Abstract
Prodrugs of dynemicin analogs were synthesized, and their activation by aldolase antibody (Ab) 38C2 was evaluated by DNA-cleaving activity, as well as tumor cell growth inhibition. Further, we provide evidence that the activated enediynes underwent covalent crosscoupling with the aldolase Ab, which appears to be a limiting factor of their tumor cell growth-inhibiting activity and should be of general interest in the field of enediyne chemotherapy. These findings might open new avenues for defined conjugations of small molecule drugs to mAbs in general and aldolase Abs in particular.
The specific elimination of cancer cells with potent chemotherapeutic agents is limited by their inability to selectively target cancerous cells. To overcome the nonselectivity of the chemotherapy, new approaches, including Ab-directed enzyme prodrug therapy, have been developed. (1-3) In the Ab-directed enzyme prodrug therapy approach, an enzyme is directed to the tumor cells by using a targeting Ab, which selectively recognizes a cancer-associated cell-surface antigen. After the enzyme-Ab conjugate has localized at the tumor site and cleared from the periphery, a prodrug of a potent chemotherapeutic agent is administered. The prodrug is then activated by the enzyme-Ab conjugate selectively at the tumor site, thereby reducing the toxicity to normal tissue. Based on well documented achievements in Ab-mediated cancer therapy, the targeting Ab component for this strategy is often readily available. The requirements for the enzyme component and complementary prodrug, however, have remained elusive.
With the discovery of catalytic Abs, (4, 5) several research groups suggested that mAbs could be used to replace the enzyme component of the Ab-directed enzyme prodrug therapy approach. (6-8) Using an mAb as the catalyst could (i) access reactions that are not catalyzed by endogenous enzymes, and (ii) reduce the immunogenicity of the catalyst through Ab humanization. (9) Thus, recent studies (10) using catalytic aldolase mAb, 38C2, to activate prodrugs of several anticancer drugs, including doxorubicin, etoposide, and camptothecin, have opened another front in the Ab-directed enzyme prodrug therapy approach. (11, 12) These prodrugs contained a linker, which involved an aldol and an oxa-Michael motif in a sequence. Both the aldol and the oxa-Michael motifs were stable in the absence of the catalyst 38C2, but they readily underwent retro-aldol and β-elimination reactions in the presence of a catalytic amount of mAb 38C2 releasing the active drugs. This chemistry by using catalyst 38C2 is further desirable for the activation of a prodrug because there are no native enzymes in vivo that catalyze this reaction, thus background activation is diminished. Hence, we are developing prodrugs of additional anticancer compounds, including enediynes, which are extremely toxic. (13, 14) In this article, we describe the synthesis of the prodrugs of the simplified analogs (2) of a highly toxic enediyne, dynemicin A (1; Fig. 1), and the in vitro evaluation of the prodrugs in the presence and absence of mAb 38C2.
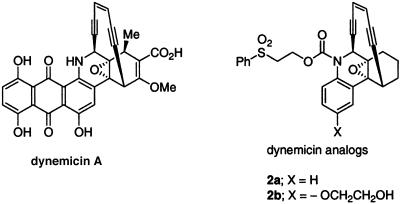
Fig. 1.
Structure of dynemicin A (1) and the corresponding simplified analogs (2a and 2b).
Materials and Methods
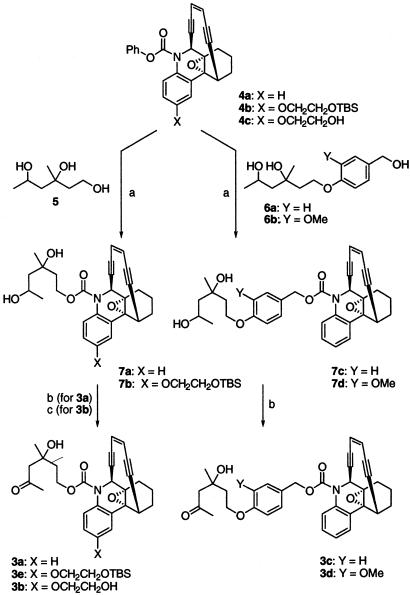
Syntheses of Prodrugs (±)3a-3d. Prodrugs 3a, 3c, and 3d were synthesized in two steps, starting with compound 4a, and triols 5 and 6, by means of the intermediates 7a, 7c, and 7d, respectively (Scheme 2). Prodrug 3b was prepared from compound 4b and triol 5 in three steps by means of intermediates 7b and 3e. Compounds 4a and 4b were prepared as described in the literature. (15, 16) The scheme describing the syntheses of triols 5 and 6 and physical data for 3a-3e and intermediates 7a-7d are incorporated in Schemes 3-6, which are published as supporting information on the PNAS web site.
Scheme 2.
Synthesis of prodrugs 3a-3d. Step a, NaH, dimethylformamide, 0°C, 10 min, then 4a or 4b, 0.5 h. Step b, Dess-Martin reagent, CH2Cl2, 2 h at room temperature. Step c, (i) step b; (ii) TBAF, tetrahydrofuran, 0°C, 1 h.
Prodrugs (±)3a, (±)3c, and (±)3d. Sodium hydride (60% in mineral oil, 5.2 mg, 0.13 mmol) was added to a solution of triol 5 (37.7 mg, 0.25 mmol) in dry dimethylformamide (3 ml) at 0°C. After the mixture was stirred for 10 min, a solution of compound 4a (50 mg, 0.13 mmol) in dry dimethylformamide (0.5 ml) was added. Stirring was continued for 0.5 h, and was then worked up by using aqueous NH4Cl solution and ether. The organic layer was washed with brine, dried over anhydrous Na2SO4, and concentrated under vacuum. The crude residue was purified over silica gel by using hexanes/EtOAc (4:1 to 1:1) as eluants to afford the corresponding diol product 7a (35 mg, 60% yield). In a similar manner, compounds 7c and 7d were prepared by reacting 4a with 6a and 6b, respectively, in a 60-65% yield.
Dess-Martin reagent (57 mg, 0.13 mmol) was added to a solution of diol 7a (30 mg, 0.067 mmol) in dry CH2Cl2 (2 ml), and the mixture was stirred at room temperature for 2 h. It was then worked up by using aqueous NaHCO3 and Na2S2O5 solutions and ether. The organic layer was dried over anhydrous Na2SO4, the solvents were removed under vacuum, and the residue was purified over silica gel by using hexanes/EtOAc (4:1 to 1:1) to afford prodrug 3a (21 mg, 70% yield). In a similar manner, oxidation of diols 7c and 7d afforded prodrugs 3c and 3d, respectively, in a 65-75% yield.
Prodrug (±)3b. Compound 4b (35 mg, 0.062 mmol) was reacted with a resultant mixture of triol 5 (18 mg, 0.12 mmol) and sodium hydride (60% in mineral oil, 2.5 mg, 0.062 mmol) in dry dimethylformamide (2 ml) as above, and the product was purified over silica gel (hexanes/EtOAc, 4:1 to 1:1) to afford diol 7b. The latter product (20 mg, 0.03 mmol) was oxidized by using Dess-Martin reagent (25 mg, 0.06 mmol) as above and purified over silica gel (hexanes/EtOAc, 4:1 to 1:2) to afford 3e (13.6 mg, 68% yield).
Tetrahydrofuran (1M, 36 μl, 0.036 mmol) was added to a solution of 3e (7.6 mg, 0.012 mmol) in tetrahydrofuran (2 ml) at 0°C and the mixture was stirred at the same temperature for 1 h. It was then worked up by using aqueous NH4Cl solution and ether. The combined ether layer was washed with brine and dried over Na2SO4. The solvents were removed under vacuum and the residue was purified over silica gel (hexanes/EtOAc, 4:1 to 0:1) to afford prodrug 3b (5 mg, 83% yield).
Ab Preparation. The generation and purification of mouse mAb 38C2 are described elsewhere. (10) Stock solutions of 15.95 and 19.00 mg/ml 38C2 in PBS (pH 7.4) were used in our experiments.
In Vitro Cell Growth Inhibition Assay. Stock solutions of 10 mM prodrugs 3a-3d, and control compound 4a in DMSO, stored at 4°C, were used. For the cell growth assay, LIM1215 human colon carcinoma cells (kindly provided by L. J. Old, Ludwig Institute for Cancer Research, New York) grown in culture flasks were trypsinized, washed with PBS, and were resuspended in cell culture medium. After reducing them to a single-cell suspension by passing through an 18-gauge needle and counting, the cells were plated at a density of 1 × 104 cells per well in 96-well tissue culture plates and maintained in culture. Prodrugs were added to the cells 24 h after plating, making 5-80 μM final concentration for prodrug 3a, 3c, and 3d, and 0.1-100 μM for 3b. For the Ab experiments, prodrugs and 38C2 IgG were mixed just before adding to the cells. After prodrug addition, the cells were maintained at 37°C in 5% CO2 for 72 h and were periodically analyzed by microscopy for cell growth inhibition. The cells were then washed with 150 μl of PBS (pH 7.5) and incubated with 100 μl of 9% (vol/vol) Triton X-100 (Sigma) for 45 min at 37°C. The supernatant was then transferred to a 96-well V-shaped plate and centrifuged to remove cell debris. In a 96-well plate, 50 μl of the supernatants were combined with 50 μl of freshly reconstituted substrate reaction mixtures containing 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyl-tetrazolium chloride that is converted into a red formazan product in the presence of lactate dehydrogenase released from the cells. The color reaction was incubated for 5-15 min at room temperature and then stopped by adding 50 μl of 1 M acetic acid. For quantification, 50-μl aliquots were transferred to another 96-well plate, and the color was read at 490 nm in an ELISA plate reader. The absorbance data were converted to percent cell density, with the untreated controls being 100% and the background of the color reaction being 0%.
DNA Cleavage Experiment. Stock solutions of 10 mM prodrug 3a and control compound 4a in DMSO, stored at 4°C, were used. Form I DNA (50 μM per base pair) was incubated with 1 mM compound 3a or 4a in the presence or absence of 0.1 mM aldolase mAb 38C2 and was analyzed by gel electrophoresis. Briefly, compound 3a or 4a (10 mM in DMSO, 5 μl) was added to a solution with the total volume of 45 μl containing 5 kb of plasmid (1.4 μg/ml, 3.5 μl), mAb 38C2 (19 mg/ml in 50 mM PBS, 39.5 μl), and buffer. Identical experiments were set up in the absence of 38C2. The mixture was incubated for 48 h and was subjected to gel electrophoresis using 1% agarose to separate the nicked DNA (form II), linear DNA (form III), and supercoiled DNA (form I). DNA bands were visualized by ethidium bromide stain and UV illumination.
Cross-Coupling of mAb 38C2 with Enediynes. An ≈5 molar equivalent of compound 3b (10 mM in DMSO, 20 μl) was added to a solution of mAb 38C2 (15.95 mg/ml, 380 μl) and the mixture was incubated at 37°C for 72 h. During this time, consumption of prodrug 3b and production of intermediate VIb was measured by liquid chromatography MS analysis conducted at 24-h intervals. After 72 h, when >90% of 3b was consumed, the mixture was dialyzed in water and analyzed by matrix-assisted laser desorption ionization-time-of-flight MS.
Results and Discussion
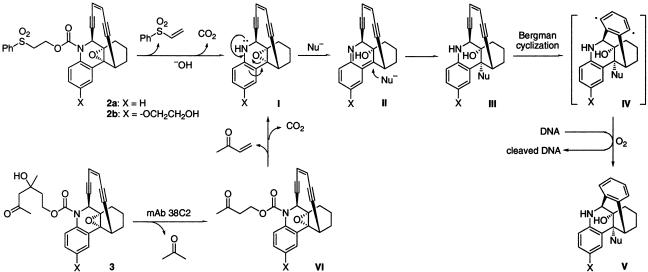
Prodrug Design and Synthesis. Dynemicin A, 1, is thought to cleave double-stranded DNA for which a plausible mechanism has been sketched. Based on this hypothesis, it can be considered that its analogs, 2a and 2b, would also function by analogous pathway, as shown in Scheme 1. The mechanism involves a base-catalyzed elimination of phenyl vinyl sulfone, leading to the carboxamic acid, which undergoes decarboxylation to produce I. The “lone pair” on the nitrogen atom in I triggers the opening of the epoxide function by addition of a nucleophile by means of intermediate II. The product III then undergoes Bergman cyclization (17) to generate phenyl diradical IV. The latter reacts with the double-stranded DNA, leading to its cleavage and production of V. Whether the cleavage of double-stranded DNA is the primary mode of action of dynemicin and its analogs is not known. In addition to the cleavage of double-stranded DNA, these enediynes have been found to damage cellular proteins. In fact, a self-sacrificing protein that mediates resistance to enediynes was described (18).
Scheme 1.
Probable pathways for the activation of prodrugs of dynemicin analogs (3a and 3b) by mAb 38C2.
Enediyne prodrugs 3a-3d were conceived (Schemes 1 and 2), based on studies with the prodrugs of doxorubicin, etoposide, and camptothecin. It was anticipated that mAb 38C2 would activate the enediyne prodrugs by catalyzing the retro-aldol reaction in the first step to produce VI followed by β-elimination reaction in the second step. The remainder of the linker in the resultant products would then become physiologically labile and would undergo self-destruction to produce the intermediate I. Thus, decarboxylation of the 38C2-catalyzed products from compounds 3a and 3b will produce I. Similarly, an elimination of 4-hydroxybenzylalcohol by means of benzoquinomethide formation, followed by decarboxylation reaction of the mAb-catalyzed products from 3c and 3d would also provide I. In any case, product I will then undergo the same transformations as described earlier leading to the diradical IV, followed by DNA cleavage (Scheme 1).
Compounds 3a-3d were synthesized as shown in Scheme 2, starting from 4a and 4b, which were prepared as described in the literature. Reaction of compounds 4a with triols 5 or 6 produced the corresponding secondary alcohol derivatives, 7a, 7c, and 7d. The latter compounds underwent oxidation by using Dess-Martin reagent to afford the prodrugs 4a, 4c, and 4d, respectively. Similarly, 4b was reacted with 5 to afford diol 7b. Secondary alcohol in 7b was oxidized by using Dess-Martin reagent, and the TBS group was deprotected by using tetrahydrofuran to afford prodrug 3b.
Prodrug Activation. Monoclonal aldolase Ab 38C2, generated against a β-diketone hapten by reactive immunization, catalyzes both aldol and retro-aldol reactions by the enamine mechanism. The highly promiscuous binding sites of 38C2 allowed production of a wide variety of aldol compounds. (19) Reactions were amenable to large-scale production of the aldol compounds (20, 21), which allowed multistep synthesis of natural products and their analogs (22-24). In general, 38C2-catalyzed aldol and retro-aldol reactions provided compounds with very high enantiomeric purity. (25) In the prodrug therapy approach, however, it was desirable that 38C2 catalyzed the retro-aldol reaction of both enantiomers of the aldol linker. This result would preclude the necessity of the enantiomerically pure linkers. Indeed, both enantiomeric forms of the linkers, which have been used to produce the 38C2-reactive prodrugs of doxorubicin, etoposide, or camptothecin, are activated by 38C2-catalyzed retro-aldol reaction. The intermediate after the retro-aldol process undergoes a β-elimination reaction, which, although highly facile under physiological condition, is also catalyzed by mAb 38C2 (11).
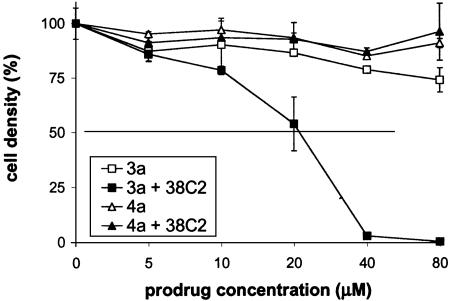
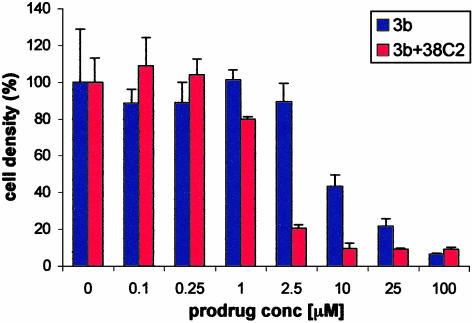
Initially, to check the 38C2-catalyzed activation of prodrugs, we attempted to analyze our experiments by using HPLC. However, we were not able to locate the product formed. In each case, only reduction in the substrate peak was noticed without showing any additional peak in the chromatogram. Hence, activation of prodrugs 3a-3d, in the presence and absence of mAb 38C2, was analyzed by in vitro cell growth inhibition assays using human colon carcinoma cell line LIM1215. From this assay it was determined that independent of the presence or absence of mAb 38C2 both 3c and 3d are toxic to the cells in low micromolar range. This indicated nonselective activation of the prodrugs, which was attributed to the instability of the benzyl carbamate function in the molecules. Prodrugs 3a and 3b, however, showed an enhanced effect in the presence of 38C2. Hence, our followup studies were focused on compounds 3a and 3b. Compound 4a was used as control. The results are summarized in Figs. 2 and 3. As shown in Fig. 2, cell growth in the presence of compound 3a and mAb 38C2 (2.5 μM) was inhibited by 50% at ≈20 μM of the prodrug. A growth inhibition of 90% was noted at 40 μM of 3a in the presence of the mAb. At the same concentration, 3a showed only 20% growth inhibition in the absence of 38C2. The control compound 4a also showed lesser toxicity independent of the presence or absence of 38C2 as expected. The other prodrug, 3b, was ≈10-fold more toxic than 3a (IC50 ≈2 μM vs. 20 μM) in the presence of 38C2 (2.5 μM). However, prodrug 3b was also significantly toxic (IC50 ≈10 μM) in the absence of the mAb (Fig. 3). At higher concentration, prodrug 3a was more selective than 3b. Presumably, mAb 38C2 activated 3a and 3b, which then initiated a series of reaction steps to produce IV. The latter caused DNA damage leading to the observed cell growth inhibition.
Fig. 2.
Growth inhibition of human colon carcinoma cells, LIM1215, by enediyne prodrug 3a in the presence and absence of 2.5 μM 38C2 IgG (72 h).
Fig. 3.
Growth inhibition of human colon carcinoma cells, LIM1215, by enediyne prodrug 3b in the presence and absence of 2.5 μM 38C2 IgG (72 h).
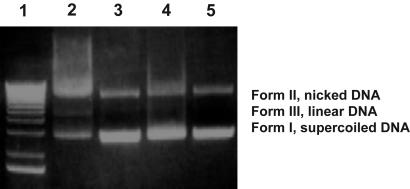
Nicolaou et al. (15) have shown that intermediate I (X = H), which was produced by LiAlH4-reduction of 4a, cleaved supercoiled double-stranded DNA to nicked and linear DNA. To show that the activated drug from 3a or 3b also functioned by this mechanism, an analogous experiment was carried out in the presence and absence of mAb 38C2. Compound 4a, which possesses a phenyl carbamate function and not the 38C2-activable linker, with or without mAb 38C2, was used in the control experiments. As shown in Fig. 4, 1 mM prodrug 3a in the presence of 0.1 mM mAb 38C2 (lane 2) clearly cleaves supercoiled DNA to both nicked and linear forms, whereas no effect was found in the absence of the mAb (lane 3). Control compound 4a did not reveal any DNA-cleaving activity in the presence (lane 4) or absence (lane 5) of mAb 38C2.
Fig. 4.
Form I DNA (50 μM per base pair) incubated for 48 h at 37°C with compound 3a or 4a in the presence or absence of aldolase mAb 38C2 (in 50 mM phosphate buffer, pH 7.4) and analyzed by gel electrophoresis (1% agarose, ethidium bromide stain). Lane 1, reference sample for MW determination; lane 2, 3a (1 mM) and 38C2 (100 μM); lane 3, 3a (1 mM) alone; lane 4, 4a (1 mM) and 38C2 (100 μM); lane 5, 4a (1 mM) alone.
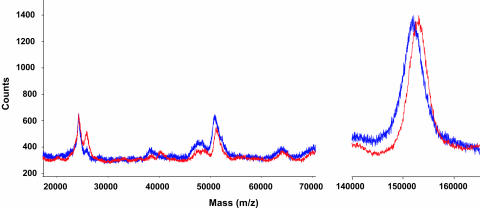
Limiting Factors. Enediynes 2a and 2b, and their analogs, which all produce III (or analogs) on activation, have been found to be toxic in less than low micromolar or submicromolar concentrations, depending on the cell lines used for the assay. However, as evident from Figs. 2 and 3, the enediyne drugs produced by 38C2-catalyzed activation of the corresponding prodrugs possessed low toxicities. Several possible explanations address the observed low toxicity. First, mAb 38C2 binds these substrates in high micromolar concentration (high Km). Thus, an excess of prodrug molecules might remain unactivated at low concentrations of prodrug. Second, it is highly probable that drug III, produced by activation of the enediyne prodrugs, might undergo Bergman cyclization extracellularly to afford diradical IV. The latter might be quenched by extracellular proteins including mAb 38C2 before it causes any effect to the cells. Earlier observations by various research groups that the diradicals from an enediyne are transferred to proteins producing aminoacyl radicals, which then lead to degradation or cross-coupling of the proteins, support this assumption. (26) Moreover, after cellular uptake of III, the produced diradical IV is exposed to another wave of quenching by intracellular proteins that might shield the DNA. Third, the covalent reaction of proteins including mAb 38C2 with reactive epoxide residues of the activated enediyne drugs II. To test this idea, a solution of mAb 38C2 with a 5-molar excess of enediyne prodrug 3b was incubated at 37°C for 72 h. During this time, consumption of prodrug 3b and production of intermediate VIb was measured by liquid chromatography MS analysis conducted at 24-h intervals. After 72 h, >90% of prodrug molecules were consumed, but only a minute amount of Bergman cyclized product was detected. This finding suggested the possible covalent coupling of mAb 38C2, probably through surface lysine residues surrounding the catalytic center, with activated enediyne drug II by an epoxide-opening reaction. That mAb 38C2 reacted with the activated drug II and not the prodrug 3b was evident from a control experiment using 4c, which is not a substrate of mAb 38C2, and remained unconsumed over a period of 96 h. The findings were further corroborated by an analysis of the matrix-assisted laser desorption/ionization-time-of-flight mass spectra of the dialyzed proteins obtained after 72-h incubation of 38C2 with enediyne prodrug 3b or 38C2 alone. A comparison of the mass spectra suggested that, on average, 2.7 molecules of the intermediate downstream of enediyne prodrug 3b reacted with mAb 38C2 (Fig. 5). Based on the 5-molar excess of enediyne prodrug with respect to 38C2 in this experiment, this finding translates into a loss of 60-65% of the activated drug molecules through covalent binding to mAb 38C2.
Fig. 5.
Comparison of mass spectra (matrix-assisted laser desorption/ionization-time-of-flight) of mAb 38C2 (average molecular weight = 151,981) in blue and mAb 38C2 incubated with enediyne 3b (average molecular weight = 152,946), showing, on average, 2.7 molecules of drug conjugated to the mAb in red.
Conclusions
In this study, we have developed enediyne prodrugs, analyzed their activation in the presence of mAb 38C2, and evaluated their in vitro activity by using a DNA-cleaving and a tumor cell growth inhibition assay. Evidently, the prodrugs were activated by a reaction cascade triggered by mAb 38C2. The free drugs were capable of cleaving DNA and inhibiting tumor cell growth in vitro. The lower activity in the tumor cell growth inhibition assay, however, is likely due to covalent cross-coupling with the activating mAb. We conclude that future studies to determine whether this coupling is site- and/or Ab-selective are warranted. A detailed molecular analysis of the observed coupling might open new avenues for defined conjugations of small-molecule drugs to mAbs in general and aldolase Abs like mAb 38C2 in particular. In contrast to previously reported conjugations of β-diketone derivatives to mAb 38C2 (27), conjugations with epoxide derivatives might preserve the reactive lysine and, thus, the catalytic activity of the aldolase Ab.
Supplementary Material
Acknowledgments
We thank The Skaggs Institute for Chemical Biology for their financial support.
References
- 1.Bagshawe, K. D., Sharma, S. K., Burke, P. J., Melton, R. G. & Knox, R. J. (1999) Curr. Opin. Immunol. 11, 579-583. [DOI] [PubMed] [Google Scholar]
- 2.Syrigos, K. N. & Epenetos, A. A. (1999) Anticancer Res. 19, 605-613. [PubMed] [Google Scholar]
- 3.Xu, G. & McLeod, H. L. (2001) Clin. Cancer Res. 7, 3314-3324. [PubMed] [Google Scholar]
- 4.Tramontano, A., Janda, K. D. & Lerner, R. A. (1986) Science 234, 1566-1570. [DOI] [PubMed] [Google Scholar]
- 5.Pollack, S. J., Jacobs, J. W. & Schultz, P. G. (1986) Science 234, 1570-1573. [DOI] [PubMed] [Google Scholar]
- 6.Miyashita, H., Karaki, Y., Kikuchi, M. & Fujii, I. (1993) Proc. Natl. Acad. Sci. USA 90, 5337-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, D. A., Gong, B., Kochersperger, L. M., Yonkovich, S., Gallop, M. A. & Schultz, P. G. (1994) J. Am. Chem. Soc. 116, 2165-2166. [Google Scholar]
- 8.Wentworth, P., Datta, A., Blakey, D., Boyle, T., Partridge, L. J. & Blackburn, G. M. (1996) Proc. Natl. Acad. Sci. USA 93, 799-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rader, C., Turner, J. M., Heine, A., Shabat, D., Sinha, S. C., Wilson, I. A., Lerner, R. A. & Barbas, C. F., III (2003) J. Mol. Biol. 332, 889-899. [DOI] [PubMed] [Google Scholar]
- 10.Wagner, J., Lerner, R. A. & Barbas, C. F., III (1995) Science, 270, 1797-1800. [DOI] [PubMed] [Google Scholar]
- 11.Shabat, D., Rader, C., List, B., Lerner, R. A. & Barbas, C. F., III (1999) Proc. Natl. Acad. Sci. USA 96, 6925-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shabat, D., Lode, H. N., Pertl, U., Reisfeld, R. A., Rader, C., Lerner, R. A. & Barbas, C. F., III (2001) Proc. Natl. Acad. Sci. USA 98, 7528-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolaou, K. C., Dai, W. M., Tsay, S. C., Estevez, V. A. & Wrasidlo, W. (1992) Science 256, 1172-1178. [DOI] [PubMed] [Google Scholar]
- 14.Hay, M. P., Wilson, W. R. & Denny, W. A. (1999) Bioorg. Med. Chem. Lett. 9, 3417-3422. [DOI] [PubMed] [Google Scholar]
- 15.Nicolaou, K. C., Smith, A. L., Wendeborn, S. V. & Hwang, C.-K. (1991) J. Am. Chem. Soc. 113, 3106-3114. [Google Scholar]
- 16.Nicolaou, K. C., Maligres, P., Suzuki, T., Wendeborn, S. V., Dai, W.-M. & Chadha, R. K. (1992) J. Am. Chem. Soc. 114, 8890-8907. [Google Scholar]
- 17.Bergman, R. G. (1973) Acc. Chem. Res. 6, 25-31. [Google Scholar]
- 18.Biggins, J. B., Onwueme, K. C. & Thorson, J. S. (2003) Science 301, 1537-1541. [DOI] [PubMed] [Google Scholar]
- 19.Barbas, C. F., III, Heine, A., Zhong, G., Hoffmann, T., Gramatikova, S., Björnstedt, R., List, B., Anderson, J., Stura, E. A., Wilson, I. A. & Lerner, R. A. (1997) Science 278, 2085-2092. [DOI] [PubMed] [Google Scholar]
- 20.Sinha, S. C., Sun, J., Miller, G., Barbas, C. F., III, & Lerner, R. A. (1999) Org. Lett. 1, 1623-1626. [DOI] [PubMed] [Google Scholar]
- 21.Turner, J. M., Bui, T., Lerner, R. A., Barbas, C. F., III, & List, B. (2000) Chemistry Eur. J. 6, 2772-2774. [DOI] [PubMed] [Google Scholar]
- 22.List, B., Shabat, D., Barbas, C. F., III, & Lerner, R. A. (1998) Chemistry Eur. J. 4, 881-885. [Google Scholar]
- 23.Sinha, S. C., Barbas, C. F., III, & Lerner, R. A. (1998) Proc. Natl. Acad. Sci. USA 95, 14603-14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha, S. C., Sun, J., Wartmann, M. & Lerner, R. A. (2001) ChemBioChem, 2, 656-665. [DOI] [PubMed] [Google Scholar]
- 25.Zhong, G., Shabat, D., List, B., Anderson, J., Sinha, S. C., Lerner, R. A. & Barbas, C. F., III (1998) Angew. Chem. Int. Ed. Engl. 37, 2481-2484. [DOI] [PubMed] [Google Scholar]
- 26.Jones, G. B., Plourde, G. W., II, & Wright, J. M. (2000) Org. Lett. 2, 811-813. [DOI] [PubMed] [Google Scholar]
- 27.Rader, C., Sinha, S. C., Popkov, M., Lerner, R. A. & Barbas, C. F., III (2003) Proc. Natl. Acad. Sci. USA, 100, 5396-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.