Abstract
HuR, a protein that binds to specific mRNA subsets, is increasingly recognized as a pivotal posttranscriptional regulator of gene expression. Here, HuR was immunoprecipitated under conditions that preserved HuR-RNA interactions, and HuR-bound target mRNAs were identified by cDNA array hybridization. Analysis of primary sequences and secondary structures shared among HuR targets led to the identification of a 17- to 20-base-long RNA motif rich in uracils. This HuR motif was found in almost all mRNAs previously reported to be HuR targets, was located preferentially within 3′ untranslated regions of all unigene transcripts examined, and was conserved in >50% of human and mouse homologous genes. Importantly, the HuR motif allowed the successful prediction and subsequent validation of novel HuR targets from gene databases. This study describes an HuR target RNA motif and presents a general strategy for identifying target motifs for RNA-binding proteins.
Keywords: mRNA stability, posttranscriptional, gene expression, ribonucleoprotein complex, cDNA array
Proteins of the Hu family of RNA-binding proteins, which comprises the ubiquitous HuR and the primarily neuronal proteins HuB, HuC, and HuD (1-5), are emerging as pivotal regulators of posttranscriptional gene expression. Hu proteins possess three RNA-recognition motifs through which they bind with high affinity and specificity to target labile mRNAs bearing AU- and U-rich sequences and modify their expression by altering their stability, translation, or both (6-14). HuR is predominantly localized in the nucleus of most unstimulated cells (>90%), but it can translocate to the cytoplasm upon cell stimulation (15-21). Export pathways and signaling cascades that regulate its cytoplasmic abundance have been described (18, 22, 23). The hypothesis that HuR exports target mRNAs to the cytoplasm remains to be definitively proven; however, its influence on target mRNA stabilization and translation is robustly linked to HuR's cytoplasmic presence (19).
In recent years, cytoplasmic HuR has been found to be a key regulator of a variety of cellular responses through its effects on specific target mRNAs. HuR has been shown to increase cellular division by enhancing the stability of mRNAs encoding cell cycle control and proliferation-associated genes such as cyclin A, cyclin B1, and c-fos (6, 24), and it has likewise been linked to carcinogenesis through its ability to regulate the expression of proteins like vascular endothelial growth factor, tumor necrosis factor-α, β-catenin, c-myc, and cyclooxygenase-2 (5, 25-29). HuR has also been implicated in regulating muscle cell differentiation by enhancing the expression of proteins such as myogenin and MyoD (30, 31), replicative senescence (24), and the activation of immune cells, an effect that likely relies on HuR's ability to regulate the expression of genes such as granulocyte macrophage colony-stimulating factor, tumor necrosis factor-α, eotaxin, and IL-2 (16, 32, 33). An additional role for HuR within the stress response was underscored by its ability to enhance either mRNA stability or translation of various stress-response genes, including p21, p53, and hsp70 (13, 20, 34).
In light of HuR's increasingly recognized role in responding to cellular stimulation, a great deal of effort has been made toward identifying the particular sets of mRNAs that are bound by HuR in different cells under specific conditions. Despite reports of about two dozen mRNA targets of HuR, the elucidation of common sequences or motifs remains a major challenge for investigators, primarily because RNA regulatory elements typically entail a combination of a loosely defined primary sequence within the context of a secondary structure; by contrast, DNA regulatory elements, for example, can often be described solely by a specific primary sequence. A growing number of examples underscore the roles that RNA structural motifs play in controlling mRNA metabolism. Significantly, many such RNA regulatory motifs have been found to be conserved among genes that are coordinately regulated and functionally related, as well as among different species (35). In this article, we use human colorectal carcinoma cells to identify a large set of endogenous HuR target mRNAs through immunoprecipitation (IP) of HuR under native conditions that preserve [HuR-mRNA] interactions (36), followed by cDNA array hybridizations. Computational analysis of the strongest HuR targets on the array allowed us to delineate the requirements of primary sequence and secondary structure in the HuR target motif. The HuR motif described here was successfully used in the prediction of novel HuR target mRNAs.
Materials and Methods
Cell Culture and Preparation of Lysates. Human colorectal carcinoma RKO cells were cultured in minimum essential medium (GIBCO/BRL), and HeLa cells were cultured in Dulbecco's modified essential medium, each supplemented with 10% FBS and antibiotics. Actinomycin D (used at 2 μg/ml for 2 h) was from Sigma. Cytoplasmic and whole-cell fractions were prepared as previously described (20).
IP Assays. IP of endogenous HuR-mRNA complexes were used to assess the association of endogenous HuR with endogenous target mRNAs. The assay was performed essentially as described (36) except that 75 million cells were used as starting material, and lysate supernatants were precleaned for 30 min at 4°C by using 15 μg of IgG1 (BD PharMingen) and 50 μl of Protein-A Sepharose beads (Sigma) that had been previously swollen in NT2 buffer (50 mM Tris, pH 7.4/150 mM NaCl/1 mM MgCl2/0.05% Nonidet P-40) supplemented with 5% BSA. Beads (100 μl) were incubated (16 h, 4°C) with 30 μg of antibody [either IgG1 (BD PharMingen) or anti-HuR (3A2, sc-5261, Santa Cruz Biotechnology)], and then for 3 h with 3 mg of cell lysate at room temperature. After extensive washes and digestion of proteins in the IP material (36), RNA was extracted and used for either hybridization of cDNA arrays (explained below), or for verification of HuR target transcripts. For the latter analysis, RNA in the IP material was used to perform RT-PCR and/or real-time RT-PCR to detect the presence of specific target mRNAs by using gene-specific primer pairs (see Data Set 1, which is published as supporting information on the PNAS web site). For RT-PCR, the following amplification conditions were used: 2 min at 94°C, then 25-30 cycles (depending on the transcripts) of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and finally 5 min at 72°C. PCR products were visualized after electrophoresis in 1% agarose, ethidium bromide-stained gels. For real-time RT-PCR analysis, amplification conditions were 2 min at 50°C, 10 min at 95°C, and then 40 cycles of 15 s at 95°C and 1 min at 60°C.
cDNA Array Analysis. RNA in the material obtained after IP reactions using either an anti-HuR antibody or IgG1 was reverse-transcribed in the presence of [α-33P]dCTP, and the radiolabeled product was used to hybridize cDNA arrays (www.grc.nia.nih.gov/branches/rrb/dna/index/dnapubs.htm#2, MGC arrays, 9,600 genes), employing previously reported methodologies (36-39). All of the data were analyzed by using the array pro software (Media Cybernetics, Carlsbad, CA), then normalized by Z score transformation (38) and used to calculate differences in signal intensities. The complete cDNA array data are available (Data Set 1).
Computational Analysis. Human unigene records were first identified from the most strongly enriched HuR targets derived from the array analysis; 57 such transcripts (see Data Set 1), from which 5′ UTR, coding region (CR), and 3′ UTR were available, served as the experimental data set from which the HuR motif was derived. The computer program foldalign (40) was used to search against the unaligned sequences of the experimental data set for common RNA motifs, including both primary sequence and secondary structure. Secondary structures identified were further cross-validated by the program mfold (41), and modeled by stochastic context-free grammars, by using the program cove (42). The stochastic context-free grammar model, capturing both the primary and secondary features of the RNA motif, was then used to search against different gene data sets using the program covels. Three gene data sets were searched: the experimental data set, the human unigene, and the mouse unigene (unigene data sets, Hs.unigene.uniq and Mm.seq.uniq, were downloaded from NCBI on May 10, 2003). In turn, each data set searched comprised three subsets: 5′ UTR, CR, and 3′ UTR sequences. Cross-genome comparisons of all genes identified as bearing the putative HuR motif revealed homologous pairs of human and mouse genes, based on blast comparisons of the protein-coding sequences. The motif was also used to search against human refseq sequences to identify additional possible targets of HuR. The motif logo was constructed using weblogo (http://weblogo.berkeley.edu). RNAPLOT was used to depict the secondary structure of the RNA motif.
Synthesis of Biotinylated Transcripts and Analysis of HuR Bound to Biotinylated RNA. For in vitro synthesis of biotinylated transcripts, reverse-transcribed total RNA was used as template for PCR reactions. All 5′ oligonucleotides contained the T7 RNA polymerase promoter sequence. All oligonucleotide pairs (5′ and 3′ primers, respectively) used to synthesize DNA templates for the production of biotinylated transcripts are listed in Data Set 1. Primers were used for the amplification of sequences 2018-2609 of MTA1 (NM_004689); 562-1195 of UBE2N (NM_003348); 533-832 of NDUFB6 (NM_002493); 1299-1501 of NDUFV1 (BC008146); 1201-1904 of ACTG1 (BC007442); 1312-2077 of DEK (NM_003472); 109-498, 1384-1864, and 3303-3992 of HLF (NM_002126); 1675-1969 of HDAC2 (NM_001527); 2930-3869 of CDH2 (NM_001792); and 981-1283 of GAPDH (NM_002046). PCR-amplified products were resolved on agarose gels, purified, and used as templates for the synthesis of corresponding biotinylated RNAs using T7 RNA polymerase and biotin-CTP; transcripts were purified as described (6).
Biotin pull-down assays (6) were carried out by incubating 40 μg of cytoplasmic fractions with 1 μg of biotinylated transcripts for 1 h at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway), and bound proteins in the pull-down material were analyzed by Western blotting by using monoclonal antibodies recognizing either HuR or β-tubulin (Santa Cruz Biotechnology). After secondary antibody incubations, signals were visualized by enhanced chemiluminescence.
Oligomers. A description of the oligomers used for in vitro synthesis of biotinylated transcripts and for assessment of endogenous [HuR-target mRNA] complexes can be found in Supporting Text, which is published as supporting information on the PNAS web site.
Results and Discussion
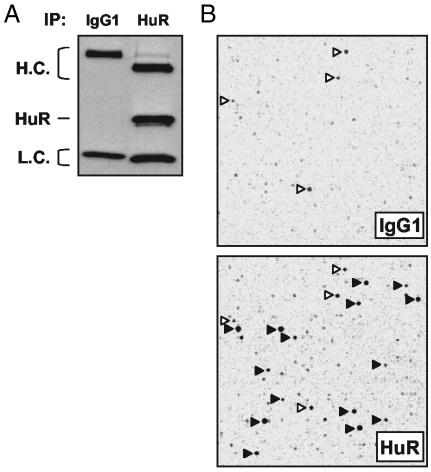
Identification of HuR Target mRNAs by Using cDNA Arrays. As a first step toward delineating the precise RNA sequences and/or structures recognized by HuR, we set out to identify a large pool of HuR targets. Using the human colorectal carcinoma cell line RKO, we isolated subsets of HuR target mRNAs by carrying out IP assays with a previously described anti-HuR antibody (34) under conditions that preserved mRNA-protein complexes (mRNPs) (Fig. 1A). HuR-bound mRNAs were then eluted and used to prepare reverse-transcribed products that were subsequently hybridized to human cDNA arrays; representative fields are shown in Fig. 1B. Array patterns obtained from untreated RKO cells were similar to those obtained from cells treated with the potent HuR activator actinomycin D (ActD), but overall signal intensities were higher in the ActD-treated groups (data not shown). The latter set of arrays was therefore used for the identification of HuR target mRNAs. Transcripts corresponding to ≈15% of all genes on the array were substantially enriched in the HuR IP compared with the IgG1 IP (see Data Set 1). The association between HuR and target mRNAs was deemed specific based on evidence that the anti-HuR antibody exhibited no appreciable cross-reactivity with other cellular proteins (data not shown); the specificity of the interactions was further ensured through the elimination of non-target mRNAs associating with either the antibody or the beads (detected in the IgG1 arrays). Among the specific HuR-bound targets, the most abundant transcripts for which full-length mRNAs were available were selected for further analysis.
Fig. 1.
Identification of HuR target mRNAs by IP of HuR-containing mRNP complexes and cDNA array hybridization with probes derived from RNA in the IP material. IP assay using 3 mg of protein lysate prepared from ActD-treated RKO cells and 30 μg of either anti-HuR antibody or IgG1, under conditions that preserved mRNP complexes. (A) HuR was detected by Western blotting in aliquots of IP material. H.C., heavy chain; L.C., light chain. (B) Representative images obtained after reverse-transcribing mRNAs in the IP material and hybridizing it to human cDNA arrays. Solid arrowheads, signals enriched in samples obtained in the HuR IP; open arrowheads, nonspecific signals, present in both IP materials (IgG1 and HuR).
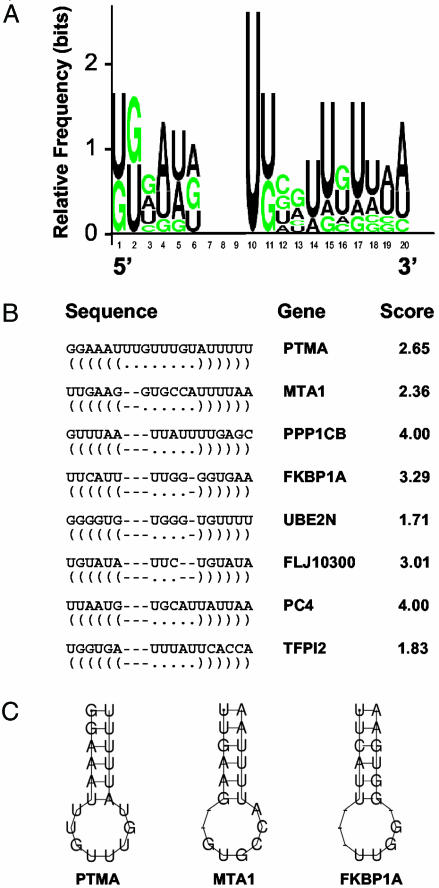
Sequence and Structure of the Predicted Binding Site. RNA sequences for the most enriched HuR targets, the experimental data set (see Data Set 1), were used in computational analyses to identify and characterize HuR motifs, based on both primary RNA sequences and secondary structures. Of the several possible candidate motifs, one motif comprising 17-20 nucleotides that was jointly recognized by several programs analyzing similarities in RNA sequence and folding (see Materials and Methods) was identified as being present in most transcripts of the experimental data set but absent from non-target transcripts. The sequence alignment and motif logo (graphic representation of the relative frequency of nucleotides at each position), as well as examples of the secondary structures of this putative HuR motif, are shown in Fig. 2. Each possible version of this somewhat flexible motif was assigned a score, which reflected the degree of conformity to the expected nucleotide(s) at each position, according to the stochastic context-free grammar model of the motif. Unexpectedly, the motif was found to be much more U-rich than AU-rich, even though traditionally HuR binding sites have been described as AU-rich elements. In this regard, it is important to note that earlier in vitro studies had defined the Hu binding sequences as consisting of short U-rich stretches that also contained nucleotides A and G, but rarely C (3, 43, 44); it is also worth remarking that the notion that AUUUA pentamers must be present within AU-rich elements has become a misleading expectation based on earlier classification methods (45). The HuR motif (Fig. 2) is also consistent with consensus sequences derived from studies by Tenenbaum and coworkers (36). Table 1 depicts a partial list of genes within the experimental data set, which encode HuR target mRNAs (a complete list is available in Data Set 1), as well as the positions of individual HuR motif hits and the score of each motif hit. It was somewhat surprising to find multiple occurrences of the HuR motif within many of the transcripts examined, as discussed below. It should be noted that several top HuR target mRNAs identified on the arrays (such as those encoding Profilin 1 and PLA2G12) did not appear to contain the 17- to 20-base motif, suggesting that additional HuR motifs may also exist that were not identified in this analysis.
Fig. 2.
Sequence and structure of the predicted HuR motif. (A) Probability matrix (graphic logo) depicting the relative frequency of finding each residue at each position within the motif, elucidated from the array-derived experimental data set. (B) Structural alignment of eight examples of the HuR motif in specific mRNAs; the corresponding gene name and motif score are shown. (C) Secondary structure of three representative examples of the HuR motif.
Table 1. HuR motif-bearing targets on cDNA arrays.
| Motif position and score | Accession no. | Gene |
|---|---|---|
| 969 (3.42) | Hs#S3322070 | Matrix metalloprotease 7 (MMP7) |
| 1714 (1.42), 1783 (0.78) | Hs#S1726209 | Actin, gamma 1 (ACTG1) |
| 537 (1.00), 750 (0.42) | NM_004181.3 | Ubiquitin thiolesterase (UCHL1) |
| 1434 (2.00), 1492 (2.42), 1515 (1.42) | Hs#S5494967 | Sorting nexin 3 (SNX3), transcript variant 3 |
| 571 (0.42), 929 (0.29), 977 (0.42) | Hs#S1727443 | Protein phosphatase 1, catalytic subunit β (PPP1CB) |
| 1417 (0.36), 1610 (1.83), 2332 (2.42) | ||
| 2414 (0.83), 2707 (0.42), 3423 (4.00) | ||
| 3439 (4.00), 3515 (0.01) | ||
| 2824 (0.42) | Hs#S1729935 | Solute carrier family 1 (SLC1A5) |
| 3848 (1.71), 1184 (0.42) | Hs#S1728144 | Ubiquitin-conjugating enzyme E2N (UBE2N) |
| 436 (0.42), 2470 (2.38) | Hs#S1729115 | Metastasis-associated 1 (MTA1) |
| 3845 (0.29) | Hs#S1824446 | Leucine zipper-EF-hand transmembrane (LETM1) |
| 2320 (1.42) | Hs#S1730282 | Nuclear factor (erythroid-derived 2)-like 2 (NFE2L2) |
| 496 (1.36), 626 (0.42), 734 (0.01) | Hs#S1727788 | Chemokine (C-X-C motif) ligand 6 (CXCL6) |
| 789 (0.29), 1311 (0.42) | ||
| 1263 (0.29), 2039 (3.42), 3111 (1.42) | Hs#S1731058 | Poliovirus receptor (PVR) |
| 2876 (0.83), 3288 (0.42), 3533 (0.01), 3564 (0.42), 3727 (0.42), 3958 (1.42), 4032 (1.42), 4078 (0.42), 4310 (2.00), 4340 (0.29), 4497 (1.88), 4566 (0.42) | Hs#S2294124 | Minichromosome maintenance deficient 10 (MCM10) |
| 791 (1.42) | Hs#S1731210 | Ribosomal protein L14 (RPL14) |
| 768 (2.65), 797 (0.88), 1031 (0.83), 1103 (0.42) | Hs#S1727552 | Prothymosin, alpha (PTMA) |
| 1102 (1.42), 1210 (0.29), 1235 (0.01) | Hs#S3438634 | RNA processing factor 1 (RPF1) |
| 1934 (1.29), 3085 (0.42), 3323 (0.01), 3359 (2.42), 3622 (1.42) | Hs#S1731856 | Karyopherin alpha 4 (importin alpha 3) (KPNA4) |
| 724 (0.42), 2028 (0.83) | Hs#S4631077 | Crystallin, zeta (quinone reductase)-like 1 (CRYZL1) |
| 580 (4.00), 828 (0.42), 985 (1.42) | Hs#S1731044 | RNA polymerase II transcription cofactor 4 (PC4) |
| 691 (3.29), 1519 (0.29) | NM_000801.2 | FK506 binding protein 1A, 12 kDa (FKBP1A) |
Partial list of genes encoding transcripts more abundantly found in HuR immunoprecipitates; these were used to create the experimental data set and derive the HuR motif. The relative position(s) of the HuR motif within each transcript and the corresponding scores (parentheses, bold) are indicated. A complete list of transcripts enriched in HuR IP compared with IgG1 IP is available as supporting information on the PNAS web site.
Stochastic context-free grammars-based models were used to search for the presence of the putative HuR motif in the human and mouse unigene and human refseq transcript databases. Table 2 lists the relative frequency with which the motif was found in those databases; sequences comprising the 5′ UTR, CR, and 3′ UTR were assessed as separate collectives. The number of motif hits for each data set was calculated with respect to the relative total size of the data set (presented as motif hits per kb). As shown, the frequency of the HuR motif in the 3′ UTR is 1.214 per kb, a markedly higher frequency than that seen for the 5′ UTR (0.536 per kb) or CR (0.158 per kb). Comparable differences were seen in the mouse unigene database, with 1.322, 0.457, and 0.308 hits per kb in the 3′ UTR, 5′ UTR, and CR, respectively. As noted in Table 2, increasing scores (>0, >1, and >3) for the HuR motif were found with progressively lower frequencies, although hit occurrences were always markedly higher in the 3′ UTR, in keeping with previous reports analyzing specific transcripts. Interestingly, more than half of motif-bearing human transcripts corresponded to genes homologous to those encoding motif-bearing mouse transcripts, indicating that the presence of this motif was significantly conserved through evolution. The complete list of homologous genes, as well as the complete list of human and mouse unigene and human refseq transcripts that contain the HuR binding motif, is available from the authors.
Table 2. Relative presence of the HuR motif in the 5′ UTR, CR, and 3′ UTR of human and mouse genes.
| Score>0
|
Score>1
|
Score>3
|
|||
|---|---|---|---|---|---|
| Total seq. searched, kb | Motif hits per kb | Motif hits per kb | Motif hits per kb | ||
| Training data set | 5′ UTR | 12.5 | 0.160 | 0.000 | 0.000 |
| CR | 59.6 | 0.386 | 0.084 | 0.000 | |
| 3′ UTR | 43.8 | 2.146 | 1.005 | 0.160 | |
| Human unigene | 5′ UTR | 4,885.5 | 0.536 | 0.175 | 0.013 |
| CR | 30,537.2 | 0.158 | 0.045 | 0.003 | |
| 3′ UTR | 18,522.9 | 1.214 | 0.434 | 0.040 | |
| Mouse unigene | 5′ UTR | 2,853.5 | 0.457 | 0.148 | 0.008 |
| CR | 24,717.5 | 0.308 | 0.086 | 0.005 | |
| 3′ UTR | 11,538.1 | 1.322 | 0.469 | 0.041 |
Number of motif hits and frequency in the human and mouse unigene databases, as well as the experimental data set. For each transcript region (5′ UTR, CR, and 3′ UTR), the total number of motif hits, as well as the frequency (hits per kb), are shown. Data are provided for the HuR motif meeting three different score thresholds (>0, >1, and >3).
Important support for the validity of this motif was obtained through the discovery that virtually all of the mRNAs reported to be targets of HuR were found to contain at least one motif (Table 3). Taken together, the motif identified from the array-based experimental data set supports the previous HuR literature, including its existence on published HuR target mRNAs, its preferential location on the 3′ UTR, and its joint presence on human and mouse homologous target mRNAs.
Table 3. Motif location and score in reported HuR targets.
| Motif position and score | Accession no. | Gene |
|---|---|---|
| 534 (0.42), 2717 (0.83), 2830 (0.42), 3134 (1.42), 3168 (0.42) | Hs#S1726566 | β-Catenin |
| 1983 (0.42), 2103 (0.42) | NM_002467.2 | Myc |
| 379 (0.42), 1662 (1.00), 1710 (0.83), 1956 (1.00), 2091 (0.29), 2660 (0.78) | M90100.1 | COX-2 |
| 813 (0.83), 867 (4.00), 956 (0.42), 1060 (3.00) | Hs#S2324 | IL-6 |
| 1015 (1.42), 2090 (0.42) | U03106.1 | p21Cip1 |
| 330 (0.42), 1020 (0.01), 1344 (0.42), 1405 (0.42), 1689 (1.42) | AY004255.1 | p27Kip1 |
| 685 (0.42), 1912 (0.42) | Hs#S1728078 | p53 |
| 2038 (0.29), 2699 (0.42), 2804 (0.01) | Hs#S1620 | TGF-β |
| 1247 (2.42) | Hs#S3218906 | TNF-α |
| 1109 (0.83), 1565 (3.00) | Hs#S1726397 | Cyclin A |
| 562 (0.42), 630 (1.42) | NM_002986.2 | Eotaxin |
| 177 (1.01) | Hs#S1728178 | VEGF |
| 734 (0.01), 1149 (3.42) | Hs#S3218902 | IL-8 |
| 1031 (0.42) | Hs#S3618391 | Cyclin B1 |
| 1517 (0.83), 1783 (1.13), 1937 (0.83) | Hs#S1732326 | Fos |
Number, score (parentheses, bold), and location of the HuR motif in known HuR mRNA targets.
Assessment of the Validity of the Motif in the Identification of Novel HuR Target Transcripts. To test the usefulness of the HuR motif in predicting novel HuR targets, we randomly selected several genes identified as having the HuR motif within the unigene database for further analysis. The formation of complexes between the encoded transcripts and HuR was tested by using two different binding assays: first, by testing the ability of endogenous HuR to bind biotinylated transcripts of interest using pull-down assays coupled with Western blotting, and second, by IP of endogenous HuR-containing mRNP complexes and assessing the presence of bound target mRNAs of interest by RT-PCR.
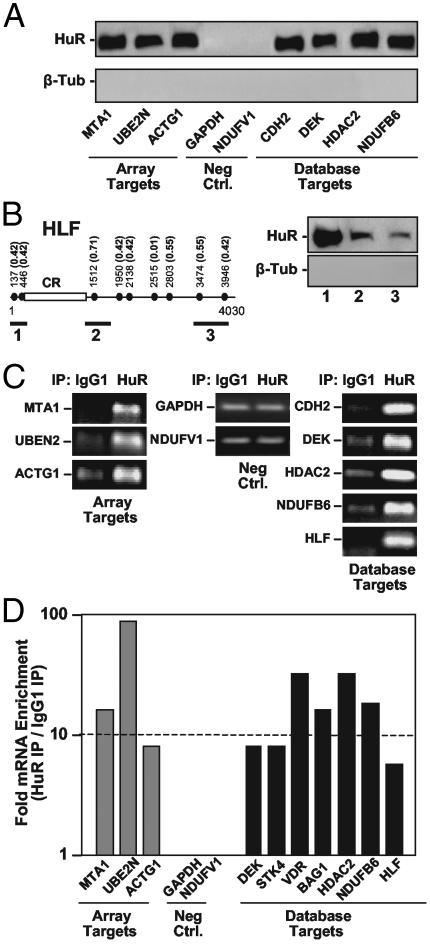
Biotinylated transcripts encompassing motif-containing sequences from the mRNAs indicated (Fig. 3A) indeed formed complexes with HuR, as such transcripts effectively pulled down HuR on streptavidin-coated beads; HuR was subsequently detected by Western blotting. Included in this validation approach were transcripts identified in the cDNA array search, as well as transcripts identified after motif searches within the human unigene database. Negative control transcripts encompassing the 3′ UTRs of GAPDH and NDUFVI, which lacked the HuR motif, failed to pull down HuR. Although the existence of unspecific contaminating proteins in the pull-down material could not be definitively ruled out, control hybridizations of the blots to detect β-tubulin, which does not bind mRNAs, yielded no signals. Of note, biotinylated transcripts BAG1 and VDR, predicted to contain an HuR motif (position 1169, score 0.42), failed to show binding (data not shown). Although the reasons for these negative results remain to be elucidated, the motif may lie within a region of secondary structure that precludes its appropriate folding into the expected stem-loop conformation; alternatively, another RNA-binding protein might compete for binding to the HuR motif within the context of the specific transcript. Additional work will be needed to discern why HuR binding to the BAG1 and VDR transcripts and possibly other HuR targets may not be validated by this in vitro assay. Interestingly, however, endogenous BAG1 and VDR mRNAs were found to bind to HuR as assessed by a different method (see below and Fig. 3D), suggesting that the association of HuR to these targets may occur through other regions in the mRNA.
Fig. 3.
Validation of novel HuR targets predicted by using the HuR motif. (A) Biotin pull-down assays to assess the ability of endogenous HuR to bind to biotinylated transcripts of interest. The indicated biotinylated transcripts were incubated with cytoplasmic protein lysate from ActD-treated RKO cells, whereupon their association with HuR was detected by Western blotting; stripping and rehybridization of blots to detect β-tubulin (β-Tub) revealed an absence of contaminating proteins in the pull-down material. (B) Three biotinylated transcripts (heavy bars) spanning different regions of HLF mRNA, which bears nine hits for the HuR motif (solid circles) with varying scores (parentheses), were used in pull-down assays to detect their association with HuR as described in A. (C) The binding of endogenous HuR with endogenous target mRNAs was further tested in RKO cells by IP, followed by detection of the target transcripts of interest by low-cycle (25-30) RT-PCR amplification of IP material. PCR products were visualized by electrophoresis in ethidium bromide-stained 1% agarose gels. (D) The abundance of transcripts present in material obtained from HeLa cells after either HuR IP or IgG1 IP was assessed by real-time RT-PCR, and fold differences were plotted on a log scale.
HLF was chosen for a more detailed study of HuR binding. As shown in Fig. 3B, all of the biotinylated HLF transcripts comprising hits of the HuR motif were shown to bind HuR, although the score values and number of motif hits were poor predictors of the strength of HuR binding to each transcript. Further work will be needed to elucidate a possible ranking of preferential association of HuR and target transcripts. Also awaiting future systematic testing are the possibilities that the clustering and relative distance of HuR motif occurrences may affect HuR's affinity for a given target transcript, and that several individual HuR proteins associate simultaneously on a transcript with multiple HuR motif hits. Finally, although it is tempting to conclude that the HuR motif is precisely the HuR binding site, based on extensive literature and a very recent report (46), formal efforts are underway to test this hypothesis in vitro and in vivo.
The association of HuR with target mRNAs was also tested by IP of mRNP complexes formed in vivo in RKO cells by using an anti-HuR antibody (or IgG1 in control IP reactions), and then performing RT-PCR assays to detect the presence of the endogenous mRNA of interest in the IP material (Fig. 3C). Of note, non-target GAPDH and NDUFV1 mRNAs could also be amplified, albeit inefficiently and to the same extent in both IP groups (Fig. 3C, Neg. Ctrl.); these findings revealed the presence of low levels of contaminating, unspecific mRNAs in all IP samples, verified the use of equal amounts of input material, and demonstrated that HuR mRNA targets were amplified from HuR IPs markedly better than from IgG1 IPs. Importantly, the transcripts shown to bind in vitro (Fig. 3A) were confirmed to be endogenous, specific mRNA targets of HuR.
To ascertain whether the identified HuR motif was suitable for the successful prediction of novel HuR target mRNAs in other cell systems, we extended our validation efforts to the human cervical carcinoma cell line HeLa. Here, a more quantitative assessment of the enrichment in HuR target mRNAs in the HuR IPs relative to the IgG1 IPs was carried out by performing real-time RT-PCR analyses. As shown, no differences in the abundance of control mRNAs lacking an HuR motif (those encoding GAPDH and NDUFVI) were seen when comparing each IP material. By contrast, all other mRNAs were enriched between 6- and 90-fold in the HuR IPs (Fig. 3D). Comparable differences were seen when assessing HuR mRNP complexes in RKO cells (data not shown). Together, these findings reveal that the HuR motif identified here can predict with a high degree of confidence if a novel mRNA will be a target of binding by HuR.
In summary, we have used an approach based on IP of HuR-containing mRNPs followed by hybridization of cDNA arrays using the immunoprecipitated mRNA material to identify a novel subset of HuR target mRNAs. Computational analysis of these sequences led to the elucidation of a U-rich common motif present in HuR target mRNAs. This motif was found in virtually all HuR targets reported thus far and was successfully used for the prediction and subsequent validation of additional novel HuR target mRNAs in genome-wide database searches. We propose that a similar approach can be used for the prediction of mRNA subsets that are targets of other RNA-binding proteins. We anticipate that such systematic identification of RNA targets for RNA-binding proteins will be extremely valuable as we pursue an increasingly thorough understanding of posttranscriptional gene regulation.
Supplementary Material
Acknowledgments
We thank M. B. Kastan for the RKO cells, and U. Atasoy, K. Mazan-Mamczarz, and K. G. Becker for guidance with experimental procedures.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ActD, actinomycin D; CR, coding region; IP, immunoprecipitation; mRNP, mRNA-protein complexes.
References
- 1.Good, P. J. (1995) Proc. Natl. Acad. Sci. USA 92, 4557-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma, W.-J., Cheng, S., Campbell, C., Wright, A. & Furneaux, H. (1996) J. Biol. Chem. 271, 8144-8151. [DOI] [PubMed] [Google Scholar]
- 3.Antic, D. & Keene, J. D. (1997) Am. J. Hum. Genet. 61, 273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King, P. H., Levine, T. D., Fremeau, R. T. & Keene, J. D. (1994) J. Neurosci. 14, 1943-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szabo, A., Dalmau, J., Manley, G., Rosenfeld, M., Wong, E., Henson, J., Posner, J. B. & Furneaux, H. M. (1991) Cell 67, 629-639. [DOI] [PubMed] [Google Scholar]
- 6.Wang, W., Lin, S., Caldwell, C. M., Furneaux, H. & Gorospe, M. (2000) EMBO J. 19, 2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy, N. S., Chung, S., Furneaux, H. & Levy, A. P. (1998) J. Biol. Chem. 273, 6417-6423. [DOI] [PubMed] [Google Scholar]
- 8.Millard, S. S., Vidal, A., Markus, M. & Koff, A. (2000) Mol. Cell. Biol. 20, 5947-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kullmann, M., Gopfert, U., Siewe, B. & Hengst, L. (2002) Genes Dev. 16, 3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antic, D., Lu, N. & Keene, J. D. (1999) Genes Dev. 13, 449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, S., Eckrich, M., Perrone-Bizzozero, N., Kohn, D. T. & Furneaux, H. (1997) J. Biol. Chem. 272, 6593-6598. [DOI] [PubMed] [Google Scholar]
- 12.Galbán, S., Martindale, J. L., Mazan-Mamczarz, K., López de Silanes, I., Fan, J., Wang, W., Decker, J. & Gorospe, M. (2003) Mol. Cell. Biol. 23, 7083-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazan-Mamczarz, K., Galbán, S., López de Silanes, I., Martindale, J. L., Atasoy, U., Keene, J. D. & Gorospe, M. (2003) Proc. Natl. Acad. Sci. USA 100, 8354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain, R. G., Andrews, L. G., McGowan, K. M., Pekala, P. H. & Keene, J. D. (1997) Mol. Cell. Biol. 17, 954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennan, C. M. & Steitz, J. A. (2001) Cell. Mol. Life Sci. 58, 266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atasoy, U., Watson, J., Patel, D. & Keene, J. D. (1998) J. Cell. Sci. 111, 3145-3156. [DOI] [PubMed] [Google Scholar]
- 17.Fan, X. C. & Steitz, J. A. (1998) Proc. Natl. Acad. Sci. USA 95, 15293-15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan, X. C. & Steitz, J. A. (1998) EMBO J. 17, 3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keene, J. D. (1999) Proc. Natl. Acad. Sci. USA 96, 5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, W., Furneaux, H., Cheng, H., Caldwell, M. C., Hutter, D., Liu, Y., Holbrook, N. J. & Gorospe, M. (2000) Mol. Cell. Biol. 20, 760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, C. Y., Xu, N. & Shyu, A.-B. (2002) Mol. Cell. Biol. 22, 7268-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallouzi, I. E. & Steitz, J. A. (2001) Science 294, 1895-1901. [DOI] [PubMed] [Google Scholar]
- 23.Wang, W., Fan, J., Yang, X., Fürer, S., López de Silanes, I., von Kobbe, C., Guo, J., Georas, S. N., Foufelle, F., Hardie, D. G., et al. (2002) Mol. Cell. Biol. 22, 3425-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, W., Yang, X., Cristofalo, V. J., Holbrook, N. J. & Gorospe, M. (2001) Mol. Cell. Biol. 21, 5889-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nabors, L. B., Gillespie, G. Y., Harkins, L. & King, P. H. (2001) Cancer Res. 61, 2154-2161. [PubMed] [Google Scholar]
- 26.Dalmau, J., Furneaux, H. M., Gralla, R. J., Kris, M. G. & Posner, J. B. (1990) Ann. Neurol. 27, 544-552. [DOI] [PubMed] [Google Scholar]
- 27.López de Silanes, I., Fan, J., Yang, X., Potapova, O., Zonderman, A. B., Pizer, E. S. & Gorospe, M. (2003) Oncogene 22, 7146-7154. [DOI] [PubMed] [Google Scholar]
- 28.Blaxall, B. C., Dwyer-Nield, L. D., Bauer, A. K., Bohlmeyer, T. J., Malkinson, A. M. & Port, J. D. (2000) Mol. Carcinog. 28, 76-83. [PubMed] [Google Scholar]
- 29.Galbán, S., Fan, J., Martindale, J. L., Cheadle, C., Hoffman, B., Woods, M. P., Temeles, G., Brieger, J., Decker, J. & Gorospe, M. (2003) Mol. Cell. Biol. 23, 2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Figueroa, A., Cuadrado, A., Fan, J., Atasoy, U., Muscat, G. E., Gorospe, M. & Muñoz, A. (2003) Mol. Cell. Biol. 23, 4991-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Der Giessen, K., DiMarco, S., Clair, E. & Gallouzi, I. (2003) J. Biol. Chem. 278, 47119-47128. [DOI] [PubMed] [Google Scholar]
- 32.McMullen, M. R., Cocuzzi, E., Hatzoglou, M. & Nagy, L. E. (2003) J. Biol. Chem., in press. [DOI] [PMC free article] [PubMed]
- 33.Atasoy, U., Curry, S., López de Silanes, I., Shyu, A.-B., Casolaro, V., Gorospe, M. & Stellato, C. (2003) J. Immunol. 171, 4369-4378. [DOI] [PubMed] [Google Scholar]
- 34.Gallouzi, I. E., Brennan, C. M., Stenberg, M. G., Swanson, M. S., Eversole, A., Maizels, N. & Steitz, J. A. (2000) Proc. Natl. Acad. Sci. USA 97, 3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keene, J. D. & Tenenbaum, S. A. (2002) Mol. Cell 9, 1161-1167. [DOI] [PubMed] [Google Scholar]
- 36.Tenenbaum, S. A., Lager, P. J., Carson, C. C. & Keene, J. D. (2002) Methods 26, 191-198. [DOI] [PubMed] [Google Scholar]
- 37.Vawter, M. P., Barrett, T., Cheadle, C., Sokolov, B. P., Wood, W. H., III, Donovan, D. M., Webster, M., Freed, W. J. & Becker, K. G. (2001) Brain Res. Bull. 55, 641-650. [DOI] [PubMed] [Google Scholar]
- 38.Fan, J., Yang, X., Wang, W., Wood, W. H., III, Becker, K. G. & Gorospe, M. (2002) Proc. Natl. Acad. Sci. USA 99, 10611-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheadle, C., Vawter, M. P., Freed, W. J. & Becker, K. G. (2003) J. Mol. Diagn. 5, 73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorodkin, J., Heyer, L. J. & Stormo, G. D. (1997) Nucleic Acids Res. 25, 3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuker, M. (2003) Nucleic Acids Res. 31, 3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eddy, S. R. & Durbin, R. (1994) Nucleic Acids Res. 22, 2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine, T. D., Gao, F., King, P. H., Andrews, L. G. & Keene, J. D. (1993) Mol. Cell. Biol. 13, 3494-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao, F. B., Carson, C. C., Levine, T. & Keene, J. D. (1994) Proc. Natl. Acad. Sci. USA 91, 11207-11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chyi-Ying, A. C. & Shyu, A.-B. (1995) Trends Biochem. Sci. 20, 465-470. [DOI] [PubMed] [Google Scholar]
- 46.Cok, S. J., Acton, S. J. & Morrison, A. R. (2003) J. Biol. Chem. 278, 36157-36162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.