Abstract
The global gene expression profiling of early T helper (Th) 1 and Th2 differentiation reveals that this process can be divided into two stages, activation and differentiation. The activation stage is manifested in coordinated mobilization of the replication machinery, a process that we hypothesize may be responsible for establishing genomewide opening of transcription loci. The molecular programs underlying the differentiation stage consist of highly regulated expression of functional groups of genes that are important for the biological properties of Th1/2 cells and transcription factors that are likely important in establishing terminal differentiation of these cells. The kinetics of expression pattern of a number of transcription factors shed new light on the molecular events that shape the outcome of Th1/2 differentiation.
Thelper (Th) 1/2 differentiation is an important paradigm in adaptive immunity (1, 2). Naïve CD4+ T cells do not secrete effector cytokines. Effector CD4+ T cells acquire the capacity to make these cytokines but do not do so without TCR triggering or activation through inflammatory cytokines such as IL-18 (3, 4). Naïve resting CD4+ cells are arrested at the G0 stage, and dramatic chromatin remodeling or even cell division is needed to establish the capacity to secrete effector cytokines (5). The opening of Th1/2 loci seems to be random at the beginning of the culture and polarity is established in later differentiation (6, 7). How this polarized pattern is established has not been systematically addressed.
In this study, we undertook a global gene profiling analysis of early Th1 and Th2 differentiation. This study has revealed key molecular circuitries that control T cell activation, Th1/2 differentiation, and functional properties of Th1 and Th2 cells.
Materials and Methods
T Cell Culture. CD4+ T cells were prepared as described (8). Briefly, naïve CD4+ T cells are cultured on 24-well plates precoated with α-CD3 (10 μg/ml) and α-CD28 (5 μg/ml). Th1 conditions were formulated by using IL-12 (3.4 ng/ml), human IL-2 (20 units/ml), and α-IL-4 (clone 11B11) antibody (2 μg/ml). Th2 conditions were formulated by using IL-4 (3000 units/ml), human IL-2 (20 units/ml), and α-IFN-γ (clone XMG2.1) antibody (2 μg/ml). Forty-eight hours after starting the culture, cells are replated, with the original culture media (including polarizing cytokines and anticytokine antibodies), to another culture dish with freshly added human IL-2 (5 units/ml). Cells were cultured further for another 2 days and then washed and stimulated with plate-bound α-CD3 for 4 h.
Expression Analysis. Total RNA purification and target labeling were done according to Affymetrix-recommended protocols. Using mas 4.0 software, the stained GeneChips were processed and calculations were made to determine the average difference values (level of gene expression), fold-change values, and detection calls (absent, present, or marginal). Genes that showed changes >2-fold in both experiments were chosen for further analysis.
Results
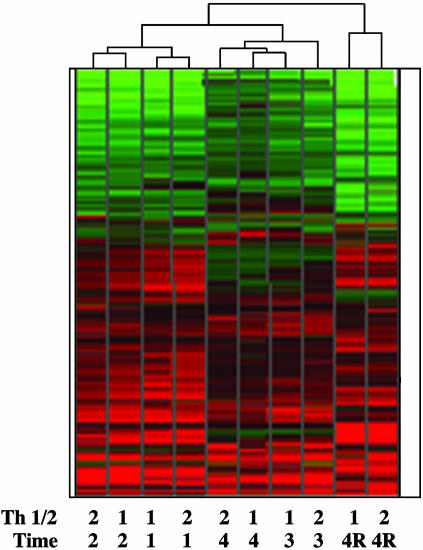
Clustering Analysis Revealed That Th1/2 Differentiation Experiences Two Stages: Activation and Differentiation. Two independent sets of RNA samples from in vitro-differentiated naïve CD4+ T cells were processed and analyzed by using the Affymetrix U74av2 mouse GeneChip. The expression pattern of a number of well known Th1 and Th2 genes revealed in this analysis was as expected and validated (Fig. 4, which is published as supporting information on the PNAS web site). More than 3,000 genes were expressed in each of the samples examined, including naïve CD4+ T cells (Table 1, which is published as supporting information on the PNAS web site). Global hierarchical clustering analysis revealed that the expression pattern of day-1 or day-2 Th1 cells is closer to day-1 or day-2 Th2 cells than to Th1 cells from day 3 or 4 (Fig. 1). The same hierarchical relationships were also true for Th2 cells, revealing that at the global gene expression level, Th1 and Th2 cells begin to diverge at day 3 after primary stimulation. These data fit the hypothesis that Th1/2 lineage establishment is contingent on proliferation/replication events. At the end of the culture, ≈300 genes were differentially expressed between Th1 and Th2 cells (Table 2, which is published as supporting information on the PNAS web site).
Fig. 1.
Hierarchical clustering analysis. Analyzed were 2,196 genes with changes of expression level >2-fold vs. naïve CD4 T cells in both of two datasets. The clustering method was performed in Spotfire software (Somerville, MA) and used the unweighted paired group averages, using city-block distance measurements, and an average value ordering function.
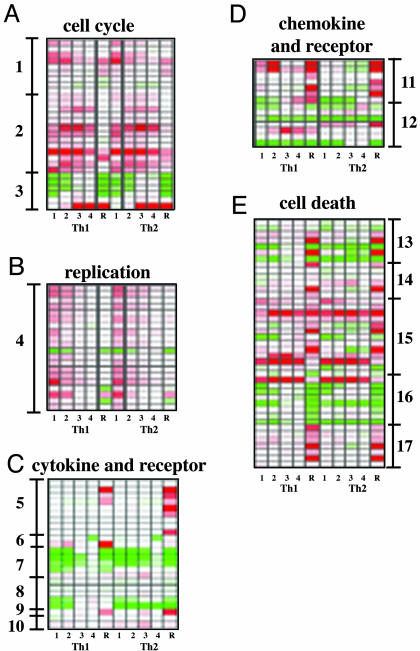
Functional and Expression Analysis of T Cell Activation and Differentiation Process. To understand the activation and differentiation process, we organized the genes that showed changes in our experiments into a number of functional categories, further grouped them according to their kinetic expression profiles, and analyzed the functional impact of these patterns to T cell activation and Th1/2 differentiation (Figs. 2 and 3, and Figs. 5 and 6 and Table 3, which are published as supporting information on the PNAS web site).
Fig. 2.
Heat map of functional group genes-separate clusters. The ratio of gene expression level of various time points to naïve cell value was used to compile the heat map representing increase (red) and decrease (green) in expression relative to native cell level. The grades of the shade of these colors were used to represent gene expression levels. The range of ratios represented is 0.01 (brightest green) to 100 (brightest red) for Fig. 3C and 0.05-20 for all other figures. In all cases, white represents the ratio of 1. Genes in various clusters are list as follows: 1 (Chek1 MGC37111 Cdc45l Tfdp1 Pin1 Nol1 Cdc7l1 Rad1 Ccne2), 2 (Ccna2 Ccnf Ccnb2 Cdc25c Ttk Mki67 Mad2l1 Mcmd7 Bub1 Cdc2a Cdkn1a Ccnb1-rs1 Gmnn), 3 (Cdk5 Ccng2 Rbl2 Cdkn2d Cdkn3 S100a6), 4 (Mcmd Cdc45l Blm Pola1 Rfc3 Mcmd Orc1 Lig1 Mcmd4 Mcmd2 Chaf1a Prim2 Rfc5 Orc6l Prim1 Pola2 Pold2 Recc1 Rpa2), 5 (Il1a Il3 Csf2 Tnfsf11 Il4 Il5 Il6 Il9 Il13), 6 (Lta Ifng), 7 (Ifnar Il7r Il10rb Il4ra Il2ra), 8 (Tgfb3 Grn Tnfrsf8 Ifngr Il18r1), 9 (Il10), 10 (Il2rb Il2ra), 11 (Ccl3 Ccl4 Ccl9 Cxcl2 Xcl1 Ccl1 Ccl5), 12 (Ccr2 Cmkbr4 Cmkbr7 Cmkbr8 Cmkbr5 Cmkar3 Cmkar4), 13 (Map3k5 Dapk2 Pdcd1 Bcl2a1d Tnfrsf1a Bcl2a1b Birc3), 14 (Perp-pending Map3k5 Bag3 Casp6 Bcl2a1d Casp11), 15 (Siva-pending Pdcd1 Casp3 Tnfsf6 Bcl2a1d Pglyrp Casp11 Bcl2a1b Gadd45g, Birc5 MGC6998 Cradd), 16 (Birc5 Bcl2 Bnip3l Dap3 Tnfrsf1a Apaf1 Cradd Birc3), 17 (Perp-pending Bcl2l Pdcd1 Bcl2a1d Casp11 Gadd45b Pdcd5). The data are also presented as graphs in Fig. 5.
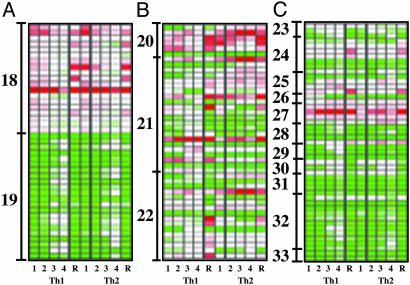
Fig. 3.
Heat map of functional group genes encoding transcriptional factors. The ratio of gene expression level of various time points to naïve cell value was used to compile the heat map representing increase (red) and decrease (green) in expression relative to native cell level. The range of ratios represented is 0.05 (brightest green) to 20 (brightest red). In all cases, white color represents the ratio of 1. The 10 columns of blocks from left to right are Th1day1, Th1day2, Th1day3, Th1day4, Th1day4 restimulated, Th2day1, Th2day2, Th2day3, Th2day4, and Th2day4 restimulated samples. Genes in various clusters are list as follows: 18 (Mcmd Tfdp1 Gtf2h4 Egr2 Taf9 Spnr Gtf2f2 Atf3 RPB6 Myc Mcmd Irf4 Polr2h Mcmd4 Mcmd2 Pou2af1 Smarca5 Gtf2a2 Gtf2e2), 19 (Klf3 Ezh1 Ddit3 Tctex3 Ifi-203 Lef1 Zfp99 Pcaf Klf7 Klf13 Elf1 Madh7 Rara Cbx4 Gtf2i Fli1 Ets1 Rbl2 Klf2 Nfatc3 Ifi204 Mad4), 20 (Gata3 Nfil3 Gfi1 Atf3 Icsbp Txk), 21 (Nfil3 Runx1 Epas1 Gata3 Rbpsuh Hic1 Rfxank Gfi1 Ddit3 Nr4a1 Trim30 Gtf2i Irf1 Gtf2i Mail-pending Bhlhb2 Klf7 Mad Ifi204 Icsbp Txk Hlx), 22 (Gata3 Irf1 Runx1 Nfil3 Stat1 Rbl1 Hmgb3 Madh7 Nr4a1 Utf1 Ifi204 Spi-B Ar Runx2 Stat4 Nr4a2), 23 (Nfatc1 Nfatc2 Nfatc3), 24 (Nfkbia Nfkbie Nfkb2 Nfkb1 Ikbke Ikbkb Ikbkg), 25 (Mad4 Mxi1 Mad Myc), 26 (Idb3 Idb2), 27 (Hif1a Bhlhb2 Hif1a Epas1), 28 (Lfng Ng7 Dtx1 Rbpsuh), 29 (Ifi204 Ifi203 Ifi203), 30 (Rbl1 Rbl2 Rb1), 31 (Lef1 Tcf7 Tcf7 Lef1), 32 (Nr1i3 Klf3 Klf3 Zfp26 Znfn1a1 Mll Klf7 Klf13 Tbx6 Klf2 Zfp292), 33 (Madh7 Madh4). The data are also presented as graphs in Fig. 6.
Cell Cycle and Replication. We first grouped cell cycle-related genes based on the similarity of their expression profile during differentiation by using clustering analysis. This type of analysis revealed several clusters of genes. One group of cell cycle-related genes were up-regulated only during the first 48 h (Fig. 2A, cluster 1). This group of genes is involved in the G1/S phase transition that initiates the first cell cycle of naïve T cells on activation. This marking of the G1/S phase transition is consistent with prior observations that it takes 20 h of TCR stimulation to commit a naïve T cell to proliferate (9). Another group of cell cycle-related genes was up-regulated throughout the 4 days of primary stimulation (Fig. 2A, cluster 2). The last four genes of this group were also highly expressed during restimulation. Within this group, there are genes involved in both G1/S and G2/M transition, such as cyclin A2 and cdc2a (10), and there are also genes that are involved in only G2/M transition, such as cyclin F and cyclin B2 (11). Another group of genes was down-regulated on primary and secondary stimulation. These are likely important in maintaining a postmitotic state (Fig. 2A, cluster 3). Among 58 replication-related genes, 21 showed significant differences between the first 48 h and last 48 h of culture. Strikingly, 20 of these genes displayed an expression pattern (Fig. 2B, cluster 4) that was induced early during T cell activation and declined thereafter. These data suggest commitment to replication early after activation.
Cytokines and Cytokine Receptors. Differentiation of naïve T cells into Th1 and Th2 cells prepares T cells for the rapid release of effector cytokines on restimulation. As expected, much of the change in Th2 cytokine mRNA occurred after secondary stimulation (Fig. 2C, cluster 5). Consistent with previous studies (5), Th1 cytokine mRNAs appeared earlier during primary stimulation, yet the bulk of the cytokine expression occurred after secondary stimulation (Fig. 2C, cluster 6). The expression of a group of cytokine receptors declined after primary and secondary stimulation (Fig. 2C, cluster 7). IL-7 has been shown to be critical for naïve cell homeostatic proliferation (12). The expression profile observed here indicates that on TCR stimulation, the survival signal provided by IL-7 receptor signaling is turned off. During the last 48 h of primary stimulation, IL-7 receptor expression was restored, thereby reinstating the survival signal for the “resting state.” Down-regulation of cytokine receptor messages on TCR signaling can be extended also to other cytokine receptors, such as IL-10 receptor β, IFN-γ receptor 1, and IL-4R α. In contrast, mRNAs for receptors such as IL-12Rβ2 were up-regulated preferentially in Th1 after TCR simulation as shown (Table 3) (13). TCR engagement therefore drastically changes responsiveness to cytokines from being homeostatic to inflammatory in nature. The expression of IL-10, the gene encoding a major negative regulatory cytokine, was slowly increased during the 4 days of culture. Therefore, IL-10 may play a negatively regulatory role for both Th1 and Th2 cell during differentiation (Fig. 2C, cluster 9). Interestingly, IL-2Rβ expression increased gradually during T cell activation. Because IL-2Rβ also mediates IL-15 signaling, IL-15 may be important for survival of effector T cells, a role similar to IL-7 (14-17) (Fig. 2C, cluster 10).
Chemokines and Chemokine Receptors. The expression of a number of T cell chemokines increased dramatically during T cell activation. Although major changes in expression occur during secondary stimulation, smaller increases were observed during primary stimulation for a number of chemokines. Although none of the chemokines were expressed in an exclusively Th1 and Th2 pattern, they all showed some degree of preference. Whereas Th1 cells expressed higher levels of mip1α, mip1β, mip1γ, lymphotoxin, and Rantes, Th2 cells expressed higher levels of mip2 and TCA3 (Fig. 2D, cluster 11). Among T cell chemokine receptors, CCR5 showed clear Th1 restriction, and CCR4 and CCR8 expression were restricted to Th2 cells. CCR7 is a secondary lymphoid organ homing receptor and was shown to be highly expressed on naïve T cells (18). CXCR4 is also a naïve T cell receptor that is down-regulated on T cell activation (19). Consistent with these observations, we saw the highest expression of CCR7 and CXCR4 in naïve T cells. CXCR3 showed an interesting, slow up-regulation without Th1 or Th2 preference (Fig. 2D, cluster 12). The change of expression of chemokine receptors suggests drastically different homing patterns after T cell activation and differentiation.
Apoptosis. Apoptosis has been defined as programmed cell death, a term implying active participation of a cell in its own demise (20). Apoptosis signaling occurs via two different pathways, namely the death receptor pathway and the mitochondrial pathway (reviewed in ref. 21).
First, we examined apoptosis-related genes that showed different expression between Th1 and Th2 cells. Although Th1 cells have been reported to be more susceptible to activation-induced cell death (AICD), resting effector Th1 cells (96 h) preferentially expressed both prodeath genes and genes that protect cells from cell death (Fig. 2E, cluster 13), including genes regulating both apoptotic pathways described above. For example, IAP3 and two members of the BCL-2 like genes (BCL2a1b and BCL2a1d), a set of genes clustered in the mouse genome (22), are up-regulated in Th1 cells versus Th2 cells. These genes likely protect cells from cell death. The mRNA encoding proapoptosis protein PD-1, FasL and TNF receptor alpha are highly expressed in Th1 cells. The expression profile seems to suggest that Th1 cells contain both suicide machinery that makes them more susceptible to apoptosis and mechanisms to counter this machinery to achieve an unstable balance. In this way, Th1 cells may be highly sensitive to external triggers for apoptosis such as TCR challenges or withdrawal of survival factors. After TCR challenge, some genes involved in apoptosis also showed a Th1/2 dichotomy (Fig. 2E, cluster 14). Several genes that favor apoptosis were expressed selectively in Th1 cells, such as Pmp22 (23) and caspase 11 (24). However, antiapoptotic genes such as Bcl2-associated athanogene 3 and BFL-1 were also higher in Th1 cells. These data suggest that both pro- and antiapoptotic machinery is coordinately up-regulated in Th1 cells, and this puts Th1 cells in a checked/controlled apoptotic state. Surprisingly, caspase 6, which is believed to mediate apoptosis, is highly expressed in Th2 cells versus Th1 cells. This finding is consistent with recent data (25) suggesting that IL-4 regulates caspase 6 expression. Our preliminary data suggest that caspase 6 is dispensable for apoptosis (B.L., T. Zheng, and R.A.F., unpublished data).
TCR stimulation triggers a type of cell death called AICD (26, 27). Effector T cells, in particular Th1 cells, are more sensitive to AICD. To understand the molecular basis of this difference, we examined the death genes that are differentially regulated between activated T cells and naïve cells. Indeed, FasL was highly expressed in Th1 cells and inducible by anti-CD3 stimulation in both Th1 and Th2 cells (Fig. 2E, cluster 14). Other prodeath genes were also up-regulated, such as caspase 3, caspase 11, GADD45γ, GADD45β, pmp22, BCL2-like 11, PD-1, and BH3 interacting domain death agonist. Bcl-2 was down-regulated, suggesting a coordinated response. On the contrary, some antiapoptotic genes were induced, such as BFL-1 and baculoviral IAP repeat-containing 5, suggesting a counterbalance mechanism even in this proapoptotic situation (Fig. 2E, clusters 15-17).
In addition to studying genes that showed dramatic changes, we also examined death-related genes that did not change their expressions in these experiments (Table 4, which is published as supporting information on the PNAS web site). The fact that the mRNA for proapoptotic members of bcl-2 family such as bax and bak did not change suggested that the transcriptional regulation of this apoptotic pathway is primarily by the antiapoptotic members of this family in T cells.
Transcription Regulation Network. To understand the mechanism of gene regulation during early Th1/2 cell differentiation, we examined expression profiles of transcription factors. This goal was achieved through two types of approaches. First, we focused on expression characteristics. We started with transcription factors that showed differences between the first 2 days of culture and last 2 days of culture; these could provide insight into transcriptional regulatory components responsible for T cell activation. We then queried for transcription factors that showed Th1/2 dichotomy. Second, we dissected most of the transcription factors according to their molecular categories and pathways.
Transcription factors differentially expressed between the first and second 2 days of culture can be clustered into two major groups. The first group of genes was expressed at high levels in early time points and was down-regulated at later time points (Fig. 3A, cluster 18). It is likely that many of these genes are related to chromosomal remodeling on early T cell activation and replication. The second group of genes showed early down-regulation on T cell activation, and their expression goes up in later differentiation times. Many genes of these group are likely responsible for establishing a resting state. Thus, these observations are consistent with the notion that T cell early differentiation commences with an activation stage during which replication-related genes are up-regulated and resting-stage transcription factors that prohibit cell cycle are down-regulated (Fig. 3A, cluster 19).
We then looked for transcription factors that displayed a Th1/2 dichotomy during these cultures. There were several transcription factors that showed differences between Th1 and Th2 as early as 24 h. These are GATA-3, NF-IL-3, GFI1, ATF-3, Icsbp, and Txk (Fig. 3B, cluster 20). Interestingly all these factors except for GATA-3 were induced in both Th1/2 cultures, although they were preferentially expressed in Th2 conditions. At least two genes, NF-IL-3 and GFI1, of this group have been demonstrated to be inducible by IL-4 (28, 29). GFI1 has been proposed to be pro-proliferative in Th2 cells (29). Therefore, it is responsible for a key property of Th2 cells. The role of NF-IL-3 is less clear in Th2 cells. Based on previous studies of this molecule, it is likely to be responsible for protecting cells from apoptosis (reviewed in ref. 30), another feature of Th2 cells. Strikingly, GATA-3 was repressed during Th1 differentiation at 24 h but up-regulated in Th2 cells, demonstrating a clear lineage affinity. Icsbp, also called IRF-8, is highly induced in Th1 cells and slightly induced in Th2 cells. Therefore, Icsbp, a factor inducible by IFN-γ, may contribute to early Th1 differentiation through mediating IFN-γ and IL-12 signals (31). The other Th1 transcription factor predominantly expressed at this time point is Txk, which has been shown to directly bind to the IFN-γ promoter and activate its transcription (32). Txk is highly expressed in resting naïve CD4+ T cells and down-regulated after TCR engagement. However, in Th2 cells, its level of expression was much more significantly reduced than in Th1 cells and never recovered to the naïve T cell level at any Th2 time point. In contrast, under Th1 conditions, the expression of this gene was reduced in early T cell activation to low but detectable levels and almost recovered to the level in naïve cells in later time points. Both of these factors are directly related to IFN-γ function. However, the expression pattern of these genes are different from a reported Th1 gene T-bet (33), which is absent in naïve cells. Further studies are needed to reveal whether they act earlier than T-bet in Th1 cells.
To study transcription regulation of Th1 and Th2 effector cells, we studied transcription factors showing a Th1/2 dichotomy at 96 h. The selected transcription factors can be further subdivided into five clusters based on their expression pattern during T cell differentiation. For Th2-specific genes, three types of expression profiles existed for these factors, while one cluster contains GATA-3, Chop-10, and EPAS-1. These genes are highly expressed in Th2 cells but reduced or not significantly changed in Th1 cells when compared with naïve CD4+ T cells. The second cluster of Th2 transcription factors consist of Rbpsuh, Hic1, and Gfi1. Although it is clear that these factors are highly expressed in differentiating Th2 cells, these factors are also inducible in Th1 differentiation conditions. The third type contains only one gene Runx1. The level of expression of this gene was reduced on TCR stimulation and is preferentially recovered in differentiating Th2 cells. Based on previous studies that identify runx1 as a tumor suppressor (34) and affecting T cell differentiation in thymus (35), it likely plays a role in limiting Th2 cells in later stage. The Th1-selective transcription factors can also be divided into two groups based on their transcription profiles. The first group contains Bhlhb2 and Nr4a1, which were induced both in Th1 and Th2 cells but were significantly higher in Th1 cells. Both genes have been associated with apoptosis in T cells. Therefore, they likely provide mechanisms to explain the proapoptotic properties of Th1 cells. The second group contains KLF7, Mad, Ifi204, and Gtf2i. All of these genes are associated with resting cells. They are down-regulated in both Th1 and Th2 conditions on TCR stimulation but return to higher levels quickly in Th1 cells but not cells cultured under Th2 conditions. These observations, therefore, may provide another mechanism, likely through cell cycle arrest, to contain Th1 cells (Fig. 3B, cluster 21).
Transcription factors that showed differential expression pattern between Th1/2 cells after restimulation were also examined (Fig. 3B, cluster 22). The expression dichotomy of genes such as GATA-3, NF-IL-3, Ifi204, and Nr4a1 was not affected by TCR restimulation. Other genes only show clear Th1/2 dichotomy after restimulation. These genes can also be further grouped by their expression pattern after clustering analysis. One group contained TCR-induced early genes such as Utf1, Ifi204, Spi-B, Ar, Runx2, and Nr4a2. These genes were rapidly induced by TCR stimulation in both Th1 and Th2 cells, but the levels are higher in Th1 cells than in Th2 cells.
To fully understand the role of different categories of transcription factors in T cell differentiation and activation, we further studied the expression pattern of several major families of transcription factors.
In our array analysis, three members of the NFAT genes (Fig. 3C, cluster 23) are present in two different expression profiles during T cell activation. This group of Rel domain-containing proteins seem to fit a profile of immediate early genes. Another group of immediate-early genes in effector T cells are the NF-κB pathway genes (Fig. 3C, cluster 24). Some of these genes, however, were up-regulated during primary stimulation and maintained during the course of Th1 and Th2 differentiation (NF-κB p105). NF-κB is known to be involved in both T cell activation and effector cytokine production. Components of NF-κB showed coordinated up-regulation after TCR rechallenge in both Th1 and Th2 effector cells. Therefore, simultaneous up-regulation of all components, both positive and negative factors, will make this module respond to signals more vigorously but can still be shut off promptly without the consequence of activation out of control.
Myc is an important transcription factor in T cell activation (36). Therefore, its function is subjected to transcription regulation by an array of transcription factors. Myc is a transcription activator. Its activity is negatively regulated by max, mxi, mad1, mad3, and mad4 (reviewed in ref. 37). Our analysis revealed that the level of myc, mxi, mad1, and mad4 were regulated in a coordinated fashion. During the early phase of the activation (24 and 48 h), myc levels were elevated, correlating with its role in facilitate proliferation. At the same time points, the level of mxi, mad1, and mad4 were all very significantly reduced compared to naïve CD4+ T cells (Fig. 3C, cluster 25). These data suggested a coordinated expression profile that allowed myc to function. In the later time points (72 and 96 h), myc levels were reduced and at the same time points, levels of mxi, mad1, and mad4 rose (Fig. 3C, cluster 25). When cells were restimulated, once again, this group of genes showed a synchronized expression profile that enabled myc activities (Fig. 3C, cluster 25). The expression profile during differentiation and activation clearly showed a coordinated expression pattern to either facilitate, after stimulation, or inhibit, before stimulation, myc function.
Helix-loop-helix (HLH) transcription factor mRNAs were also analyzed and an interesting expression pattern was observed. One subfamily of HLH proteins containing HIF-1 α and EPAS-1, as well as their downstream genes, also showed interesting expression patterns. These include HIF-1α, EPAS-1, and Stra13 (Fig. 3C, cluster 27). HIF-1 α is induced (38) on T cell activation. Consistent with this pattern, its downstream target gene Stra13 (39, 40) is also induced on T cell activation. Although no differences were observed in the expression of HIF-1α between Th1/2 culture conditions, the downstream gene Stra13 was higher in Th1 than Th2. This finding correlated well with the expression pattern of EPAS-1, which is present in Th2 but very low in Th1 cells. EPAS-1 and HIF-1α interact with the common heterodimerization partner ARNT-1 (41). Elimination of Stra13 by gene knockout favored a Th2-like autoimmune disorder (42). It remains to be elucidated whether EPAS regulates Stra13 expression and whether it has other functions in Th2 cells. A genetic pathway that targets some HLH proteins, mainly the Hes family of genes is the Notch pathway (Fig. 3C, cluster 28). Notch 1 is present in naïve CD4+ T cells and maintains a constant level during T cell activation and differentiation. Interestingly, deltex1 is down-regulated during T cell activation or TCR rechallenge. Because deltex1 has been shown to potently inhibit notch signaling (43), down-regulation of this gene might allow activation of Notch1 in activated T cells, suggesting that Notch1 may play a role in activating T cells. Recombining binding protein suppressor of hairless, also called CBF-1, is the only known Notch direct downstream gene. Interestingly, this gene was highly expressed in Th2 cells and induced in Th1 cells only on TCR rechallenge. These observations suggest that Notch may play a role in regulating Th2 cell differentiation as well as a role in T cell activation in general.
Our analysis also revealed many “resting stage” transcription factors. For example, HIN200 family (Fig. 3C, cluster 29) genes were down-regulated on T cell activation and recovered to naïve cells level at 96 h in Th1 cells but remained low in Th2 cells. This finding is consistent with their role in controlling cell cycle, given that Th1 cells are more restricted in proliferation. The RB family genes, including RB1, RBL1, and RBL2, were down-regulated on TCR restimulation in effector Th1 and Th2 cells, consistent with their roles in cell cycle arrest (Fig. 3C, cluster 30).
Another example is the HMG family of genes in which whole levels were down-regulated during the first 48 h of T cell activation and recovered in the last 48 h. TCF-1 and LEF-1 are known transcription factors mediating WNT signals. It is likely that the WNT pathway is important in regulating the physiology of naïve cells, and in particular in maintaining the expression of TCRα (44) (Fig. 3C, cluster 31). Their down-regulation after TCR stimulation may represent a mechanism that restricts excessive TCR signaling.
Interestingly, a group of zinc finger-containing genes showed an expression pattern similar to the Kruppel family transcription factors. This group of genes was highly expressed in resting naïve cells but was down-regulated on T cell activation. Their level of expression was restored as cells returned to a resting state during longer culture of T cells or cessation of stimulation. This type of expression profile has been demonstrated before for LKLF in CD8+ T cells (45), suggesting a role for these factors in regulating resting stage T cells (Fig. 3C, cluster 32).
SMAD family proteins are phosphorylated in response to transforming growth factor (TGF)-β and are implicated in control of cell growth. SMADs can be divided into three distinct classes (reviewed in ref. 46), namely receptor-regulated SMADs (Smads1, -2, -3, -5, and -8), Co-Smads (Smad4), and the inhibitory SMADs (Smad6 and -7). In all of the experimental points examined, Smad1 expression remained at the same level (Table 4). Interestingly, although Smad4 and Smad7 antagonize each other, the levels of their mRNA were simultaneously reduced in activated cells (Fig. 3C, cluster 33). These data suggest that after T cell activation, the responsiveness to TGF-β signaling may be reduced. The coordinated reduction of Smad7 suggests that it is a member of a stringent negative feedback loop that regulates TGF-β1 signaling. The extrapolation of these data may actually suggest that TGF-β1 constantly acts on naïve CD4+ T cells but not TCR-activated T cells and regulates their naïve T cell homeostasis.
A final group of transcription factors are known to or have the potential to play a role in Th1 and Th2 differentiation, yet their mRNA level did not change in our study (Table 4). These include Jun-B (47, 48), Bcl-6 (49-53), FOG-1 (54), Stat-1, IRF-1, and IRF-7 (13, 55, 56). These results are consistent with the notion that naïve CD4+ T cells are intrinsically capable of both Th1 and Th2 differentiation. External instructive signals manipulate the balance of the functions of these factors to achieve polarized differentiation.
Discussion
This study covers gene expression of T cells from the naïve stage to 4 days of differentiation and restimulation, and provides a comprehensive picture of the kinetics of gene expression. Two prior analyses of Th1/2 gene profiling have been reported. Our study, however, provides a kinetic and global view of early Th1/2 differentiation rather than a “snapshot” single time point-like analysis (57, 58). The clustering analysis of expression profiles has revealed that the polarized development of the Th1/2 phenotypes is subsequent to and likely dependent on an activation phase, consistent with several recent studies on this subject using conventional approaches (5, 59). We found that in the activation phase many replication-related genes were turned on with coordinate shutting down of resting-state genes. These events likely reorganize chromatin structure and derepress silent loci and provide access to transcription factors and their associated machineries such as methylases, acetylases, deacetylases, etc. During this process, we have observed coordinated expression of many genes with similar functions. A striking case is that of genes that are highly expressed in naïve CD4+ T cells and likely play an important role in resting cells and establishment of terminal differentiation. These genes include the HIN200 family, RB family, HMG family, and Kruppel family of transcription factors. The expression patterns of all these resting genes are very similar, thereby forming a large cluster in our clustering analysis. These genes are down-regulated early after activation, but their expression is later recovered. This finding correlates with changes in histone acetylation patterns observed in IFN-γ and IL-4 loci, i.e., IL-4 in Th1 and IFN-γ locus in Th2 cultures (6, 7). We therefore postulate a possible role of these factors or some of these factors in maintaining chromatin silencing, in naïve CD4+ T cells, of genes expressed in effector cells. The lineage-specific gene expression is achieved by the break of silencing, in resting cells, by specific transcription activators such as GATA-3 and T-bet in terminal differentiated memory T cells.
The mechanisms that underlie Th1 and Th2 differentiation, especially the origination of Th1 and Th2 cells, have been under intense investigation over the past 10 years or so, and both stochastic and instructive models have been proposed (59, 60). However, questions still remain about the mechanism underlying early events driving Th1/2 differentiation. The transcription factor GATA-3 provides a satisfactory instructive mechanism for Th2 differentiation (13, 60-65). Furthermore, a recent study identified T-bet as a Th1-specific transcription factor directly working on the IFN-γ promoter (33, 66). However, GATA-3 reaches polarized expression as early as 24 h differentiation, whereas it takes T-bet at least 48 h to reach peak expression (unpublished data). Therefore, there may be factors that are expressed earlier than T-bet that regulate or instruct Th1 differentiation. Here we proposed, based on expression profiling that factors such as Icsbp, Txk, and IRF-1, may play a role in early Th1 differentiation. Consistent with this idea, it has been shown recently that T-bet induction depends on Stat-1, a transcription factor used by both IFN-γ and IFN-α/β (67). It is possible, that IFN-γ, IFN-α/β, or both are important in maintaining the levels of expression of transcription factors, such as IRF-1 and ICSBP, in naïve CD4+ T cells. Therefore, an instructive model is also likely for Th1 cells.
Supplementary Material
Acknowledgments
We thank Drs. Eileen Elliot, Patrick Fields, and Lin Liu for helpful suggestions and critical reading of the manuscript; Anthony Ferrandino for technical assistance; G. Chenell for secretarial assistance; and F. Manzo for help with manuscript preparation. This work was supported by National Institutes of Health Grant 1 P01 AI36529 and the Howard Hughes Medical Institute. B.L. is supported by National Institutes of Health Grant 1 K01 AR048854. R.A.F. is an Investigator of the Howard Hughes Medical Institute.
Abbreviation: Th, T helper.
References
- 1.Mosmann, T. R., Cherwinski, H., Bond, M. W., Giedlin, M. A. & Coffman, R. L. (1986) J. Immunol. 136, 2348-2357. [PubMed] [Google Scholar]
- 2.Bottomly, K. (1989) Semin. Immunol. 1, 21-31. [PubMed] [Google Scholar]
- 3.Okamura, H., Tsutsi, H., Komatsu, T., Yutsudo, M., Hakura, A., Tanimoto, T., Torigoe, K., Okura, T., Nukada, Y., Hattori, K., et al. (1995) Nature 378, 88-91. [DOI] [PubMed] [Google Scholar]
- 4.Robinson, D., Shibuya, K., Mui, A., Zonin, F., Murphy, E., Sana, T., Hartley, S. B., Menon, S., Kastelein, R., Bazan, F. & O'Garra, A. (1997) Immunity 7, 571-581. [DOI] [PubMed] [Google Scholar]
- 5.Bird, J. J., Brown, D. R., Mullen, A. C., Moskowitz, N. H., Mahowald, M. A., Sider, J. R., Gajewski, T. F., Wang, C. R. & Reiner, S. L. (1998) Immunity 9, 229-237. [DOI] [PubMed] [Google Scholar]
- 6.Avni, O., Lee, D., Macian, F., Szabo, S. J., Glimcher, L. H. & Rao, A. (2002) Nat. Immunol. 3, 643-651. [DOI] [PubMed] [Google Scholar]
- 7.Fields, P. E., Kim, S. T. & Flavell, R. A. (2002) J. Immunol. 169, 647-650. [DOI] [PubMed] [Google Scholar]
- 8.Lu, B., Yu, H., Chow, C., Li, B., Zheng, W., Davis, R. J. & Flavell, R. A. (2001) Immunity 14, 583-590. [DOI] [PubMed] [Google Scholar]
- 9.Iezzi, G., Karjalainen, K. & Lanzavecchia, A. (1998) Immunity 8, 89-95. [DOI] [PubMed] [Google Scholar]
- 10.Morgan, D. O. (1997) Annu Rev. Cell Dev. Biol. 13, 261-291. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, D. G. & Walker, C. L. (1999) Annu. Rev. Pharmacol. Toxicol. 39, 295-312. [DOI] [PubMed] [Google Scholar]
- 12.Tan, J. T., Dudl, E., LeRoy, E., Murray, R., Sprent, J., Weinberg, K. I. & Surh, C. D. (2001) Proc. Natl. Acad. Sci. USA 98, 8732-8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy, K. M. & Reiner, S. L. (2002) Nat. Rev. Immunol. 2, 933-944. [DOI] [PubMed] [Google Scholar]
- 14.Prlic, M., Lefrancois, L. & Jameson, S. C. (2002) J. Exp. Med. 195, F49-F52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieper, W. C., Tan, J. T., Bondi-Boyd, B., Gapin, L., Sprent, J., Ceredig, R. & Surh, C. D. (2002) J. Exp. Med. 195, 1533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan, J. T., Ernst, B., Kieper, W. C., LeRoy, E., Sprent, J. & Surh, C. D. (2002) J. Exp. Med. 195, 1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldrath, A. W., Sivakumar, P. V., Glaccum, M., Kennedy, M. K., Bevan, M. J., Benoist, C., Mathis, D. & Butz, E. A. (2002) J. Exp. Med. 195, 1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forster, R., Schubel, A., Breitfeld, D., Kremmer, E., Renner-Muller, I., Wolf, E. & Lipp, M. (1999) Cell 99, 23-33. [DOI] [PubMed] [Google Scholar]
- 19.Bermejo, M., Martin-Serrano, J., Oberlin, E., Pedraza, M. A., Serrano, A., Santiago, B., Caruz, A., Loetscher, P., Baggiolini, M., Arenzana-Seisdedos, F. & Alcami, J. (1998) Eur. J. Immunol. 28, 3192-3204. [DOI] [PubMed] [Google Scholar]
- 20.Lakhani, S., Lu, B. & Flavell, R. A. (2002) in Apoptosis and Autoimmunity, eds. Kalden, R. J. & Herrmann, M. (Wiley, Weinheim, Germany), pp. 13-35.
- 21.Green, D. R. & Reed, J. C. (1998) Science 281, 1309-1312. [DOI] [PubMed] [Google Scholar]
- 22.Hatakeyama, S., Hamasaki, A., Negishi, I., Loh, D. Y., Sendo, F. & Nakayama, K. (1998) Int. Immunol. 10, 631-637. [DOI] [PubMed] [Google Scholar]
- 23.Fabbretti, E., Edomi, P., Brancolini, C. & Schneider, C. (1995) Genes Dev. 9, 1846-1856. [DOI] [PubMed] [Google Scholar]
- 24.Hisahara, S., Yuan, J., Momoi, T., Okano, H. & Miura, M. (2001) J. Exp. Med. 193, 111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schroder, A. J., Pavlidis, P., Arimura, A., Capece, D. & Rothman, P. B. (2002) J. Immunol. 168, 996-1000. [DOI] [PubMed] [Google Scholar]
- 26.Ashwell, J. D., Cunningham, R. E., Noguchi, P. D. & Hernandez, D. (1987) J. Exp. Med. 165, 173-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, Y. F., Sahai, B. M. & Green, D. R. (1989) Nature 339, 625-626. [DOI] [PubMed] [Google Scholar]
- 28.Chu, C. C. & Paul, W. E. (1998) Mol. Immunol. 35, 487-502. [DOI] [PubMed] [Google Scholar]
- 29.Zhu, J., Guo, L., Min, B., Watson, C. J., Hu-Li, J., Young, H. A., Tsichlis, P. N. & Paul, W. E. (2002) Immunity 16, 733-744. [DOI] [PubMed] [Google Scholar]
- 30.Cowell, I. G. (2002) BioEssays 24, 1023-1029. [DOI] [PubMed] [Google Scholar]
- 31.Contursi, C., Wang, I. M., Gabriele, L., Gadina, M., O'Shea, J., Morse, H. C., III, & Ozato, K. (2000) Proc. Natl. Acad. Sci. USA 97, 91-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeba, Y., Nagafuchi, H., Takeno, M., Kashiwakura, J. & Suzuki, N. (2002) J. Immunol. 168, 2365-2370. [DOI] [PubMed] [Google Scholar]
- 33.Szabo, S. J., Kim, S. T., Costa, G. L., Zhang, X., Fathman, C. G. & Glimcher, L. H. (2000) Cell 100, 655-669. [DOI] [PubMed] [Google Scholar]
- 34.Silva, F. P., Morolli, B., Storlazzi, C. T., Anelli, L., Wessels, H., Bezrookove, V., Kluin-Nelemans, H. C. & Giphart-Gassler, M. (2003) Oncogene 22, 538-547. [DOI] [PubMed] [Google Scholar]
- 35.Taniuchi, I., Osato, M., Egawa, T., Sunshine, M. J., Bae, S. C., Komori, T., Ito, Y. & Littman, D. R. (2002) Cell 111, 621-633. [DOI] [PubMed] [Google Scholar]
- 36.Trumpp, A., Refaeli, Y., Oskarsson, T., Gasser, S., Murphy, M., Martin, G. R. & Bishop, J. M. (2001) Nature 414, 768-773. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, Z. Q. & Hurlin, P. J. (2001) Trends Cell Biol. 11, S10-S14. [DOI] [PubMed] [Google Scholar]
- 38.Lukashev, D., Caldwell, C., Ohta, A., Chen, P. & Sitkovsky, M. (2001) J. Biol. Chem. 276, 48754-48763. [DOI] [PubMed] [Google Scholar]
- 39.Miyazaki, K., Kawamoto, T., Tanimoto, K., Nishiyama, M., Honda, H. & Kato, Y. (2002) J. Biol. Chem. 277, 47014-47021. [DOI] [PubMed] [Google Scholar]
- 40.Ivanova, A. V., Ivanov, S. V., Danilkovitch-Miagkova, A. & Lerman, M. I. (2001) J. Biol. Chem. 276, 15306-15315. [DOI] [PubMed] [Google Scholar]
- 41.Peng, J., Zhang, L., Drysdale, L. & Fong, G. H. (2000) Proc. Natl. Acad. Sci. USA 97, 8386-8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, H., Lu, B., Li, R. Q., Flavell, R. A. & Taneja, R. (2001) Nat. Immunol. 2, 1040-1047. [DOI] [PubMed] [Google Scholar]
- 43.Izon, D. J., Aster, J. C., He, Y., Weng, A., Karnell, F. G., Patriub, V., Xu, L., Bakkour, S., Rodriguez, C., Allman, D. & Pear, W. S. (2002) Immunity 16, 231-243. [DOI] [PubMed] [Google Scholar]
- 44.Okamura, R. M., Sigvardsson, M., Galceran, J., Verbeek, S., Clevers, H. & Grosschedl, R. (1998) Immunity 8, 11-20. [DOI] [PubMed] [Google Scholar]
- 45.Schober, S. L., Kuo, C. T., Schluns, K. S., Lefrancois, L., Leiden, J. M. & Jameson, S. C. (1999) J. Immunol. 163, 3662-3667. [PubMed] [Google Scholar]
- 46.Attisano, L. & Wrana, J. L. (2002) Science 296, 1646-1647. [DOI] [PubMed] [Google Scholar]
- 47.Li, B., Tournier, C., Davis, R. J. & Flavell, R. A. (1999) EMBO J. 18, 420-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hartenstein, B., Teurich, S., Hess, J., Schenkel, J., Schorpp-Kistner, M. & Angel, P. (2002) EMBO J. 21, 6321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris, M. B., Chang, C. C., Berton, M. T., Danial, N. N., Zhang, J., Kuehner, D., Ye, B. H., Kvatyuk, M., Pandolfi, P. P., Cattoretti, G., et al. (1999) Mol. Cell. Biol. 19, 7264-7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye, B. H., Cattoretti, G., Shen, Q., Zhang, J., Hawe, N., de Waard, R., Leung, C., Nouri-Shirazi, M., Orazi, A., Chaganti, R. S., et al. (1997) Nat. Genet. 16, 161-170. [DOI] [PubMed] [Google Scholar]
- 51.Dent, A. L., Doherty, T. M., Paul, W. E., Sher, A. & Staudt, L. M. (1999) J. Immunol. 163, 2098-2103. [PubMed] [Google Scholar]
- 52.Dent, A. L., Hu-Li, J., Paul, W. E. & Staudt, L. M. (1998) Proc. Natl. Acad. Sci. USA 95, 13823-13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dent, A. L., Shaffer, A. L., Yu, X., Allman, D. & Staudt, L. M. (1997) Science 276, 589-592. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, M., Ouyang, W., Gong, Q., Katz, S. G., White, J. M., Orkin, S. H. & Murphy, K. M. (2001) J. Exp. Med. 194, 1461-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Y., Apilado, R., Coleman, J., Ben-Sasson, S., Tsang, S., Hu-Li, J., Paul, W. E. & Huang, H. (2001) J. Exp. Med. 194, 165-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu, B., Ebensperger, C., Dembic, Z., Wang, Y., Kvatyuk, M., Lu, T., Coffman, R. L., Pestka, S. & Rothman, P. B. (1998) Proc. Natl. Acad. Sci. USA 95, 8233-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogge, L., Bianchi, E., Biffi, M., Bono, E., Chang, S. Y., Alexander, H., Santini, C., Ferrari, G., Sinigaglia, L., Seiler, M., et al. (2000) Nat. Genet. 25, 96-101. [DOI] [PubMed] [Google Scholar]
- 58.Chtanova, T., Kemp, R. A., Sutherland, A. P., Ronchese, F. & Mackay, C. R. (2001) J. Immunol. 167, 3057-3063. [DOI] [PubMed] [Google Scholar]
- 59.Mullen, A. C., High, F. A., Hutchins, A. S., Lee, H. W., Villarino, A. V., Livingston, D. M., Kung, A. L., Cereb, N., Yao, T. P., Yang, S. Y. & Reiner, S. L. (2001) Science 292, 1907-1910. [DOI] [PubMed] [Google Scholar]
- 60.Farrar, J. D., Ouyang, W., Lohning, M., Assenmacher, M., Radbruch, A., Kanagawa, O. & Murphy, K. M. (2001) J. Exp. Med. 193, 643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee, G. R., Fields, P. E. & Flavell, R. A. (2001) Immunity 14, 447-459. [DOI] [PubMed] [Google Scholar]
- 62.Ouyang, W., Lohning, M., Gao, Z., Assenmacher, M., Ranganath, S., Radbruch, A. & Murphy, K. M. (2000) Immunity 12, 27-37. [DOI] [PubMed] [Google Scholar]
- 63.Ouyang, W., Ranganath, S. H., Weindel, K., Bhattacharya, D., Murphy, T. L., Sha, W. C. & Murphy, K. M. (1998) Immunity 9, 745-755. [DOI] [PubMed] [Google Scholar]
- 64.Ranganath, S., Ouyang, W., Bhattarcharya, D., Sha, W. C., Grupe, A., Peltz, G. & Murphy, K. M. (1998) J. Immunol. 161, 3822-3826. [PubMed] [Google Scholar]
- 65.Zheng, W. & Flavell, R. A. (1997) Cell 89, 587-596. [DOI] [PubMed] [Google Scholar]
- 66.Szabo, S. J., Dighe, A. S., Gubler, U. & Murphy, K. M. (1997) J. Exp. Med. 185, 817-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Afkarian, M., Sedy, J. R., Yang, J., Jacobson, N. G., Cereb, N., Yang, S. Y., Murphy, T. L. & Murphy, K. M. (2002) Nat. Immunol. 3, 549-557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.