyze small numbers of p53 mutated cells from early passages of treated Hupki cell populations. In distinction to established mutation assays employing selectable markers in mammalian or yeast cells, the Hupki model has the advantage of addressing the cancer-relevant p53 gene in its natural mammalian context. The limitations for mutation spectrum studies of the elegant yeast/p53 reporter system were illustrated by a recent study on solar UV- and B[a]P-induced p53 mutations, which showed that preferentially mutated p53 sequences in the yeast assay were entirely different from those observed in human skin tumors and in smokers' lung cancers (22). To accelerate mutation analysis in Hupki cells, it will be necessary to examine how soon after crisis cell populations can be screened for the presence of mutations.
To refine the application of HUFs to the study of spontaneously arising and mutagen-induced p53 mutations, it would be desirable to know whether and how mouse strain and cell culture immortalization protocol parameters may influence outgrowth and clonal expansion of postcrisis cells that harbor p53 mutations. Studies with primary MEFs have revealed that the rare cell (1 cell per 106, or fewer) that survives the initial crisis period has lost either p53 function (typically by mutation) or INK4a sequences (typically by biallelic deletion), and, in BALB/c murine embryonic fibroblasts passaged according to a defined subculturing schedule, immortalization through the p53 pathway is the more common route (51-55). The p19ARF and p53 tumor suppressor pathways intersect, and it has been argued that p53 has stronger tumor suppressor activity than p19ARF in mice (56). The conditions or cell attributes of WT MEFs that regulate the relative frequencies of spontaneous p19ARF allele loss on the one hand, or p53 mutation on the other, are incompletely understood.
To exert its mutagenic effect, AAI requires metabolic activation through reduction of the nitro group to form DNA adducts that result predominantly in A to T transversions if unrepaired. Enzymes capable of bioactivating AAI are cytosolic nitroreductases, cytochrome P450 enzymes, and peroxidases (33). The preponderance in AAI-treated HUFs of the A to T substitution, an infrequent type of substitution in spontaneous mutation spectra, is consistent with the prevalent type of DNA adduct and mutation observed from AAI treatment in different experimental systems. We conclude that the A to T transversion in the HUFs we identified probably were induced by the AAI treatment. The RAL of dA-AAI in Hupki cells cultured for 48 or 72 h in medium containing 50 μM AAI was comparable with levels we found previously in the kidneys and ureters of CHN cancer patients from Belgium (36). Interestingly, urothelial tumor cells from the one CHN/AAI-exposed patient available to us at the time of this study harbored an A to T transversion on the nontranscribed strand at c.139, leading to a stop mutation (57).
It is noteworthy that the 12 p53 DBD mutant HUFs selected for in vitro in our protocol harbored nonsilent base substitutions, and 6 of the mutations are at sites of the p53 gene that are among the 15 most commonly mutated codons in human cancers (c.248, rank 1; c.273, rank 2; c.179, rank 7; c.176, rank 10; c.158, rank 12; and c.280, rank 13; IARC TP53 Database). This finding suggests that in vitro selective pressures leading to outgrowth of p53 DBD mutant Hupki fibroblasts in culture may be a surprisingly good approximation of selection for mutant p53 in tissues in vivo.
Acknowledgments
The technical assistance of K.-R. Muehlbauer and A. Weninger is gratefully acknowledged. We also thank D. Belharazem for preparation of primary HUFs.
Abbreviations: HUF, Hupki embryonic fibroblast cell line; DBD, DNA-binding domain; AAI, aristolochic acid I; dA-AAI, 7-(deoxyadenosine-N6-yl) aristolactam I; RAL, relative adduct labeling; Hupki, human p53 knock-in; CHN, Chinese herbs nephropathy; MEF, mouse embryonic fibroblast; c.n, codon n; p.n, passage n.
References
- 1.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 2.Olivier, M., Eeles, R., Hollstein, M., Khan, M. A., Harris, C. C. & Hainaut, P. (2002) Hum. Mutat. 19, 607-614. [DOI] [PubMed] [Google Scholar]
- 3.Hollstein, M., Rice, K., Greenblatt, M. S., Soussi, T., Fuchs, R., Sorlie, T., Hovig, E., Smith-Sorensen, B., Montesano, R. & Harris, C. C. (1994) Nucleic Acids Res. 22, 3551-3555. [PMC free article] [PubMed] [Google Scholar]
- 4.Hollstein, M., Sidransky, D., Vogelstein, B. & Harris, C. C. (1991) Science 253, 49-53. [DOI] [PubMed] [Google Scholar]
- 5.Hainaut, P. & Hollstein, M. (2000) Adv. Cancer Res. 77, 81-137. [DOI] [PubMed] [Google Scholar]
- 6.Jones, P. A., Buckley, J. D., Henderson, B. E., Ross, R. K. & Pike, M. C. (1991) Cancer Res. 51, 3617-3620. [PubMed] [Google Scholar]
- 7.Hussain, S. P. & Harris, C. C. (1998) Cancer Res. 58, 4023-4037. [PubMed] [Google Scholar]
- 8.Miller, J. H. (1982) Cell 31, 5-7. [DOI] [PubMed] [Google Scholar]
- 9.Greenblatt, M. S., Grollman, A. P. & Harris, C. C. (1996) Cancer Res. 56, 2130-2136. [PubMed] [Google Scholar]
- 10.Hainaut, P. & Pfeifer, G. P. (2001) Carcinogenesis 22, 367-374. [DOI] [PubMed] [Google Scholar]
- 11.Ziegler, A., Leffell, D. J., Kunala, S., Sharma, H. W., Gailani, M., Simon, J. A., Halperin, A. J., Baden, H. P., Shapiro, P. E., Bale, A. E., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 4216-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brash, D. E., Rudolph, J. A., Simon, J. A., Lin, A., McKenna, G. J., Baden, H. P., Halperin, A. J. & Ponten, J. (1991) Proc. Natl. Acad. Sci. USA 88, 10124-10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tommasi, S., Denissenko, M. F. & Pfeifer, G. P. (1997) Cancer Res. 57, 4727-4730. [PubMed] [Google Scholar]
- 14.Pfeifer, G. P., Denissenko, M. F., Olivier, M., Tretyakova, N., Hecht, S. S. & Hainaut, P. (2002) Oncogene 21, 7435-7451. [DOI] [PubMed] [Google Scholar]
- 15.Tornaletti, S., Rozek, D. & Pfeifer, G. P. (1993) Oncogene 8, 2051-2057. [PubMed] [Google Scholar]
- 16.Gee, P., Maron, D. M. & Ames, B. N. (1994) Proc. Natl. Acad. Sci. USA 91, 11606-11610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishioka, C., Frebourg, T., Yan, Y. X., Vidal, M., Friend, S. H., Schmidt, S. & Iggo, R. (1993) Nat. Genet. 5, 124-129. [DOI] [PubMed] [Google Scholar]
- 18.Moshinsky, D. J. & Wogan, G. N. (1997) Proc. Natl. Acad. Sci. USA 94, 2266-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fronza, G., Inga, A., Monti, P., Scott, G., Campomenosi, P., Menichini, P., Ottaggio, L., Viaggi, S., Burns, P. A., Gold, B., et al. (2000) Mutat. Res. 462, 293-301. [DOI] [PubMed] [Google Scholar]
- 20.Cariello, N. F., Cui, L. & Skopek, T. R. (1994) Cancer Res. 54, 4436-4441. [PubMed] [Google Scholar]
- 21.Schmezer, P. & Eckert, C. (1999) IARC Sci. Publ. 146, 367-394. [PubMed] [Google Scholar]
- 22.Yoon, J. H., Lee, C. S. & Pfeifer, G. P. (2003) Carcinogenesis 24, 113-119. [DOI] [PubMed] [Google Scholar]
- 23.Hahn, W. C. & Weinberg, R. A. (2002) Nat. Rev. Cancer 2, 331-341. [DOI] [PubMed] [Google Scholar]
- 24.Luo, J. L., Yang, Q., Tong, W. M., Hergenhahn, M., Wang, Z. Q. & Hollstein, M. (2001) Oncogene 20, 320-328. [DOI] [PubMed] [Google Scholar]
- 25.Clarke, A. R. & Hollstein, M. (2003) Cell Death. Differ. 10, 443-450. [DOI] [PubMed] [Google Scholar]
- 26.Luo, J. L., Tong, W. M., Yoon, J. H., Hergenhahn, M., Koomagi, R., Yang, Q., Galendo, D., Pfeifer, G. P., Wang, Z. Q. & Hollstein, M. (2001) Cancer Res. 61, 8158-8163. [PubMed] [Google Scholar]
- 27.Chao, C., Hergenhahn, M., Kaeser, M. D., Wu, Z., Saito, S., Iggo, R., Hollstein, M., Appella, E. & Xu, Y. (2003) J. Biol. Chem. 278, 41028-41033. [DOI] [PubMed] [Google Scholar]
- 28.Torres, R. & Kühn, R. (1997) Laboratory Protocols for Conditional Gene Targeting (Oxford Univ. Press, Oxford).
- 29.Biramijamal, F., Allameh, A., Mirbod, P., Groene, H. J., Koomagi, R. & Hollstein, M. (2001) Cancer Res. 61, 3119-3123. [PubMed] [Google Scholar]
- 30.Choudhary, D., Jansson, I., Schenkman, J. B., Sarfarazi, M. & Stoilov, I. (2003) Arch. Biochem. Biophys. 414, 91-100. [DOI] [PubMed] [Google Scholar]
- 31.Pan, J., Xiang, Q. & Ball, S. (2000) Drug Metab. Dispos. 28, 709-713. [PubMed] [Google Scholar]
- 32.Kwak, M. K., Itoh, K., Yamamoto, M., Sutter, T. R. & Kensler, T. W. (2001) Mol. Med. 7, 135-145. [PMC free article] [PubMed] [Google Scholar]
- 33.Bieler, C. A., Stiborova, M., Wiessler, M., Cosyns, J. P., van Ypersele, D. S. & Schmeiser, H. H. (1997) Carcinogenesis 18, 1063-1067. [DOI] [PubMed] [Google Scholar]
- 34.Brash, D. E. (1997) Trends Genet. 13, 410-414. [DOI] [PubMed] [Google Scholar]
- 35.O'Connor, P. M., Jackman, J., Bae, I., Myers, T. G., Fan, S., Mutoh, M., Scudiero, D. A., Monks, A., Sausville, E. A., Weinstein, J. N., et al. (1997) Cancer Res. 57, 4285-4300. [PubMed] [Google Scholar]
- 36.Nortier, J. L., Martinez, M. C., Schmeiser, H. H., Arlt, V. M., Bieler, C. A., Petein, M., Depierreux, M. F., De Pauw, L., Abramowicz, D., Vereerstraeten, P., et al. (2000) N. Engl. J. Med. 342, 1686-1692. [DOI] [PubMed] [Google Scholar]
- 37.Arlt, V. M., Stiborova, M. & Schmeiser, H. H. (2002) Mutagenesis 17, 265-277. [DOI] [PubMed] [Google Scholar]
- 38.Schmeiser, H. H., Janssen, J. W., Lyons, J., Scherf, H. R., Pfau, W., Buchmann, A., Bartram, C. R. & Wiessler, M. (1990) Cancer Res. 50, 5464-5469. [PubMed] [Google Scholar]
- 39.Kohara, A., Suzuki, T., Honma, M., Ohwada, T. & Hayashi, M. (2002) Mutat. Res. 515, 63-72. [DOI] [PubMed] [Google Scholar]
- 40.Ahrendt, S. A., Halachmi, S., Chow, J. T., Wu, L., Halachmi, N., Yang, S. C., Wehage, S., Jen, J. & Sidransky, D. (1999) Proc. Natl. Acad. Sci. USA 96, 7382-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buters, J. T., Sakai, S., Richter, T., Pineau, T., Alexander, D. L., Savas, U., Doehmer, J., Ward, J. M., Jefcoate, C. R. & Gonzalez, F. J. (1999) Proc. Natl. Acad. Sci. USA 96, 1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleiner, H. E., Vulimiri, S. V., Reed, M. J., Uberecken, A. & DiGiovanni, J. (2002) Chem. Res. Toxicol. 15, 226-235. [DOI] [PubMed] [Google Scholar]
- 43.Nesnow, S., Davis, C., Nelson, G. B., Lambert, G., Padgett, W., Pimentel, M., Tennant, A. H., Kligerman, A. D. & Ross, J. A. (2002) Mutat. Res. 521, 91-102. [DOI] [PubMed] [Google Scholar]
- 44.Vogel, C. F. & Matsumura, F. (2003) Biochem. Pharmacol. 66, 1231-1244. [DOI] [PubMed] [Google Scholar]
- 45.Nelson, D. R. (2002) Methods Enzymol. 357, 3-15. [DOI] [PubMed] [Google Scholar]
- 46.Nelson, D. R. (1999) Arch. Biochem. Biophys. 369, 1-10. [DOI] [PubMed] [Google Scholar]
- 47.Sobol, R. W., Watson, D. E., Nakamura, J., Yakes, F. M., Hou, E., Horton, J. K., Ladapo, J., Van Houten, B., Swenberg, J. A., Tindall, K. R., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 6860-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortellino, S., Turner, D., Masciullo, V., Schepis, F., Albino, D., Daniel, R., Skalka, A. M., Meropol, N. J., Alberti, C., Larue, L., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 15071-15076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett, S. E., Sung, J. S. & Mosbaugh, D. W. (2001) J. Biol. Chem. 276, 42588-42600. [DOI] [PubMed] [Google Scholar]
- 50.Hanawalt, P. C. (2002) Oncogene 21, 8949-8956. [DOI] [PubMed] [Google Scholar]
- 51.Kamijo, T., Zindy, F., Roussel, M. F., Quelle, D. E., Downing, J. R., Ashmun, R. A., Grosveld, G. & Sherr, C. J. (1997) Cell 91, 649-659. [DOI] [PubMed] [Google Scholar]
- 52.Harvey, D. M. & Levine, A. J. (1991) Genes Dev. 5, 2375-2385. [DOI] [PubMed] [Google Scholar]
- 53.Serrano, M., Lee, H., Chin, L., Cordon-Cardo, C., Beach, D. & DePinho, R. A. (1996) Cell 85, 27-37. [DOI] [PubMed] [Google Scholar]
- 54.Zindy, F., Quelle, D. E., Roussel, M. F. & Sherr, C. J. (1997) Oncogene 15, 203-211. [DOI] [PubMed] [Google Scholar]
- 55.Zindy, F., Eischen, C. M., Randle, D. H., Kamijo, T., Cleveland, J. L., Sherr, C. J. & Roussel, M. F. (1998) Genes Dev. 12, 2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore, L., Venkatachalam, S., Vogel, H., Watt, J. C., Wu, C. L., Steinman, H., Jones, S. N. & Donehower, L. A. (2003) Oncogene 22, 7831-7837. [DOI] [PubMed] [Google Scholar]
- 57.Lord, G., Hollstein, M., Arlt, V. M., Roufosse, C., Pusey, C., Cook, T. & Schmeiser, H. H. (2004) Am. J. Kidney Dis., in press. [DOI] [PubMed]