Abstract
Several mouse models have already proved valuable for investigating hypertrophic responses to cardiac stress. Here, we characterize one caused by a well defined single copy transgene, RenTgMK, that genetically clamps plasma renin and thence angiotensin II at high levels. All of the transgenic males develop concentric cardiac hypertrophy with fibrosis but without dilatation. Over half die suddenly aged 6-8 months. Telemetry showed disturbances in diurnal rhythms a few days before death and, later, electrocardiographic disturbances comparable to those in humans with congestive heart failure. Expression of seven hypertrophy-related genes in this and two categorically different models (lack of atrial natriuretic peptide receptor A; overexpression of calsequestrin) were compared. Statistical analyses show that ventricular expressions of the genes coding for atrial natriuretic peptide, β myosin heavy chain, medium chain acyl-CoA dehydrogenase, and adrenomedullin correlate equally well with the degree of hypertrophy, although their ranges of expression are, respectively, 50-, 30-, 10-, and 3-fold.
Hypertrophy of cardiac myocytes, a response to increased cardiac stress, can be induced by many different physiological and molecular stimuli (1), as illustrated by a variety of experimental models in mice, including genetic modification (2), pressure overload (3), drug treatment (4), and strain-dependent genetic modifiers (5, 6). Despite the variety of methods used to induce the hypertrophy, the resulting animals share a common feature: re-expression of a fetal-like gene program. This program shift includes changes in secreted and contractile proteins, ion channels, and energy metabolism. Yet recent studies using differential display (7) and microarray technology (4, 8, 9) have shown no single program of gene expression common to all models, making further comparative studies desirable.
Toward this end, we have developed and characterized several genetic models of cardiac hypertrophy mediated through different pathways and of differing severity. Two have been described previously. First, mice that lack the guanylyl cyclase-type atrial natriuretic peptide receptor (NPRA-/-) develop mild hypertension, ≈9 mmHg (1 mmHg = 133 Pa) greater than WT controls (10); and dilated cardiac hypertrophy that is independent of and disproportionate to their elevated blood pressure, demonstrating a direct role of NPRA in the progression of cardiac hypertrophy (11). A considerably more severe cardiac hypertrophy is generated in mice with transgenic overexpression of a cDNA coding for the sarcomeric calcium binding protein, calsequestrin (CSQ) (12). The CSQ transgenic mice develop severe dilated cardiomyopathy and all die within 16 weeks of age (13).
In the present paper we describe and characterize a third model of (intermediate) cardiac hypertrophy without dilatation in which about half of the males die between 6 and 8 months of age. The hypertrophy is caused by a high plasma level of active renin, and thence of angiotensin II (Ang II), resulting from a “genetically clamped” transgene (RenTgMK). This transgene is clamped because it produces physiologically active renin at a constant level that is insensitive to cardiovascular homeostatic signals and is expressed in the liver, an organ that is uninvolved in blood pressure regulation (14). We compare the RenTgMK model with the two previous models by using precision quantitative RT-PCR to determine the expression of a panel of genes, each relevant to cardiac myocyte function. The high sensitivity of this molecular phenotyping procedure allows precise data to be obtained with samples from individual animals rather than pooled samples. Consequently we have been able to determine the degree to which differences in gene expression correlate with differences in hypertrophy between the three models and with differences between individual mice within the same model.
Experimental Procedures
Our procedure for generating renin transgenic mice has been described (14). The transgene, RenTgMK, used in the present work consists of (i) a liver-specific albumin promoter/enhancer (15), (ii) a 20-bp oligonucleotide insert in the 5′ UTR to increase its length (16), (iii) a synthetic mouse renin cDNA, and (iv) a rabbit β-globin 3′ UTR. The synthetic renin gene includes an N-glycosylation site for increased stability and is engineered so that processing from prorenin to renin can be efficiently achieved in hepatic or other cells by the ubiquitous enzyme furin (17). Single-copy chosen-site gene targeting (18) was used to insert the RenTgMK into the apolipoprotein locus between the Apoa1 and Apoc3 genes.
For PCR-based genotyping of the RenTgMK mice we use three primers: primer 1, 5′-TGGGATTCTAACCCTGAGGACC-3′; primer 2, 5′-CACAGATTGTAACTGCAAATCTGTCG-3′; and primer 3, 5′-GTTCTTCTGAGGGGATCGGC-3′.
The mice used in the present study had one copy of the RenTgMK and were 4- to 8-month-old males. For comparative studies between lines, age-matched male mice were used.
The NPRA-/- mice (10) and CSQ transgenic mice (12, 13) have been described.
Blood pressures were measured on unanesthetized mice by a computerized tail cuff system (19). Transthoracic M-mode echocardiography was performed on unanesthetized 6 month old mice by using a HDI 5000CV echocardiograph machine with a 10.5-MHz frequency probe (ATL Ultrasound).
In vivo physiological monitoring of 4-month-old RenTgMK and WT mice was performed by using biosensors (TA10ETA-F20, Data Sciences International, Minneapolis) to measure temperature, activity, and electrocardiographic waveforms. The biosensor is surgically placed in the peritoneal area and the attached s.c. insulated wire electrodes are positioned in the upper and left lower thorax analogous to a lead II configuration. Data were collected at 1,000 samples per s for 10 consecutive seconds at intervals of 5 min and collected by using physiostat 3.22 software (Data Sciences International). The electrocardiographic waveforms were analyzed for rhythm, morphology, and intervals [QRS, R-R, QT, and SD between normal intervals (SDNN)] by using physiostat 3.22 software. Microvolt T wave alternans were detected by using custom designed software based on the method described by Nearing and Verrier (20).
Quantitative RT-PCR was performed with an Applied Biosystems 7700 Sequence Detection System by using total RNA purified from the left ventricle with the primers and probes listed in Table 2, which is published as supporting information on the PNAS web site; some of these have been described (21).
For general histology, 5-μm-thick paraffin sections were stained with hematoxylin and eosin or with Masson's trichrome reagents.
All experiments were conducted with 4- to 8-month-old male mice and were approved by the Institutional Animal Care and Use Committee of the University of North Carolina, Chapel Hill.
Statistical analyses were performed with jmp software (SAS Institute, Cary, NC).
Results
Cardiac Hypertrophy in RenTgMK Mice. We have previously reported a method for generating mice with genetically clamped renin transgene expression in their livers (14). The transgene is inserted as a single copy into the ApoA1/ApoC3 locus that is active in the liver. Our previous publication focused on the blood pressures and renal phenotypes of mice carrying our simplest transgene, designated RenTgKC. The present study focuses on cardiac hypertrophy with male mice that have a modified form of this renin transgene. The modification consists of the insertion of a random 20-bp oligonucleotide sequence into the 5′ UTR of the transgene to increase its translation efficiency, as described by Marilyn Kozak (16). We refer to this modified transgene and mouse line as RenTgMK. The RenTgMK mice have increased production of renin relative to mice with the unmodified RenTgKC. Their natural renin genes are WT.
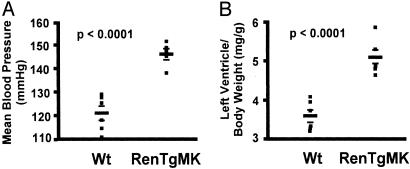
Fig. 1A shows that the RenTgMK mice have significantly elevated blood pressure compared to WT littermates (146 ± 3 mmHg for the RenTgMK vs. 121 ± 4 mmHg for the WT, P < 0.0001). Fig. 1B shows that the mice develop cardiac hypertrophy as judged by an increase in left ventricle/body weight (BW) ratio compared to controls (5.2 ± 0.2 for the RenTgMK vs. 3.5 ± 0.2 for the WT, P < 0.0001).
Fig. 1.
(A) Mean blood pressure of 6-month-old RenTgMK and WT control mice measured by a computerized tail cuff method. (B) Left ventricle/BW ratio of 8-month-old RenTgMK and WT control mice.
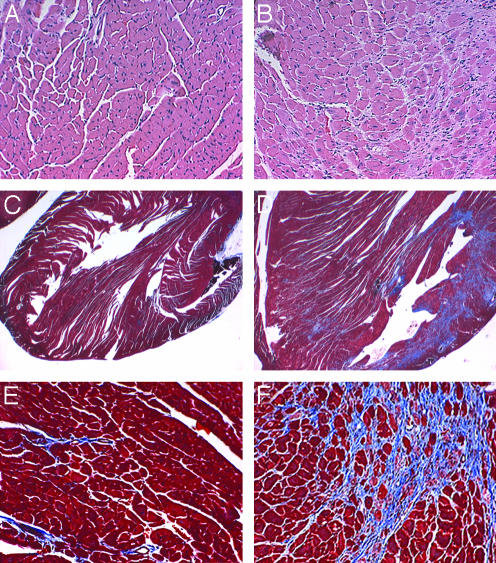
Histological examination of the hearts of the RenTgMK mice confirms the cardiac hypertrophy (Fig. 2 A and B), as judged by an increase in cardiac myocyte cross-sectional area. Masson's trichrome staining revealed generalized fibrosis that was more predominant in the posterior wall of the left ventricle. The WT controls had no pathological fibrosis (Fig. 2 C-F).
Fig. 2.
Hematoxylin/eosin stained sections of the left ventricle of 8-month-old WT (A) and RenTgMK (B) mice. (C-F) Masson's trichrome stained sections of left ventricle of 8-month-old WT (C and E) and RenTgMK (D and F) mice.
To assess the cardiac function of the RenTgMK mice, we performed echocardiography on conscious 6-month-old animals. As shown in Table 1, the RenTgMK mice display a significant decrease in left ventricular end systolic dimension (LVESD) coupled with an increase in cardiac function (as assessed by fractional shortening). Heart rate was also increased in the RenTgMK mice and likely accounts for the modest, yet significant increase in fractional shortening. Taken together, these parameters indicate that at 6 months these RenTgMK mice have concentric hypertrophy, without ventricular dilation or overt heart failure.
Table 1. Echocardiography in WT and RenTgMK mice.
| WT | RenTgMK | P value | |
|---|---|---|---|
| LVEDD, mm | 3.3 ± 0.1 | 3.1 ± 0.1 | 0.192 |
| LVESD, mm | 1.3 ± 0.1 | 0.9 ± 0.1 | 0.022* |
| FS, % | 61.2 ± 2.1 | 69.5 ± 2.7 | 0.036* |
| IVS, mm | 1.0 ± 0.1 | 1.2 ± 0.1 | 0.059 |
| PW, mm | 0.9 ± 0.1 | 1.0 ± 0.1 | 0.109 |
| HR, bpm | 600 ± 26 | 692 ± 26 | 0.034* |
| VcFc, circ per s | 4.33 ± 0.2 | 6.17 ± 0.5 | 0.010* |
LVEDD, left ventricular end diastolic dimension; LVESD, left ventricular end systolic dimension; FS, fractional shortening (LVEDD-LVESD)/LVEDD; IVS, intraventricular septal thickness; PW, posterior wall thickness; HR, heart rate; bpm, beats per minute; circ/sec, circumference per second; VcFc, heart rate-corrected mean velocity of circumferential fiber shortening calculated as FS divided by ejection time multiplied by the square root of the R-R interval. n = 6 for each group.
Denotes a significant difference between RenTgMK and WT mice.
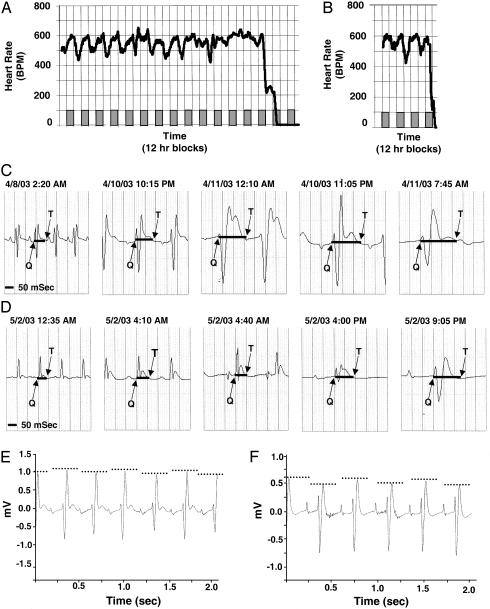
Electrocardiographic Monitoring of Sudden Death in RenTgMK Mice. Approximately 60% of RenTgMK male mice die suddenly between 6 and 8 months of age. To investigate the potential cause of these deaths, we used continuous telemetric recording of electrocardiographic waveforms, activity, and body temperature in four 4-month-old RenTgMK mice and age-matched WT controls. During the course of our studies, one of the RenTgMK mice (mouse 1) died at age 5 months while being monitored, and a second (mouse 2) was monitored until moribund. Fig. 3 A and B illustrates the continuous and diurnal heart rates that were recorded in each of these mice, respectively. The normal diurnal variation in heart rate in mouse 1 subsided approximately 4 days before death (Fig. 3A) and was correlated with a decrease in body temperature and activity (data not shown). Changes in the diurnal patterns of the recorded parameters were not observed in the age-matched WT mice. In both mice, we found a significant lengthening of the QT interval that corresponded to a prolonged and progressive bradycardia (Fig. 3 C and D). We calculated the R-R interval, the P-R interval, and the SD between normal intervals (SDNN) at the times depicted in Fig. 3 C and D and at equivalent times in WT mice. We found a significant lengthening of the R-R interval in the RenTgMK mice as compared to WT controls (144.8 ms ± 11.6 for RenTgMK versus 112.3 ms ± 1.9 for WT, P < 0.02). In both RenTgMK mice, we observed a prolongation of the P-R interval and progressive QRS widening, indicating conduction abnormalities in the atrioventricular node and ventricles (Fig. 3 C and D). The RenTgMK mice also showed a significant decrease in heart rate variability compared to WT controls. The SDNN values were 1.54 ± 0.32 for RenTgMK mice compared to 3.27 ± 0.43 for WT mice (P < 0.004). A few hours before the terminal death event, alternating electrical conduction patterns similar to microvolt T wave alternans in humans were recorded during sinus rhythm (Fig. 3 E and F).
Fig. 3.
Electrocardiographic measurements in RenTgMk mice. (A) Mouse 1; 1-h moving averages of heart rate waveforms recorded for 15 consecutive days before death. Diurnal heart rate changes are detectable until ≈4 days before death. Shaded boxes represent dark cycle, and open boxes represent light cycle. (B) Mouse 2; 1-h moving averages of heart rate wave forms recorded until mouse was moribund. (C and D) Mouse 1 (C) and mouse 2 (D); individual electrocardiographic recordings monitored before death depicting progressive bradycardia. Solid black lines depict lengthening QT interval. (E and F) Mouse 1 (E) and mouse 2 (F); presence of alternating patterns of electrical activation and recovery.
Molecular Phenotype of Cardiac Hypertrophy. To characterize the molecular phenotype of the RenTgMK mice, we used quantitative RT-PCR on left ventricular total RNA to determine the expression of seven genes previously shown or suspected to be involved in the progression of cardiac hypertrophy. These include genes that change expression during normal development [α myosin heavy chain (α-MHC), β-MHC, and medium chain acyl-CoA dehydrogenase (MCAD)], genes that are widely recognized as markers of cardiac hypertrophy [atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP)], and genes that have recently been implicated in cardiac hypertrophy [transforming growth factor β 1 (TGF-β1) and adrenomedullin (AM)]. We used β-actin as an internal standard.
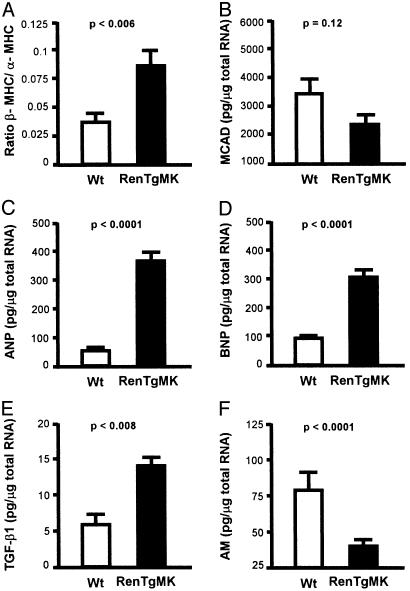
As expected and shown in Fig. 4A, the cardiac myocytes of the RenTgMK mice exhibit a switch toward the fetal-like state as indicated by a significant (P < 0.006) increase in the ratio of expression of the fetal type MHC, β-MHC, relative to that of the adult-type MHC, α-MHC [expression of α-MHC alone decreased but did not reach significance; P = 0.15]. Expression of MCAD, which decreases when cells shift their energy source from fatty acids to carbohydrates, was decreased in the RenTgMK mice relative to WT but did not reach statistical significance (Fig. 3B; P = 0.12). The RenTgMK mice show striking increases in the expression of classical markers of cardiac hypertrophy, ANP (Fig. 3C; P < 0.0001), and BNP (Fig. 3D; P < 0.0001).
Fig. 4.
Quantitative gene expression determined by an Applied Biosystems 7700 real-time RT-PCR. Total RNA from the left ventricle of 8-month-old WT and RenTgMK mice was used with the primer and probes sets depicted in a Table 2. (A) Ratio of β-MHC/α-MHC. (B) MCAD. (C) ANP. (D) BNP. (E) TGFβ-1. (F) AM.
TGF-β1 is a multifunctional growth factor that becomes elevated in pathological fibrosis (22, 23), and recent studies have shown that TGF-β1 is an important mediator of cardiac hypertrophy induced by Ang II (23, 24). Because, in the RenTgMK mice, active renin overexpression results in increased levels of Ang II (14), we expected and found that TGF-β1 expression in the ventricles of the RenTgMK mice was increased, to almost three times WT controls (Fig. 3E; P < 0.008).
AM is a recently identified potent vasodilator that is required for normal cardiovascular development (25). This peptide hormone also has growth regulatory properties in cultured cardiac myocytes and has been implicated in the homeostatic response to hypertension and cardiac hypertrophy in both rodent models and human patients (26). It is strongly expressed in the heart and vasculature and its absence in a gene targeted knockout model leads to embryonic lethality associated with hydrops fetalis and cardiovascular defects (25). We found a significant decrease (Fig. 3F; P < 0.0001) in AM gene expression in the left ventricle of the RenTgMK mice to about one-third compared to WT controls.
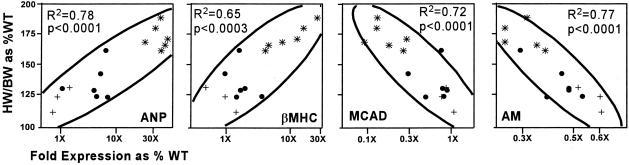
Comparative Molecular Phenotyping. For reasons considered in more detail in the discussion, we next compared expression of the same seven genes in the RenTgMK mice to expression in mice lacking the natriuretic peptide receptor A (NPRA-/-) and in transgenic mice overexpressing calsequestrin (CSQ) in their hearts. The mice lacking NPRA develop hypertrophy generally less than but overlapping that in the RenTgMK mice (heart weight/BW ratio is 127% of WT for NPRA-/- mice compared to 136% of WT for the RenTgMK mice). The CSQ transgenic mice develop hypertrophy generally greater than but again overlapping that of the RenTgMK mice (heart weight/BW ratio is 174% of WT). Because of the sensitivity of quantitative RT-PCR, the molecular phenotypes (21) of the 15 individual mice that we used in this comparison could be determined without pooling samples. The comparison showed that four of the seven genes tested had left ventricular expression changes significantly and proportionality correlated with the degree of hypertrophy regardless of the method of induction (Fig. 5). [The other three genes did not show significant correlations across all of the three models: α-MHC, P = 0.21; BNP, P = 0.82; TGF-β1, P = 0.26.] Not surprisingly, ventricular ANP expression was significantly (P < 0.0001, r2 = 0.78) and positively correlated proportionately over a 100-fold range with the degree of hypertrophy, as was expression of β-MHC (P = 0.0003, r2 = 0.65). Left ventricular expression of MCAD (P = 0.0001, r2 = 0.72) was proportionately correlated over a 10-fold range with the degree of hypertrophy but in the opposite (negative) direction. AM (P < 0.0001, r2 = 0.77) was likewise proportionately and negatively correlated over a 3-fold range with the degree of hypertrophy. We conclude that the proportionality of the response to hypertrophy of the four genes is equivalent (as judged by r2) and independent of the inducer although the magnitude of the responses range from 100-fold to 3-fold.
Fig. 5.
Multivariate correlations of the degree of hypertrophy [assessed by heart weight (HW)/BW ratio as percent of WT] with gene expression (expressed as fold expression over WT) for ANP (A), β-MHC (B), MCAD (C), and AM (D). Circles, RenTgMK; crosses, NPRA-/-; asterisks, CSQ transgene. The horizontal axes are logarithmic scale.
Discussion
We describe here a genetically clamped renin transgene mouse, RenTgMK, in which active renin is secreted from the liver independently of renal or other homeostatic cardiovascular control mechanisms. The RenTgMK mice are consequently the genetic equivalent of chronic physiological infusion of Ang II with a minipump, but with the advantage of being caused by a genetic change that is lifelong and noninvasive. We show that the RenTgMK mice have high blood pressure and concentric cardiac hypertrophy. By 8 months of age, severe cardiac remodeling and fibrosis accompany the cardiac hypertrophy, as evidenced by histological examination.
Several genetically engineered animal models that directly target the mechanical, contractile, or conduction properties of cardiomyocytes exhibit sudden cardiac death and lethal arrhythmias (27-29). In contrast, the cardiac hypertrophy and sudden death of the RenTgMK mice is hormonally induced through elevated circulating levels of Ang II. Telemetric recordings suggest that the sudden death we observe in some of the RenTg1MK mice is presaged by disturbances in the normal daily decreases in heart rate, body temperature, and activity. The electrocardiograms also demonstrated progressive slowing of the heart, increased intramyocardial conduction abnormalities, lengthening of the R-R and QT interval, decreased SD between normal intervals (SDNN), and the appearance of T-wave alternans. These changes indicate a rapidly progressing myocardial dysfunction and are analogous to clinical features shared by humans with congestive heart failure at high risk for death (30, 31).
To further characterize the cardiac hypertrophy of the RenTgMK mice we used quantitative RT-PCR assay to quantitatively assess changes in the expression of a chosen panel of seven genes variously implicated with hypertrophy. These studies were focused on male mice. However, we note that our previous molecular phenotyping studies (21) and current preliminary studies with RenTgMK female mice show that, as in humans, some genes are sexually dimorphic in their responses to cardiac hypertrophy. In male RenTgMK mice, we find five of the seven genes tested (β-MHC, ANP, BNP, TGF-β1, and AM) show significant changes (P < 0.01) in ventricular expression in the RenTgMK mice. The other two (α-MHC and MCAD) show changes but they do not reach significance (P > 0.1). However, the data clearly show that the cardiac hypertrophy in the RenTgMK mice is accompanied by the well known shift to a fetal-like pattern of gene expression.
The RenTgMK mice develop a substantial level of cardiac hypertrophy as a result of an enhanced synthesis of active renin. Consequently, their cardiomyocytes are exposed to two hypertrophic stimuli: an increased mechanical load (≈25 mmHg) and an increased level of plasma Ang II. As an approach to identifying and comparing cellular responses that are present in all hypertrophic cardiomyocytes irrespective of the means of inducing the hypertrophy, we therefore compared the molecular phenotypes of the RenTgMK mice with those of two other categorically different models. Mice lacking NPRA were chosen as one of these models because they have modest hypertrophy due to an increased mechanical load (≈9 mmHg) combined with absence of the moderating effects of the natriuretic peptides. The CSQ transgenic mice were chosen because they have more severe hypertrophy although they have blood pressures lower than WT; their stimulus is an intracellular disturbance in calcium signaling pathways. The inducers of the hypertrophy are therefore categorically different in the three models.
The results of the comparison are clear: the ventricular expressions of ANP (P < 0.0001) and β-MHC (P = 0.0003) are positively correlated with the degree of cardiac hypertrophy in the three models. Ventricular expressions of MCAD (P = 0.0001) and of AM (P < 0.0001) are likewise highly correlated in all three models, although in this case negatively. A caveat is necessary, however, for the AM gene, because our preliminary unpublished work shows that it does not change expression with the hypertrophy that develops in RenTgMK females. Nevertheless, we conclude that these four genes in males (three in females) are likely to be obligate responders to any hypertrophic stimuli. The proportionality of the responses of these four genes to the three inducers are equivalent (as judged by their very similar r2 values following bivariate analysis of hypertrophy versus expression). This equal proportionality suggests that they all receive and respond to some common signals. On the other hand, the magnitude of the induced responses are not equivalent, because they range from 50-fold (for ANP) to 30-fold (for β-MHC) to 10-fold (for MCAD) to 3-fold (for AM), suggesting that the genes are differently sensitive to these signals.
In conclusion, we emphasize that RenTgMK is a simple transgene with a well defined mode of action that is useful for inducing concentric cardiac hypertrophy, accompanied in males by fibrosis and a shift of cardiomyocytes to a fetal-like pattern of gene expression. Moreover, we conclude that the pathological manifestation of the sudden death in the RenTgMK mice, which includes loss of diurnal rhythms, lengthening of the QT interval, loss of heart rate variability, and the appearance of T-wave alternans, is similar to what is seen in the failing human heart.
Supplementary Material
Acknowledgments
We thank Drs. Nobuyo Maeda and Toshio Nishikimi for helpful advice and discussions and Gleb Rozanov for technical help. This work was supported by National Institutes of Health Grants HL1266 and HL49277 (to O.S.), HL10344 (to K.M.I.C.), and HL61558 (to H.R.) and a Biomedical Fellowship from the Kidney Foundation of Canada (to L.R.J.).
Abbreviations: AM, adrenomedullin; Ang, angiotensin; ANP, atrial natriuretic peptide; BNP, brain natriuretic peptide; BP, blood pressure; BW, body weight; MCAD, medium chain acyl-CoA dehydrogenase; MHC, myosin heavy chain; TGF, transforming growth factor.
References
- 1.Frey, N. & Olson, E. N. (2003) Annu. Rev. Physiol. 65, 45-79. [DOI] [PubMed] [Google Scholar]
- 2.Takeishi, Y. & Walsh, R. A. (2001) Acta Physiol. Scand. 173, 103-111. [DOI] [PubMed] [Google Scholar]
- 3.Takaoka, H., Esposito, G., Mao, L., Suga, H. & Rockman, H. A. (2002) Am. J. Physiol. Heart Circ. Physiol. 282, H2190-H2197. [DOI] [PubMed] [Google Scholar]
- 4.Friddle, C. J., Koga, T., Rubin, E. M. & Bristow, J. (2000) Proc. Natl. Acad. Sci. USA 97, 6745-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marian, A. J. (2002) Curr. Opin. Cardiol. 17, 242-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki, M., Carlson, K. M., Marchuk, D. A. & Rockman, H. A. (2002) Circulation 105, 1824-1829. [DOI] [PubMed] [Google Scholar]
- 7.Ihara, Y., Suzuki, Y. J., Kitta, K., Jones, L. R. & Ikeda, T. (2002) Cell Calcium 32, 21-29. [DOI] [PubMed] [Google Scholar]
- 8.Aronow, B. J., Toyokawa, T., Canning, A., Haghighi, K., Delling, U., Kranias, E., Molkentin, J. D. & Dorn, G. W. (2001) Physiol. Genomics 6, 19-28. [DOI] [PubMed] [Google Scholar]
- 9.Hwang, J. J., Allen, P. D., Tseng, G. C., Lam, C. W., Fananapazir, L., Dzau, V. J. & Liew, C. C. (2002) Physiol. Genomics 10, 31-44. [DOI] [PubMed] [Google Scholar]
- 10.Oliver, P. M., Fox, J. E., Kim, R., Rockman, H. A., Kim, H. S., Reddick, R. L., Pandey, K. N., Milgram, S. L., Smithies, O. & Maeda, N. (1997) Proc. Natl. Acad. Sci. USA 94, 14730-14735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knowles, J. W., Esposito, G., Mao, L., Hagaman, J. R., Fox, J. E., Smithies, O., Rockman, H. A. & Maeda, N. (2001) J. Clin. Invest. 107, 975-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, L. R., Suzuki, Y. J., Wang, W., Kobayashi, Y. M., Ramesh, V., Franzini-Armstrong, C., Cleemann, L. & Morad, M. (1998) J. Clin. Invest. 101, 1385-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho, M. C., Rapacciuolo, A., Koch, W. J., Kobayashi, Y., Jones, L. R. & Rockman, H. A. (1999) J. Biol. Chem. 274, 22251-22256. [DOI] [PubMed] [Google Scholar]
- 14.Caron, K. M., James, L. R., Kim, H. S., Morham, S. G., Sequeira Lopez, M. L., Gomez, R. A., Reudelhuber, T. L. & Smithies, O. (2002) Proc. Natl. Acad. Sci. USA 99, 8248-8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinkert, C. A., Ornitz, D. M., Brinster, R. L. & Palmiter, R. D. (1987) Genes Dev. 1, 268-276. [DOI] [PubMed] [Google Scholar]
- 16.Kozak, M. (1991) Gene Expression 1, 117-125. [PMC free article] [PubMed] [Google Scholar]
- 17.Methot, D., vanKats, J. P., Lochard, N., Tremblay, F., Silversides, D. W. & Reudelhuber, T. L. (2001) Am. J. Hypertens. 14, 38S-43S. [DOI] [PubMed] [Google Scholar]
- 18.Bronson, S. K., Plaehn, E. G., Kluckman, K. D., Hagaman, J. R., Maeda, N. & Smithies, O. (1996) Proc. Natl. Acad. Sci. USA 93, 9067-9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krege, J. H., Hodgin, J. B., Hagaman, J. R. & Smithies, O. (1995) Hypertension 25, 1111-1115. [DOI] [PubMed] [Google Scholar]
- 20.Nearing, B. D. & Verrier, R. L. (2002) J. Appl. Physiol. 92, 541-549. [DOI] [PubMed] [Google Scholar]
- 21.Kim, H. S., Lee, G., John, S. W., Maeda, N. & Smithies, O. (2002) Proc. Natl. Acad. Sci. USA 99, 4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lijnen, P. J., Petrov, V. V. & Fagard, R. H. (2000) Mol. Genet. Metab. 71, 418-435. [DOI] [PubMed] [Google Scholar]
- 23.Williams, B. (2001) Am. J. Cardiol. 87, 10C-17C. [DOI] [PubMed] [Google Scholar]
- 24.Schultz, J. J., Witt, S. A., Glascock, B. J., Nieman, M. L., Reiser, P. J., Nix, S. L., Kimball, T. R. & Doetschman, T. (2002) J. Clin. Invest. 109, 787-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caron, K. M. & Smithies, O. (2001) Proc. Natl. Acad. Sci. USA 98, 615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinson, J. P., Kapas, S. & Smith, D. M. (2000) Endocr. Rev. 21, 138-167. [DOI] [PubMed] [Google Scholar]
- 27.Wehrens, X. H., Lehnart, S. E., Huang, F., Vest, J. A., Reiken, S. R., Mohler, P. J., Sun, J., Guatimosim, S., Song, L. S., Rosemblit, N., et al. (2003) Cell 113, 829-840. [DOI] [PubMed] [Google Scholar]
- 28.Kuo, H. C., Cheng, C. F., Clark, R. B., Lin, J. J., Lin, J. L., Hoshijima, M., Nguyen-Tran, V. T., Gu, Y., Ikeda, Y., Chu, P. H., et al. (2001) Cell 107, 801-813. [DOI] [PubMed] [Google Scholar]
- 29.Chien, K. R. (2000) Nature 407, 227-232. [DOI] [PubMed] [Google Scholar]
- 30.Nolan, J., Batin, P. D., Andrews, R., Lindsay, S. J., Brooksby, P., Mullen, M., Baig, W., Flapan, A. D., Cowley, A., Prescott, R. J., et al. (1998) Circulation 98, 1510-1516. [DOI] [PubMed] [Google Scholar]
- 31.Gang, Y., Ono, T., Hnatkova, K., Hashimoto, K., Camm, A. J., Pitt, B., Poole-Wilson, P. A. & Malik, M. (2003) Pacing Clin. Electrophysiol. 26, 394-400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.