Abstract
Before each division, eukaryotic cells face the daunting task of completely and accurately replicating a heterogeneous, chromatinized genome and repackaging both resulting daughters. Because replication requires strand separation, interactions between the DNA and its many associated proteins – including histones – must be transiently broken to allow the passage of the replication fork. Here, we will discuss the disruption and re-establishment of chromatin structure during replication, and the consequences of these processes for epigenetic inheritance.
Introduction: the epigenetic challenge
In our understanding, the term epigenetics is used to describe the inheritance of patterns of gene expression that are not based on changes in DNA sequence [1]. Epigenetic processes need not rely on histones or post translational modifications [2], but they should fulfill three key requirements: first, to regulate gene expression; second, to be maintained through cell division; and third, to template their own duplication; thus, epigenetic factors must be heritable in the absence of ongoing inducing signals [3].
Chromatin is frequently associated with epigenetics; chromatin structure is determined by myriad histone modifications, histone variants, transcription factors, remodeling enzymes, and RNAs [4–8]. The primary repeating unit of chromatin is the nucleosome, which contains ~147 bp of DNA wrapped around an octameric core composed of two copies of each of the core histone proteins: H2A, H2B, H3, and H4 [9]. The core histones interact to form H2A–H2B and H3–H4 dimers; two H3–H4 dimers associate to form the (H3–H4)2 tetramer, and H2A–H2B dimers bind on either side of the tetramer to form the octamer. The large number of histone post-translational modifications found on histones within transcriptionally active or repressed chromatin [10,11], and the associated ‘histone code’ hypothesis [8], leads intuitively to the attractive possibility that histones may act as global carriers of regulatory information to control gene expression through generations. However, if histone modifications are to serve as epigenetic vectors they must surmount the challenge of DNA replication, in which chromatin is disassembled ahead of the replication fork and reassembled onto two daughter genomes.
Eukaryotic replisomes contain up to five distinct histone chaperones (extensively reviewed [12]), which together minimize the amount of non-nucleosomal DNA at the replication fork and ensure timely redeposition of nucleosomes on nascent DNA. After displacement, H3–H4 dimers and/or (H3–H4)2 tetramers are passed to PCNA-associated CAF-1 [13,14] via FACT [15–17], Asf1 [18,19] or other chaperones [5] for deposition. Electron microscopy has suggested the disruption of 1–2 nucleosomes in front of the replication machinery [20], and we have shown that ongoing lagging-strand synthesis is intrinsically linked to the immediate redeposition of histones in Saccharomyces cerevisiae [21••], suggesting that the location of parental histones on the daughter strand to which they have segregated should closely correspond to their position within the parental genome. Consistent with this, a clever epitope switching method that allows parental and nascent histone H3 to be distinguished has shown that the parental histones are generally redeposited within ~400 bp of their prereplication position in S. cerevisiae [22•]. The reassembly of nucleosomes close to their parental locations renders theoretically feasible the inheritance of histone post-translational modifications within relatively discrete domains (H2A and H2B are deposited after H3 and H4 [23,24] and will not be discussed further).
Importantly, (H3–H4)2 tetramers generally remain intact during replication fork passage [25•,26,27•] and appear to segregate randomly to the two daughter genomes [28]. Some tetramers, containing variants of H3 whose deposition is independent of S-phase [29], split [27] but the significance of this is presently unclear. It is notable that key histone chaperones [30,31] — including CAF-1 [32] — interact with two H3–H4 dimers most likely in the form of a (H3–H4)2 tetramer [32]. The production of two daughter genomes necessitates that nucleosomes containing parental (H3–H4)2 cores are intermixed with an equal number containing newly synthesized H3–H4, devoid of parental modifications but carrying characteristic deposition-related marks [33,34]. Trans-acting maintenance modification complexes (see below), targeted by marks on parental histones, can subsequently modify newly synthesized histones to harmonize the chromatin modification state across domains in the two daughter genomes.
Histone-mediated inheritance of chromatin domains
Histone modifications are clearly maintained in specific domains through the action of factors that target modification enzymes. Studies have shown that methylation of histone H3 lysine-9 (H3K9me) and lysine-27 (H3K27me) is catalyzed by methyltransferases that reside in complexes containing subunits that also bind methylated histone tails [35–38]. This provides a simple mechanism by which maintenance modification complexes can propagate a modification to nearby histones. The prototypic example of this behavior is provided by H3K9me3: this is bound by HP1, which recruits H3K9-specific methyltransfereases and, by oligomerizing, can spread this modification across a domain [39]. Thus, provided that modifications recognized in cis can be propagated in trans to nearby nucleosomes, domains of modified chromatin can potentially be maintained via simple mechanisms that do not rely on signaling or sequence-specific DNA binding proteins, and that are robust in the face of twofold dilution during replication [40].
Recently, a series of elegant experiments by the Crabtree laboratory [41••] has lent experimental support to the assertion that H3K9 methylation can be transmitted via truly epigenetic mechanisms. Through the small-molecule-induced recruitment of HP1α, Hathaway and coworkers were able to induce H3K9me3 at the Oct4 locus in mouse embryonic stem cells (ES) and embryonic fibroblasts (MEFs); the ensuing methylation spanned ~10 kb, making it remarkably similar in size to endogenous H3K9me3 domains. Critically, in MEFs, which do not express ES cell pluripotency factors, the induced H3K9me3 domain was heritably transmitted through DNA replication and persisted through multiple cell divisions after removal of the HP1α stimulus. Further in silico modeling demonstrated that a simple equilibrium between modification and replication-independent histone turnover or demethylation could give rise to stable, inherently bounded domains where spreading of the mark along the chromosome was opposed by removal of the modification [41,42] (see Figure 1a). According to the model proposed by the authors, steady state levels of H3K9me3 are the result of the opposing influences of histone methylation and turnover; this equilibrium can clearly be altered, which sits well with experiments in Drosophila that have shown that pericentromeric heterochromatin domains containing H3K9me3 are sensitive to the dosage of factors that either promote or oppose heterochomatin [43]. Consistent with this, targeting an activator to the ectopically methylated chromatin generated by Crabtree and colleagues was sufficient to disrupt the steady state and favor unmethylated H3K9 [41••]. Earlier studies in human fibroblasts have described a similar scenario for the persistence of H3K27me3. Hansen et al. [36] showed that H3K27me3, which is catalyzed by the Polycomb Repressive Complex 2 (PRC2), is also bound by PRC2; and, using a heterologous reporter system, found that transient recruitment of PRC2 led to H3K27me3 and gene repression that persisted through cell divisions. Importantly, methylation of H3K27 appears to be important for the long-term persistence of the repressed state as targeting a catalytically inactive methyltransferase greatly diminished the long-term silencing of the reporter.
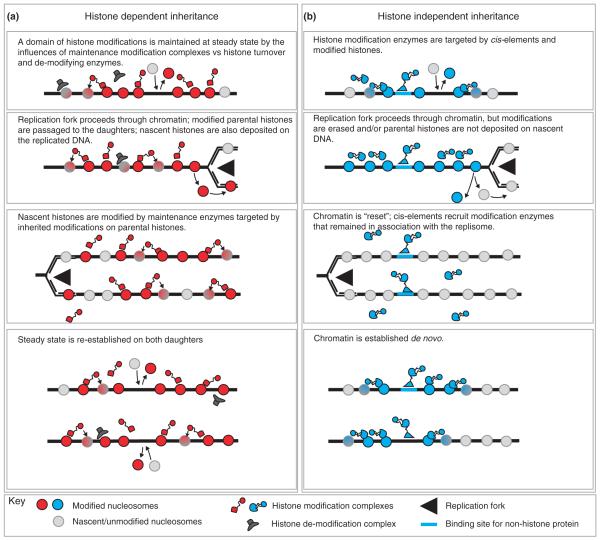
Figure 1.
Simplified schematic depiction of mechanisms by which chromatin domains may be maintained through DNA replication. (a) Histone-dependent inheritance: parental histones and modifications are retained on the daughter genomes following replication fork passage, but are diluted by nascent histones. Inheritance of modifications is mediated by maintenance enzymes targeted to inherited modifications. (b) Histone-independent inheritance: parental histones or modifications are not maintained; the chromatin state is re-established de novo by factors targeted to cis-acting elements in the DNA.
Euchromatin and heterochromatin are spatially distinct, and are replicated at different times in S-phase [44,45]. Recent work has indicated that the spatial organization of chromatin within the nucleus directly impacts the time at which it replicates, and that proximity to early-replicating or late-replicating domains [46] is by far the best predictor of when a given region of the genome will be replicated [47•• ,48••]. The broad division of the genome into early-replicating and late-replicating regions based on spatial proximity presumably provides a simple means to increase the robustness with which domains of common modifications can be inherited, as the binding of euchromatin-specific or heterochromatin-specific maintenance factors to the replisome can be both spatially and temporally controlled. Consistent with this hypothesis, microinjection experiments have shown that the assembly of transcriptionally competent chromatin is dependent upon the timing of the injection, with DNA injected early in S-phase being assembled into acetylated chromatin and expressed at higher levels [49,50]. Temporally separating the replication of active and repressed chromatin can therefore be sufficient to establish distinct chromatin types, even on naked DNA. Spatial segregation of chromatin domains likely adds a layer of redundancy, ensuring that chromatin domains are properly inherited by providing a high local concentration of certain modifying factors; replication in this context will likely ensure that the resulting chromatin on the daughters will be modified by physically associated factors, regardless of the fate of parental histones and their modifications.
Inheritance mediated by nonhistone proteins
Although histones and their modifications are often described as epigenetic, in most cases there is in fact no direct evidence to support the assertion that modifications themselves are the key determinants of epigenetic processes. Indeed, several observations make it difficult to reconcile the generality of histones and epigenetics: first, modifications themselves appear to have little instructive capability and are often correlative rather than causative of basic processes such as gene transcription [4,51]; second, histone modifications such as acetylation and phosphorylation have very short half-lives [52]. Longer-lived modifications such as methylation may not necessarily persist through generations because the histones themselves could be replaced before the next cell cycle via replication-independent turnover [53•,54]. On a global level, it has long been recognized that parental histones are passaged to the daughter genomes during DNA replication, but whether such passaging is consistent across the genome, and whether all histones maintain their modifications, often remain open questions. Recent experiments suggest that the assumption that all parental histone modifications are transferred to daughter genomes may be overly simplistic. Petruk et al., asked whether H3K4me3, which is catalyzed by the trithorax protein Trx, or H3K27me3, catalyzed by the polycomb group protein E(z), was associated with replicating DNA in Drosophila embryos [55••]. Surprisingly, the methylated histones in question were not detected during S-phase or found to be in proximity to the replication fork. Intriguingly, however, the methyltransferases Trx and E(z) were observed to colocalize with the replisome. These findings are consistent with other recent work that shows that the polycomb group complex PRC1 remains continuously associated with chromatin replicated in vitro [56,57]. Although one should not discount the possibilities that chromatin assembly in Drosophila embryos may be unusual, or that methylated histones may be present during S-phase in low quantities, these studies highlight important questions regarding the necessity of histones as mediators of epigenetic processes: propagation of histones and their modifications may not be required, provided that factors capable of establishing chromatin structures de novo are continually present (see Figure 1b).
The results of Hathaway et al. [41] and Petruk et al. [55] may represent two ends of a spectrum: if an appropriate balance between histone methylation and turnover is struck, then modifications can persist without establishment factors; however, if turnover/demethylation is dominant then establishment factors will be required. It is unclear why one mechanism should be favored over another, but clues may lie in the size and stability of the chromatin domain. Computational modeling has shown that a chromatin domain can persist if the propagation rate (i.e. methylation) is greater than the turnover rate [34]; a necessary consequence of this is a chromatin domain that may expand tens of kb. Indeed, in Drosophila, large domains of H3K27me3 spanning hundreds of kb are found in the bithorax complex region [58], which is a region of comparatively low histone turnover [59]. However, while such domains may be stable and easily persist through DNA replication, they may be too robust to allow rapid changes in gene expression or may simply be too large to be compatible with the intricate aspects of gene regulation mediated by fine-scale chromatin structure.
Evidently the association of chromatin modifying and remodeling factors with the replication fork is likely to aid the inherence of chromatin structures through DNA replication. Petruk et al. suggest that Trx and E(z) may persist at the replication fork through interactions with single stranded DNA; HP1α is also found at the replication fork in complex with the histone chaperone CAF-1 and the H3K9 methyltransferase SetDB1 [60] as are other remodeling factors such as SMARCAD1 and histone deacetylases [61]. Furthermore, modifying complexes can travel with DNA polymerase ε to progressively modify histones at the fork [62]. However, we should not discount the role of sequence-specific transcription factors, which may be retained through DNA replication and are often fundamental to the establishment of chromatin states.
Transcription factors and the limits of epigenetics
Recently, van Steensel and colleagues employed a DamID approach to map the localization of 53 broadly selected factors in Drosophila, identifying five distinct chromatin types that each contain a characteristic mixture of chromatin-associated factors [63]. Interestingly, many proteins are not confined to a single chromatin type, demonstrating that physiologically relevant chromatin structures are unlikely to be specified by the action of a single exclusive factor, but rather result from the combinatorial and/or redundant action of many activities. In addition, many regions of histone modification span only a few nucleosomes and are asymmetric [64]. Increased turnover (or restricted, hyperaccurate propagation by maintenance complexes) at these sites may be necessary to restrict the spread of the mark, but this also diminishes its ability to persist in the absence of the ongoing presence of inducing signals such as transcription factor binding, reducing the potential for epigenetic inheritance.
Similarly to nucleosomes, transcription factors must be removed from DNA before its replication. By analyzing Okazaki fragments in budding yeast, we have recently found that some transcription factors influence how Okazaki fragments are processed, which suggests that these factors are able to bind as rapidly as histones behind the replication fork [21]. Interestingly, the three proteins for which we observed rapid rebinding — Abf1, Reb1, and Rap1 — are key determinants of chromatin structure and help organize nucleosomes in gene promoters [65,66]. A recent study by the Lieb laboratory found that the occupancy times for even high-turnover Rap1 binding sites in S. cerevisiae are of the order of 30 min [67•] — almost as long as S-phase. Given that transcription factors, unlike histones, can be present at one local copy per two daughter genomes, asymmetric transcription factor binding behind the fork coupled with long residence could represent a means to establish distinct patterns of gene expression in daughter cells. In metazoan systems, rapid transcription factor reassociation with chromatin might play key regulatory roles at the promoters of tissue-specific genes during differentiation. For example, recent work has shown that the hematopoietic transcription factor GATA1 dissociates from DNA during S-phase, but rebinds to regulators of tissue-specific gene expression before M-phase. This ‘mitotic bookmarking’ [68] facilitates the rapid re-activation of these GATA1 targets in the subsequent G1, anchoring a complex of multiple transcription factors that do not reassociate in M after S-phase dissociation. The establishment of chromatin asymmetry by transcription factors, however, requires that protein binding be stable over time periods comparable with the duration of S-phase, and is thus likely to be restricted to a subset of transcription factors with unusually long residence times on DNA.
Conclusions and perspective
As the precise roles of histone modifications and sequence-specific DNA binding proteins in the perpetuation or alteration of gene expression state are further delineated, the answers to many important questions in the field of epigenetics may become clear. What is the fate during chromatin disruption and reassembly of the many histone modifications that are currently presumed to be directly inherited? For modifications such as H3K4me3 and H3K27me3 in Drosophila embryos, which are apparently erased during S-phase, it will be interesting to ascertain how this removal is accomplished and what, if any, is its biological purpose. Furthermore, the division of some cells — stem cells, for example — is accompanied by changes in expression state in a specific daughter. To what extent is this mediated by the asymmetric rebinding of transcription factors and/or modified histones behind the replication fork?
Acknowledgements
We have highlighted only papers published in the past three years and apologize to colleagues whose work could not be cited due to space limitations. This work was supported by National Institute of Health Grant R01 GM102253 and an Alfred Bressler Scholars Endowment Award to I.W.; D.J.S. is a HHMI fellow of the Damon Runyon Cancer Research Foundation (DRG-#2046-10).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- 3.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 4.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 6.Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 7.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 9.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 10.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 13.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 14.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 15.Abe T, Sugimura K, Hosono Y, Takami Y, Akita M, Yoshimura A, Tada S, Nakayama T, Murofushi H, Okumura K, et al. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem. 2011;286:30504–30512. doi: 10.1074/jbc.M111.264721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wittmeyer J, Formosa T. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol. 1997;17:4178–4190. doi: 10.1128/mcb.17.7.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wittmeyer J, Joss L, Formosa T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry. 1999;38:8961–8971. doi: 10.1021/bi982851d. [DOI] [PubMed] [Google Scholar]
- 18.Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 19.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 20.Gruss C, Wu J, Koller T, Sogo JM. Disruption of the nucleosomes at the replication fork. EMBO J. 1993;12:4533–4545. doi: 10.1002/j.1460-2075.1993.tb06142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. •• The deposition of nucleosomes and select transcription factors behind the replication fork impacts lagging-strand synthesis in vivo in yeast, suggesting immediate chromatin reassembly behind the fork.
- 22.Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 2011;9:e1001075. doi: 10.1371/journal.pbio.1001075. •This work demonstrates that parental histones, when transferred to daughter genomes, remain close to their original positions: computational models to fit these data suggest that most nucleosomes move by <400 bp.
- 23.Almouzni G, Clark DJ, Mechali M, Wolffe AP. Chromatin assembly on replicating DNA in vitro. Nucleic Acids Res. 1990;18:5767–5774. doi: 10.1093/nar/18.19.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katan-Khaykovich Y, Struhl K. Splitting of H3–H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange. Proc Natl Acad Sci U S A. 2011;108:1296–1301. doi: 10.1073/pnas.1018308108. •Two papers suggest that splitting of (H3.1–H4)2 tetramers during replication-coupled chromatin disruption and reassembly is rare compared to the maintenance of intact tetramers. Splitting is more widespread for (H3.3–H4)2 tetramers, however.
- 26.Prior CP, Cantor CR, Johnson EM, Allfrey VG. Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: a system for studying changes in nucleosomes assembled in vivo. Cell. 1980;20:597–608. doi: 10.1016/0092-8674(80)90306-2. [DOI] [PubMed] [Google Scholar]
- 27.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. •See annotation to [25•].
- 28.Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 29.Ahmad K, Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowman A, Ward R, Wiechens N, Singh V, El-Mkami H, Norman DG, Owen-Hughes T. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Mol Cell. 2011;41:398–408. doi: 10.1016/j.molcel.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fazly A, Li Q, Hu Q, Mer G, Horazdovsky B, Zhang Z. Histone chaperone Rtt106 promotes nucleosome formation using (H3–H4)2 tetramers. J Biol Chem. 2012;287:10753–10760. doi: 10.1074/jbc.M112.347450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler DD, Zhou H, Dar MA, Zhang Z, Luger K. Yeast CAF-1 assembles histone (H3–H4)2 tetramers prior to DNA deposition. Nucleic Acids Res. 2012;40:10139–10149. doi: 10.1093/nar/gks812. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 34.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 36.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 37.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 38.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, Voigt r, Martin P, Taylor SR, De Marco WR, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu B, Reinberg D. Epigenetic inheritance: uncontested? Cell Res. 2011;21:435–441. doi: 10.1038/cr.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. •• The authors describe a system in which heterochromatin can be established by ectopic HP1a recruitment to a specific locus; in certain cell types, the resulting heterochromatic domain is epigenetically stable, and is used to inform computational models of heterochromatin maintenance.
- 42.Hodges C, Crabtree GR. Dynamics of inherently bounded histone modification domains. Proc Natl Acad Sci U S A. 2012;109:13296–13301. doi: 10.1073/pnas.1211172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 44.Lima-de-Faria A, Jaworska H. Late DNA synthesis in heterochromatin. Nature. 1968;217:138–142. doi: 10.1038/217138a0. [DOI] [PubMed] [Google Scholar]
- 45.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. •• Two tours de force of replication timing analysis, demonstrating that early-replicating and late-replicating regions of the genome are conserved through evolution, change with differentiation, and occupy spatially distinct regions of the nucleus.
- 48.Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet. 2010;6:e1001011. doi: 10.1371/journal.pgen.1001011. •• See annotation to [47••].
- 49.Lande-Diner L, Zhang J, Cedar H. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol Cell. 2009;34:767–774. doi: 10.1016/j.molcel.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]
- 51.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem Sci. 2010;35:618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 53.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. •An innovative approach to measure nucleosome turnover across the Drosophila genome is developed; turnover is most rapid over active gene bodies, epigenetic regulatory elements, and replication origins.
- 54.Zee BM, Levin RS, Dimaggio PA, Garcia BA. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin. 2010;3:22. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. •• Histone H3 methylated on lysine 4 or 27 is not observed at the replication fork, or on replicated DNA: trithorax and polycomb group protein complexes – responsible for establishing and maintaining these modifications remain associated with the DNA throughout.
- 56.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lo S, Follmer N, Lengsfeld BM, Madamba E, Seong S, Grau D, Francis N. A bridging model for persistence of a polycomb group protein complex through DNA Replication in vitro. Molecular Cell. 2012;46:784–796. doi: 10.1016/j.molcel.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 59.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 60.Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, Nakatani Y, Dent SY, Almouzni G. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rowbotham SP, Barki L, Neves-Costa A, Santos F, Dean W, Hawkes N, Choudhary P, Will WR, Webster J, Oxley D, et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell. 2011;42:285–296. doi: 10.1016/j.molcel.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 62.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1–Dos2 complex. Nature. 2011;475:244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kundaje A, Kyriazopoulou-Panagiotopoulou S, Libbrecht M, Smith CL, Raha D, Winters EE, Johnson SM, Snyder M, Batzoglou S, Sidow A. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res. 2012;22:1735–1747. doi: 10.1101/gr.136366.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bai L, Ondracka A, Cross FR. Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol Cell. 2011;42:465–476. doi: 10.1016/j.molcel.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein? DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484:251–255. doi: 10.1038/nature10985. • Occupancy and residence time for Rap1 are shown to be largely independent in S. cerevisiae.
- 68.Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell. 2012;150:725–737. doi: 10.1016/j.cell.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]