Abstract
The myeloid cell leukemia-1 (MCL1) gene encodes antiapoptotic Mcl-1L and proapoptotic Mcl-1S proteins. In cancer, the Mcl-1L/Mcl-1S ratio is very high, accounting for the antiapoptotic nature of cancer cells. As such, reducing this ratio can render the cancer cells prone to apoptosis. The Mcl-1L/Mcl-1S ratio is determined in the alternative pre-mRNA splicing step that is regulated by splicing factor 3B1 (SF3B1). Here, we report that meayamycin B, a potent inhibitor of SF3B1, reversed the dominant isoform from Mcl-1L to Mcl-1S at the mRNA and protein levels. The resulting proapoptotic cellular environment was further exploited; when meayamycin B was combined with Bcl-xL inhibitor ABT-737, the combination treatment triggered apoptosis in non-small cell lung cancer A549 and H1299 cells that were otherwise resistant to ABT-737. These results demonstrate that perturbation of the MCL1 splicing with small molecule inhibitors of SF3B1 provides a means to sensitize cancer cells toward Bcl-xL inhibitors.
Introduction
Many B-cell lymphoma-2 (Bcl-2) family genes undergo alternative pre-mRNA splicing and produce both proapoptotic and antiapoptotic protein isoforms. For example, BCL2L1 and myeloid cell leukemia-1 (MCL1) genes are alternatively spliced into proapoptotic Bcl xS and Mcl-1S, antiapoptotic Bcl-xL and Mcl-1L, and the unannotated Mcl-1ES, respectively.1-3 In cancer, antiapoptotic isoforms are predominant.4 Although other antiapoptotic Bcl-2 family proteins such as Bcl-2 and Bcl-w are present in cancer cells, inhibition of both Bcl-xL and Mcl-1L is necessary and sufficient to trigger massive cell death.5 In support of this notion, ABT-737, a small molecule that selectively binds to and antagonizes Bcl-2, Bcl-xL and Bcl-w, but not Mcl-1l ,6, 7 encountered resistance in cancer cells that overexpressed Mcl-1L .8-12 To circumvent the drug resistance, Mcl-1 expression was inhibited by biological or pharmacological means, which restored the anticancer activity of ABT-737.8, 10, 13, 14 However, the only endeavor successfully sequestering Mcl-1L by perturbing the alternative splicing of Mcl-1 pre-mRNA was antisense morpholino oligonucleotides.15 So far, there is no small molecule reported to have such activity.
The expression of Mcl-1S and Bcl-xS mRNAs was upregulated when splicing factor 3B 1 (SF3B1; a.k.a. SAP155) was knocked down, indicating that SF3B1 is involved in the alternative splicing of these apoptosis related genes.16 SF3B1, an essential subunit of U2 snRNP, is critical for the faithful selection of the 3′ splice site in homeostatic cells.17 SF3B1 has also been identified as a trans-acting splicing factor that enhances the production of Bcl-xL in A549 cells.18 Meanwhile, natural product FR901464 has been found to be a low nanomolar inhibitor of SF3B1.19 Subsequently, we have developed a low to middle picomolar analog of FR901464, meayamycin B (Figure 1).20-23 Hence, we hypothesized that meayamycin B could switch the alternative splicing of Bcl-x and/or Mcl-1 pre-mRNAs towards the overexpression of Bcl-xS and Mcl-1S.
Figure 1.
Structures of meayamycin B and ABT-737.
Here, we report that meayamycin B upregulates the proapoptotic Mcl-1S and downregulates Mcl-1L at the pre-mRNA splicing level. The resulting dominance of Mcl-1S at the protein level overcame the ABT-737 resistance and induced cell death in non-small cell lung cancer A549 and H1299 cell lines when both of these compounds were present. Meayamycin B did not perturb the apparent alternative splicing of Bcl-x within 24 h of treatments in our settings.
RESULTS AND DISCUSSION
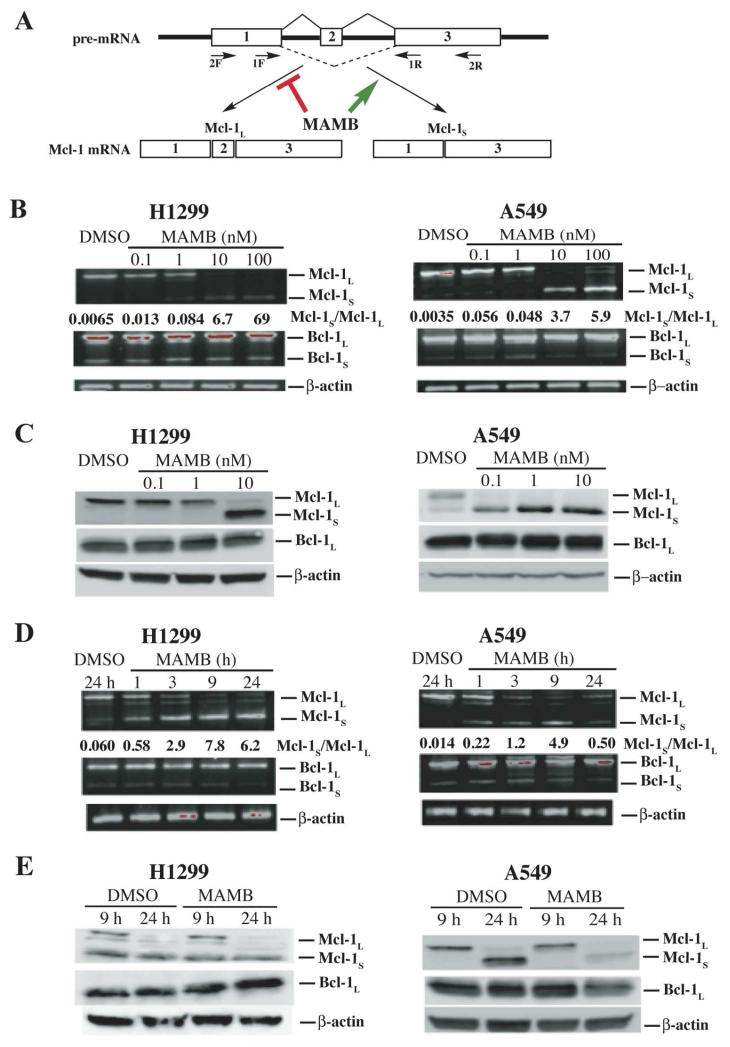
Meayamycin B regulates the alternative splicing of Mcl-1 in A549 and H1299 in a dose-dependent manner
Mcl-1L is generally overexpressed in non small cell lung cancer cells including A549 and H1299 cells.24, 25 To test the aforementioned hypothesis about meayamycin B, A549 and H1299 cell lines were exposed to meayamycin B at various concentrations. The exposure time was only 9 h, which was sufficient to generate clear-cut data because the half-life of Mcl-1 mRNA is approximately 40 min.26, 27 The short assay also minimizes the downstream secondary effects of meayamycin B. After the 9-h exposure, the relative mRNA levels of Mcl-1 splicing variants including Mcl-1L and Mcl-1S were evaluated by means of semi-quantitative RT-PCR (Figure 2A). For the RT-PCR experiment, we used primers (see Table S1 in the Supporting Information) that were designed to reverse-transcribe multiple sites of the gene. Meayamycin B dose-dependently downregulated and upregulated Mcl-1L and Mcl-1S, respectively (NCBI GenBank accession number NM_021960 and NM_182763) (Figure 2B).1 For example, the ratio of Mcl-1S/Mcl-1L increased from 0.0065 (H1299 cells) and 0.035 (A549 cells) with DMSO to 69 (H1299 cells) and 5.9 (A549 cells) with 100 nM meayamycin B. In accordance with the change at the mRNA level, immunoblotting showed that meayamycin B abrogated the expression of the full length Mcl-1 protein isomer Mcl-1L (40 kDa) and simultaneously increased Mcl-1S (35 kDa) (Figure 2C). Mcl-1L protein was more prevalent in H1299 than in A549, which might explain why 10 nM meayamycin B was required to completely remove Mcl-1L in H1299 cells. Mcl-1S protein became the dominant Mcl-1 isoform in A549 cells with only 0.1 nM meayamycin B.
Figure 2.
Dose- and time-dependent regulation of Mcl-1 splicing by meayamycin B (MAMB) in H1299 and A549 cells. (A): The alternative splicing of Mcl-1 regulated by meayamycin B. Horizontal arrows depict the locations of the primers for semi-quantitative RT-PCR. Meayamycin B treatment increased Mcl-1S and decreased Mcl-1L splice variants. (B) and (C): A549 and H1299 cells were exposed to DMSO or various concentrations of meayamycin B for 9 h before total RNA and protein were analyzed by semi-quantitative RT-PCR (B) and immunoblotting (C) for the alternative splicing of Mcl-1. The same samples were sequentially analyzed by semi-quantitative RT-PCR using primer pair 1F and 1R shown in Figure 2A. (D) and (E): Cells were incubated with 10 nM meayamycin B for indicated periods of time, and the total RNA and protein were extracted and analyzed by semi-quantitative RT-PCR (D) and immunoblotting (E) for the alternative splicing of Mcl-1. Data represent results from at least three separate experiments.
Meayamycin B regulates the alternative splicing of Mcl-1 in A549 and H1299 in a time-dependent manner
Mcl-1 has a very short half life at the mRNA and protein levels.26, 27 Therefore, we thought it would be possible to observe changes in the expression of MCL1 soon after the addition of meayamycin B to the aforementioned cells. Thus, H1299 and A549 cells were exposed to 10 nM meayamycin B for 1, 3, 9, and 24 h before relative expression of Mcl-1 splicing variants were determined at the mRNA and protein levels. The semi quantitative RT-PCR analysis revealed that the increase of the Mcl-1S mRNA was detectable after 1 h of treatment (Figure 2D). In addition, the suppression of the Mcl-1L mRNA by meayamycin B was complete in 9 h and remained as such for the next 15 h. We also observed larger RT-PCR products that increased over time. These products were partially spliced Mcl-1 pre-mRNA retaining both intron 1 and intron 2 (Figure S1 in the Supporting Information), indicating that meayamycin B acted as both a constitutive splicing inhibitor and an alternative splicing modulator for Mcl-1 pre-mRNA. At the protein level (Figure 2E), Mcl-1S was the dominant Mcl-1 isoform in both A549 and H1299 cell lines after 9 h of exposure to meayamycin B.
Meayamycin B does not regulate the alternative splicing of Bcl-x in non-small cell lung cancer cells
SF3B1 is also a trans-acting splicing factor for the Bcl-x gene.28 In neither dose- nor time-dependent treatments did meayamycin B significantly increase Bcl-xS (Figures 2C and 2D). This is in agreement with the results from the Webb group using sudemycins (analogs of FR901464) in pediatric rhabdomyosarcoma cell line Rh18 for 24 h.29 The different responses between the Mcl-1 system and the Bcl-x system toward meayamycin B might be due to the much longer half-life of Bcl-x mRNA. Therefore, although biological means to inhibit SF3B1 resulted in the perturbation of the alternative splicing of both Bcl-x and Mcl-1 systems, the chemical approach was selective for the Mcl-1 system in a short time-frame. These results warrant further investigation of the role of SF3b (possibly SF3B1) in these alternative splicing systems.
Meayamycin B sensitizes A549 and H1299 cells to ABT-737
Mcl-1L expression has been recognized as the culprit for ABT-737 resistance in various cancers, including lung cancer cell monolayer cultures and spheroids.11, 14, 30 Therefore, we hypothesized that meayamycin B could overcome the ABT-737 resistance in cancer cells that rely on Mcl-1L to survive. To test this hypothesis, A549 and H1299 cells were exposed to serial dilutions of meayamycin B and ABT-737 as single agents or in simultaneous combination at a constant ratio (meayamycin B:ABT-737 = 1:500) based on the Chou and Talalay method for 72 h.31 Post-treatment cell viability was evaluated using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt (MTS) method. ABT-737 did not affect cell viability as a single agent even at a 100 μM concentration that is the upper limit due to its solubility in a cell culture medium (Figures 3A and 3B). Meayamycin B inhibited cell growth as a single agent with GI50 values of 0.14 ± 0.008 nM and 0.15 ± 0.008 nM (n = 3) in A549 and H1299 cells, respectively. With meayamycin B by itself, the treated cells did not undergo cell death. In sharp contrast, a combination of meayamycin B and ABT-737 induced cell death at doses (≥10 nM and ≥2.5 μM, respectively) that were not cytotoxic with either of the two compounds as single agents. When the treated cells were examined under a microscope, only the combination treatment caused apoptosis-like cell shrinkage (data not shown). Although full cell-killing curves from each compound as a single agent could not be generated due to the poor solubility of ABT-737, preventing us from calculating the combination index values,31 the remarkable cytotoxic effect from the meayamycin B-ABT-737 combination indicated a strong synergism. Interestingly, under a microscope, H1299 cells displayed more prominent apoptotic morphology than A549 cells upon meayamycin B treatment. This might be related to the different p53 gene status: A549 expresses wild-type p53 protein while H1299 is p53-deficient.32 Further studies are warranted since generally the p53-null genotype in H1299 affords them stronger resistance to apoptotic stimuli.33 Nonetheless, the sensitivity of H1299 cells indicated that the apoptosis triggered by the combination of meayamycin B and ABT-737 does not require the expression of wild-type p53.
Figure 3.
72-h antiproliferation (viability) assays and basal expression of antiapoptotic Bcl-2 family proteins. (A) and (B): 72-h antiproliferation (viability) assays in H1299 and A549 cells. (C) and (D): Basal antiapoptotic Bcl-2 family protein expression of Mcl-1, Bcl-x and Bcl-2 evaluated by immunoblotting. (E) and (F): 72-h antiproliferation (viability) assays in PCI-13 and 93-UV-147T cells. Data represent results from at least three separate experiments.
Mcl-1 abundance correlates with meayamycin B-sensitivity
After examining the potency of meayamycin B in H1299 and A549, we used immunoblotting to assess the basal expression of antiapoptotic Bcl-2 family proteins in these cell lines. It was found that H1299, expressing higher level of Mcl-1L, was also more responsive to single-agent meayamycin B. The basal Mcl-1L level, as measured by the Mcl-1L/β-actin ratio, was 1.32 in H1299 and 0.41 in A549 (Figure 3C). Meayamycin B reduced the cell viability to approximately 50% in H1299 cells (Figure 3A) and 75% in A549 cells (Figure 3B). The same pattern was also observed in head-and-neck squamous cell carcinoma (HNSCC) cell lines. As an example, the data from PCI-13 and 93-UV-147T showed that the Mcl-1L/β-actin ratios were 1.24 and 0.07 (Figure 3D). Possibly due to these changes, meayamycin B reduced the cell viability to 0% in PCI-13 (Figure 3E) but only to 40% in 93-UV-147T (Figure 3F). These data indicate the potential use of meayamycin B for cancer types that overexpress Mcl-1L.
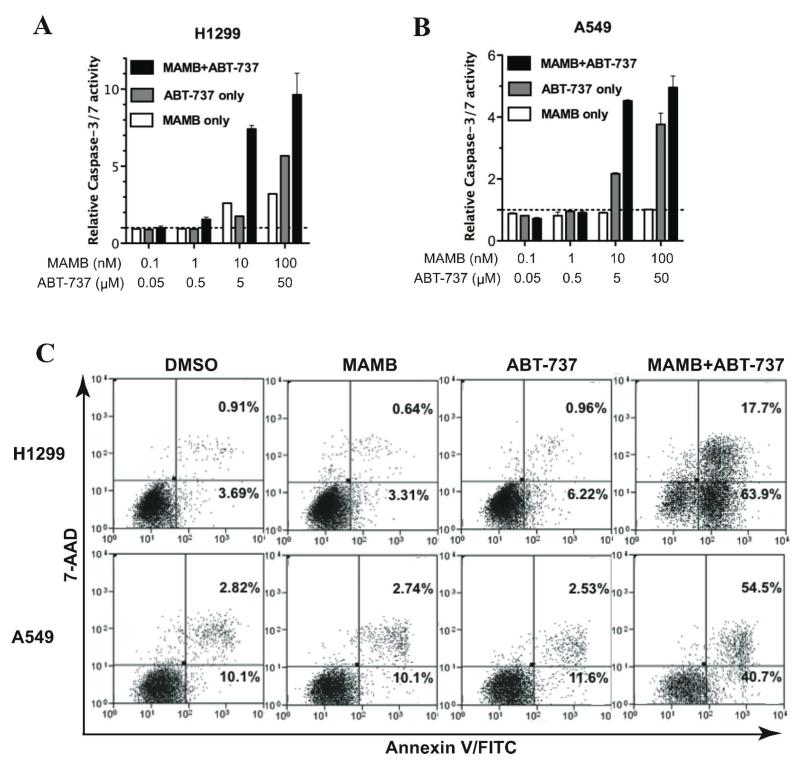
The combination of meayamycin B and ABT-737 synergistically caused apoptosis
To determine whether the combination induced apoptosis, we measured the caspase 3/7 activity of the treated H1299 and A549 cells. The cells were exposed to the combination (0.1, 1, 10, and 100 nM meayamycin B; constant meayamycin B:ABT-737 ratio at 1:500) or each agent for 9 h. Although single agent ABT-737 increased caspase 3/7 activity in a dose dependent manner, the combination treatment induced a significantly higher level of caspase 3/7 activity in both H1299 cells (Figure 4A) and A549 cells (Figure 4B). Meayamycin B by itself was a weak apoptosis inducer for these two cell lines. The rapid activation of caspase 3/7 observed in the combination-treated cells indicated a strong synergism between meayamycin B and ABT-737 in inducing apoptosis.
Figure 4.
Meayamycin B (MAMB) and ABT-737 synergistically caused apoptosis. (A) and (B): Caspase 3/7 activity assays with H1299 and A549 cells. (C): Effects of meayamycin B and ABT-737 on induction of early apoptosis (Annexin V-FITC staining) and late apoptosis (7-AAD staining) in H1299 and A549 cells as measured by flow cytometry. A representative of three separate experiments is shown.
To better understand the stages of apoptosis in the cells exposed to the combination of meayamycin B and ABT-737, we stained the cells with fluorescein isothiocyanate (FITC), annexin V, and 7-aminoactinomycin D (7-AAD) and monitored the fluorescence with flow cytometry. Annexin V stains early apoptotic cells by detecting the externalization of phosphatidylserine,34 and 7-AAD intercalates double stranded DNA to detect dead or late-stage apoptotic cells.35 The cells treated with both meayamycin B and ABT-737 displayed a significantly higher population of annexin V+/7-AAD−, indicating that these cells were in an early stage of apoptosis (Figure 4C, lower right quadrants) and annexin V+/7-AAD+ (late apoptotic/necrotic; Figure 4C upper right quadrants), indicating synergistic apoptosis stimulation.
In conclusion, meayamycin B switches the alternative splicing of Mcl-1 in a dose- and time dependent manner in non small cell lung cancer A549 and H1299 cell lines. This is, to the best of our knowledge, the first report of the modulation of Mcl-1 alternative splicing by a single small molecule through spliceosome inhibition. Although both A549 and A1299 cells were resistant to cell death in the presence of either meayamycin B or ABT-737, treatment of these cells with both meayamycin B and ABT-737 induced cell death, presumably through the meayamycin B mediated modulation of Mcl-1 pre-mRNA splicing. These results support the feasibility of using the combinations of Mcl-1L and Bcl-xL inhibitors for both research and therapeutic purposes.
METHODS
Cell lines
A549 and H1299 cells were obtained from the American Type Culture Collection (ATCC). PCI-13 and 93-UV-147T cell lines are kind gifts from Dr. Robert Ferris (University of Pittsburgh). The A549 and H1299 cells were maintained in Roswell Park Memorial Institute-1640 (RPMI-1640) medium, PCI-13 were grown in Dulbecco’s Modified Eagle’s Medium and 93-VU-147T cell line was grown in DMEM/F12 medium. All media were supplemented with 10% (v/v) fetal bovine serum, 4.5 g L−1 D-glucose, 2.0 mM L-glutamine, 100 units mL−1 penicillin, and 100 Lg mL−1 streptomycin. The cells were cultured at 37 °C in a humidified incubator with 5% CO2.
Supplementary Material
Acknowledgments
This work was in part supported by the US National Cancer Institute (R01 CA120792 and 3P50 CA097190 0751). We thank S. Osman in our group for providing meayamycin B and R. L. Ferris (University of Pittsburgh) for providing PCI-13 and 93 UV 147T cells.
Footnotes
Associated Content
Supporting information includes: Materials and Methods. Table S1: Primer sequences for RT-PCR. Figure S1: Meayamycin B inhibits the constitutive splicing of Mcl-1 in H1299 and A549 cells. This material is free via the Internet at http://pubs.acs.org.
References
- (1).Bae J, Leo CP, Hsu SY, Hsueh AJ. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J. Biol. Chem. 2000;275:25255–25261. doi: 10.1074/jbc.M909826199. [DOI] [PubMed] [Google Scholar]
- (2).Kim JH, Sim SH, Ha HJ, Ko JJ, Lee K, Bae J. MCL-1ES, a novel variant of MCL-1, associates with MCL-1L and induces mitochondrial cell death. FEBS Lett. 2009;583:2758–2564. doi: 10.1016/j.febslet.2009.08.006. [DOI] [PubMed] [Google Scholar]
- (3).Boise LH, González-García M, Postema CE, Ding LY, Lindsten T, Turka LA, Mao XH, Nuñez G, Thompson CB. bcl-x, a bcl-2 related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- (4).Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- (5).Varin E, Denoyelle C, Brotin E, Meryet-Figuiere M, Giffard F, Abeilard E, Goux D, Gauduchon P, Icard P, Poulain L. Downregulation of Bcl-xL and Mcl-1 is sufficient to induce cell death in mesothelioma cells highly refractory to conventional chemotherapy. Carcinogenesis. 2010;31:984–993. doi: 10.1093/carcin/bgq026. [DOI] [PubMed] [Google Scholar]
- (6).Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- (7).Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O’Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- (8).Huang S, Sinicrope FA. BH3 mimetic ABT-737 potentiates TRAIL-mediated apoptotic signaling by unsequestering Bim and Bak in human pancreatic cancer cells. Cancer Res. 2008;68:2944–2951. doi: 10.1158/0008-5472.CAN-07-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wesarg E, Hoffarth S, Wiewrodt R, Kroll M, Biesterfeld S, Huber C, Schuler M. Targeting BCL-2 family proteins to overcome drug resistance in non-small cell lung cancer. Int. J. Cancer. 2007;121:2387–2394. doi: 10.1002/ijc.22977. [DOI] [PubMed] [Google Scholar]
- (10).Tahir SK, Yang X, Anderson MG, Morgan-Lappe SE, Sarthy AV, Chen J, Warner RB, Ng SC, Fesik SW, Elmore SW, Rosenberg SH, Tse C. Influence of Bcl-2 family members on the cellular response of small-cell lung cancer cell lines to ABT-737. Cancer Res. 2007;67:1176–1183. doi: 10.1158/0008-5472.CAN-06-2203. [DOI] [PubMed] [Google Scholar]
- (11).Lin X, Morgan-Lappe S, Huang X, Li L, Zakula DM, Vernetti LA, Fesik SW, Shen Y. ‘Seed’ analysis of off-target siRNAs reveals an essential role of Mcl-1 in resistance to the small-molecule Bcl-2/Bcl-XL inhibitor ABT-737. Oncogene. 2007;26:3972–3979. doi: 10.1038/sj.onc.1210166. [DOI] [PubMed] [Google Scholar]
- (12).Chen S, Dai Y, Harada H, Dent P, Grant S. Mcl-1 down-regulation potentiates ABT-737 lethality by cooperatively inducing Bak activation and Bax translocation. Cancer Res. 2007;67:782–791. doi: 10.1158/0008-5472.CAN-06-3964. [DOI] [PubMed] [Google Scholar]
- (13).Kang MH, Wan Z, Kang YH, Sposto R, Reynolds CP. Mechanism of synergy of N-(4- hydroxyphenyl)retinamide and ABT-737 in acute lymphoblastic leukemia cell lines: Mcl-1 inactivation. J. Natl. Cancer Inst. 2008;100:580–595. doi: 10.1093/jnci/djn076. [DOI] [PubMed] [Google Scholar]
- (14).van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, Willis SN, Scott CL, Day CL, Cory S, Adams JM, Roberts AW, Huang DC. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Shieh JJ, Liu KT, Huang SW, Chen YJ, Hsieh TY. Modification of alternative splicing of Mcl-1 pre-mRNA using antisense morpholino oligonucleotides induces apoptosis in basal cell carcinoma cells. J. Invest. Dermatol. 2009;129:2497–2506. doi: 10.1038/jid.2009.83. [DOI] [PubMed] [Google Scholar]
- (16).Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Corrionero A, Minana B, Valcarcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Massiello A, Roesser JR, Chalfant CE. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 2006;20:1680–1682. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- (19).Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- (20).Osman S, Albert BJ, Wang Y, Li M, Czaicki NL, Koide K. Structural requirements for the antiproliferative activity of pre-mRNA splicing inhibitor FR901464. Chem.-Eur. J. 2011;17:895–904. doi: 10.1002/chem.201002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Osman S, Waud WR, Gorman GS, Day BW, Koide K. Evaluation of FR901464 analogues invitro and invivo. Med. Chem. Commun. 2011;2:38–43. [Google Scholar]
- (22).Gao Y, Vogt A, Forsyth CJ, Koide K. Comparison of splicing factor 3b inhibitors in human cells. ChemBioChem. 2013;14:49–52. doi: 10.1002/cbic.201200558. [DOI] [PubMed] [Google Scholar]
- (23).Lo CW, Kaida D, Nishimura S, Matsuyama A, Yashiroda Y, Taoka H, Ishigami K, Watanabe H, Nakajima H, Tani T, Horinouchi S, Yoshida M. Inhibition of splicing and nuclear retention of pre-mRNA by spliceostatin A in fission yeast. Biochem. Biophys. Res. Commun. 2007;364:573–577. doi: 10.1016/j.bbrc.2007.10.029. [DOI] [PubMed] [Google Scholar]
- (24).Zhang H, Guttikonda S, Roberts L, Uziel T, Semizarov D, Elmore SW, Leverson JD, Lam LT. Mcl-1 is critical for survival in a subgroup of non-small-cell lung cancer cell lines. Oncogene. 2011;30:1963–1968. doi: 10.1038/onc.2010.559. [DOI] [PubMed] [Google Scholar]
- (25).Song L, Coppola D, Livingston S, Cress D, Haura EB. Mcl-1 regulates survival and sensitivity to diverse apoptotic stimuli in human non-small cell lung cancer cells. Cancer Biol. Ther. 2005;4:267–276. doi: 10.4161/cbt.4.3.1496. [DOI] [PubMed] [Google Scholar]
- (26).Yang T, Kozopas KM, Craig RW. The intracellular distribution and pattern of expression of Mcl-1 overlap with, but are not identical to, those of Bcl-2. J. Cell Biol. 1995;128:1173–1184. doi: 10.1083/jcb.128.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Massiello A, Roesser JR, Chalfant CE. SAP155 binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 2006;20:1680–1682. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- (29).Fan L, Lagisetti C, Edwards CC, Webb TR, Potter PM. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem. Biol. 2011;6:582–589. doi: 10.1021/cb100356k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yang TM, Barbone D, Fennell DA, Broaddus VC. Bcl-2 family proteins contribute to apoptotic resistance in lung cancer multicellular spheroids. Am. J. Respir. Cell Mol. Biol. 2009;41:14–23. doi: 10.1165/rcmb.2008-0320OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Chou TC, Talalay P. Generalized equations for the analysis of inhibitions of Michaelis-Menten and higher-order kinetic systems with two or more mutually exclusive and nonexclusive inhibitors. Eur. J. Biochem. 1981;115:207–216. doi: 10.1111/j.1432-1033.1981.tb06218.x. [DOI] [PubMed] [Google Scholar]
- (32).Yu J, Yue W, Wu B, Zhang L. PUMA sensitizes lung cancer cells to chemotherapeutic agents and irradiation. Clin. Cancer Res. 2006;12:2928–2936. doi: 10.1158/1078-0432.CCR-05-2429. [DOI] [PubMed] [Google Scholar]
- (33).Rho JK, Choi YJ, Ryoo BY, Na, Yang SH, Kim CH, Lee JC. p53 enhances gefitinib induced growth inhibition and apoptosis by regulation of Fas in non small cell lung cancer. Cancer Res. 2007;67:1163–1169. doi: 10.1158/0008-5472.CAN-06-2037. [DOI] [PubMed] [Google Scholar]
- (34).Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- (35).Schmid I, Krall WJ, Uittenbogaart CH, Braun J, Giorgi JV. Dead cell discrimination with 7-amino-actinomycin D in combination with dual color immunofluorescence in single laser flow cytometry. Cytometry. 1992;13:204–208. doi: 10.1002/cyto.990130216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.