Abstract
Rationale
Multiple progenitors derived from the heart and bone marrow have been utilized for cardiac repair. Despite this, not much is known about the molecular identity and relationship among these progenitors. To develop a robust stem cell therapy for the heart, it is critical to understand the molecular identity of the multiple ‘cardiogenic progenitor cells’ (CPCs). This study is the first report of high throughput transcriptional profiling of CPCs carried out on an identical platform.
Method and Results
Microarray based transcriptional profiling was carried out for three cardiac (ckit+, Sca1+, side population) and two bone marrow (ckit+ , mesenchymal stem cell) progenitors, obtained from age- and sex-matched wild type C57BL/6 mice. Analysis indicated that cardiac-derived ckit+ population was very distinct from Sca1+ and SP cells in the downregulation of genes encoding for cell-cell and matrix adhesion proteins, and in the upregulation of developmental genes. Significant enrichment of transcripts involved in DNA replication and repair was observed in bone marrow (BM)-derived progenitors. The BM ckit+ cells appeared to have the least correlation with the other progenitors, with enrichment of immature neutrophil specific molecules.
Conclusion
Our study indicates that cardiac ckit+ cells represent the most primitive population in the rodent heart. Primitive cells of cardiac versus BM origin differ significantly with respect to stemness and cardiac lineage-specific genes, and molecules involved in DNA replication and repair. The detailed molecular profile of progenitors reported here will serve as a useful reference to determine the molecular identity of progenitors used in future preclinical and clinical studies
Keywords: cardiac progenitor cells, bone marrow cells, transcriptomics, cardiovascular diseases
INTRODUCTION
For nearly a century, the heart was viewed as a terminally differentiated post-mitotic organ characterized by a predetermined number of myocytes, which is established at birth and largely preserved throughout life until death of the organism. More than 10 years ago, the identification of male cells in female hearts transplanted in sex-mismatched male recipients provided the first evidence that the heart is a stem cell-regulated organ1. Although the magnitude of chimerism is controversial2, 3, the presence of Y-chromosome positive myocytes and coronary vessels in the female donor heart documented that primitive cells of recipient origin had colonized the female heart and had differentiated into cardiovascular lineages.
In the last decade, multiple classes of stem and progenitor cells have been identified and characterized in the adult myocardium by utilizing surface markers and functional assays. Cells expressing the surface marker CD117 (ckit) was the first stem cell population to be identified in the heart of animals and humans4. ckit identifies a population of resident cardiogenic progenitor cells (CPCs) that are self-renewing, clonogenic, and multipotent in vitro, capable of replacing necrotic and scarred tissue with functional myocardium in vivo and improving ventricular performance. This was followed by identification of a primitive population expressing the Stem Cell Antigen (Sca1)5, which represent 0.5–2% of heart cells and 10–15% of the myocyte-depleted fraction. A small fraction of ckit+ and Sca1+ CPCs (1–2%) express the pan leukocyte marker, CD45. Small subsets of Sca1+ cells also express the endothelial epitope CD315. The ability of stem cells to expel toxic compounds and dyes through an ATP-binding cassette surface transporter, which was initially used to isolate a rare ‘side population’ representing stem cells in the hematopoietic system, has been utilized to identify a cardiac resident ‘side population’6. Side population (SP) cells express the P-glycoproteins Abcg2 and Mdr17 in a developmentally regulated manner. Importantly, only the Sca1+CD31− subset of cardiac SP is characterized by a high cardiomyogenic potential. The discovery of cardiac resident stem cells in the heart generated tremendous excitement about the potential to activate these cells in situ and to mediate endogenous cardiac repair in MI patients. In fact, cardiac resident ckit+ cells are already under evaluation in a Phase 1 clinical trial and showing encouraging preliminary results8.
In addition, several preclinical and clinical studies over more than a decade have shown that progenitors from diverse adult tissues such as skeletal myoblasts, hematopoietic progenitors, and bone marrow (BM)-derived mesenchymal cells (MSCs) can repopulate the injured myocardium and improve cardiac function9–12. With respect to safety and improvement in cardiac function, the most widely used extracardiac cells in clinical trials are the BM-derived cells13, 14.
Given that most tissues possess a single unique stem cell population, the discovery of multiple cardiogenic progenitors is intriguing. By definition, stem cells possess well-defined growth properties, and it may be unrealistic to expect the heart to contain such a variety of primitive cells all performing the same biological function. As an added complexity, the multiple reports described above used different animal models, strains, lineage marker cocktails, and isolation/culture methods, thereby making it very difficult to compare the molecular relationships among different progenitors. In this current study, we have isolated multiple cardiogenic progenitor cells from age- and sex-matched mice of the same strain and utilized a common platform to analyze the molecular relationship among these primitive cells using whole genome transcriptional profiling. This study is an attempt to define whether ckit+, Sca1+, and SP cells are distinct categories of undifferentiated cells with diverse functional behavior, or whether they represent different phenotypic stages of the same cell population. In addition, we analyzed the molecular relationship between the cardiac-derived progenitors (ckit+, Sca1+, and SP) and the extracardiac BM-derived progenitors (ckit+ cells and MSCs). Differentially expressed genes were classified in functional categories and signaling pathways to define the molecular identity and relationship among the multiple cardiogenic progenitors.
METHODS
Additional information is available in the Online Supplemental Methods section.
Isolation of cardiac and bone marrow cells
In order to eliminate the variability introduced by in vitro culture, freshly isolated and minimally expanded cells were used for this study. All cell types were derived from age- (eight weeks old) and sex-matched (male) mice of the same strain (C57BL/6) obtained from the same source (Charles River Laboratory; www.criver.com), using protocols described in Supplementary Methods. The absolute number of cells obtained from one mouse heart for each cell type is as follows: Sca1+ cells: 5×104−1×105; SP: 2×103−7×103; ckit+ cells: 1×105−3×105; cardiomyocytes: 5×105−1×106; BM ckit+ cells: 5×105−8×105; and BM MSCs: 8×105−1×106
RNA isolation, amplification, and microarray
RNA from all samples (6 cell types; 3 replicates for each; total 18 samples) was isolated using the RNAeasy microkit (Qiagen) with a few changes in the protocol as detailed in Supplementary Methods. The microarray data have been submitted to Gene Expression Omnibus (GEO) database (Accession No. GSE41175).
Statistical analysis
The Rosetta Biosoftware statistical analysis package was used to analyze the raw microarray data. Differentially expressed genes among multiple groups of samples were detected by one-way analysis of variance. T test was used to identify differentially expressed genes for two individual samples. Real-time PCR validations were analyzed using Graphpad Prism (GraphPad Software, Inc., CA, USA). Differences were considered significant at P values of <0.05.
RESULTS
Isolation of cardiogenic progenitor cells
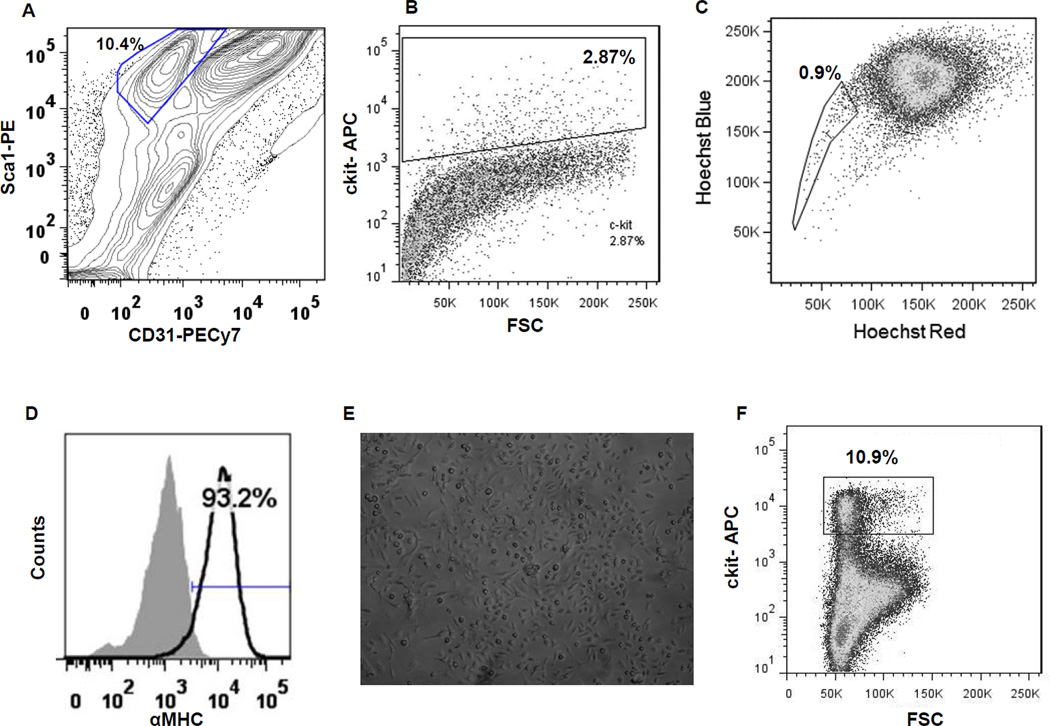
The six cell types utilized for this study were harvested from the hearts of 2 month-old male C57BL/6 mice in triplicates (Online Figure I) and characterized extensively (Figure 1 and Online Figure II). To eliminate potential inter-sample variability dictated by prolonged culture in vitro, freshly isolated and minimally expanded cells were studied. Four distinct cardiac derived cells were analyzed; the three progenitors (Sca1+, ckit+, and SP) and cardiomyocytes as the control population. Prior to analysis, the Sca1+ cell pool was depleted for CD31 and CD45 to exclude cells of endothelial lineage and bone marrow origin (Figure 1A & Online Figure IIA and B). Sca1+ CPCs accounted for ~8–10% of the unfractionated cardiac cell population. By contrast, ckit+ CPCs comprised at most 3% of the non-myocyte small cell pool (Figure 1B & Online Figure IIC). However, ckit-labeled cardiac cells included 1–2% of cells co-expressing the panleukocyte marker CD45 (Online Figure III), and the ckit+CD45+ double positive cells represent mastocytes and cells of bone marrow origin. Flow cytometry showed that SP included 0.9% of the cells enzymatically dissociated from the mouse heart (Figure 1C). Cardiomyocytes were obtained by mechanical and enzymatic digestion of the heart and purified by differential centrifugation (Figure 1D). The presence of α-myosin heavy chain was detected by flow cytometry, documenting a 90% degree of enrichment for cardiomyocytes (Online Figure IID & E). BM-derived mesenchymal stem cells were enriched and harvested after expansion in MSC media for five passages and characterized (Figure 1E & Online Figure IIF)15. BM-derived ckit+ cells comprised ~10% of the bone marrow isolate (Figure 1F & Online Figure IIG).
Figure 1. Isolation of progenitor populations from heart and bone marrow for whole genome transcriptional profiling.
(A) Profile of cardiac-derived Sca1+CD45−CD31− cells. (B) Isolation of cardiac-derived ckit+ cells. (C) Hoechst 33342 exclusion assay based isolation of ‘side population’ (SP) from heart. (D) Characterization of isolated cardiomyocytes by staining for alpha myosin heavy chain (αMHC). (E) Representative photomicrograph of enriched mesenchymal stem cells (MSCs) isolated from bone marrow and cultured for 5 passages; magnification 10×. (F) Isolation of ckit+ cells derived from the bone marrow.
Total RNA was isolated simultaneously from all cell preparations, converted to cDNA, subjected to a single round of amplification in order to obtain sufficient yield of single stranded DNA (Online Figure IV), followed by whole genome cDNA microarray profiling. Transcriptional profiling of three independently isolated biological replicates was carried out. However, after conducting the Pearson’s correlation test, one replicate of CSP and one replicate of cardiomyocyte had to be excluded from further analysis due to the low correlation coefficient.
Transcriptional profiling of CPCs and cardiomyocytes: cardiac-specific genes
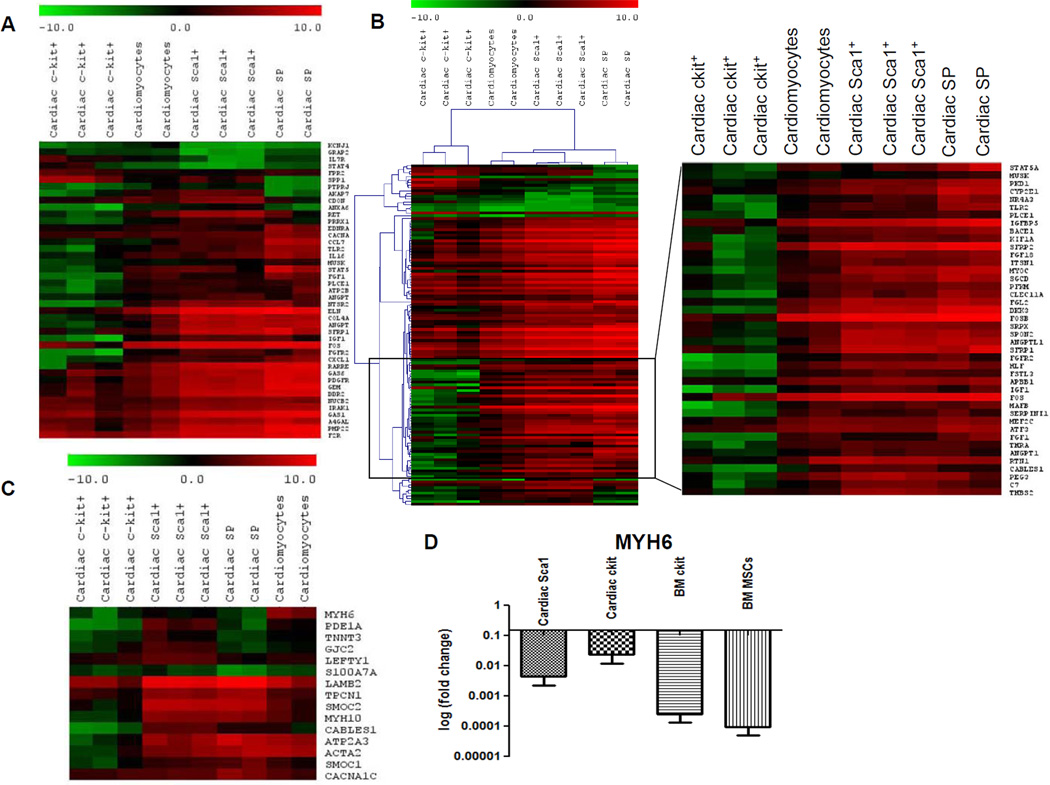
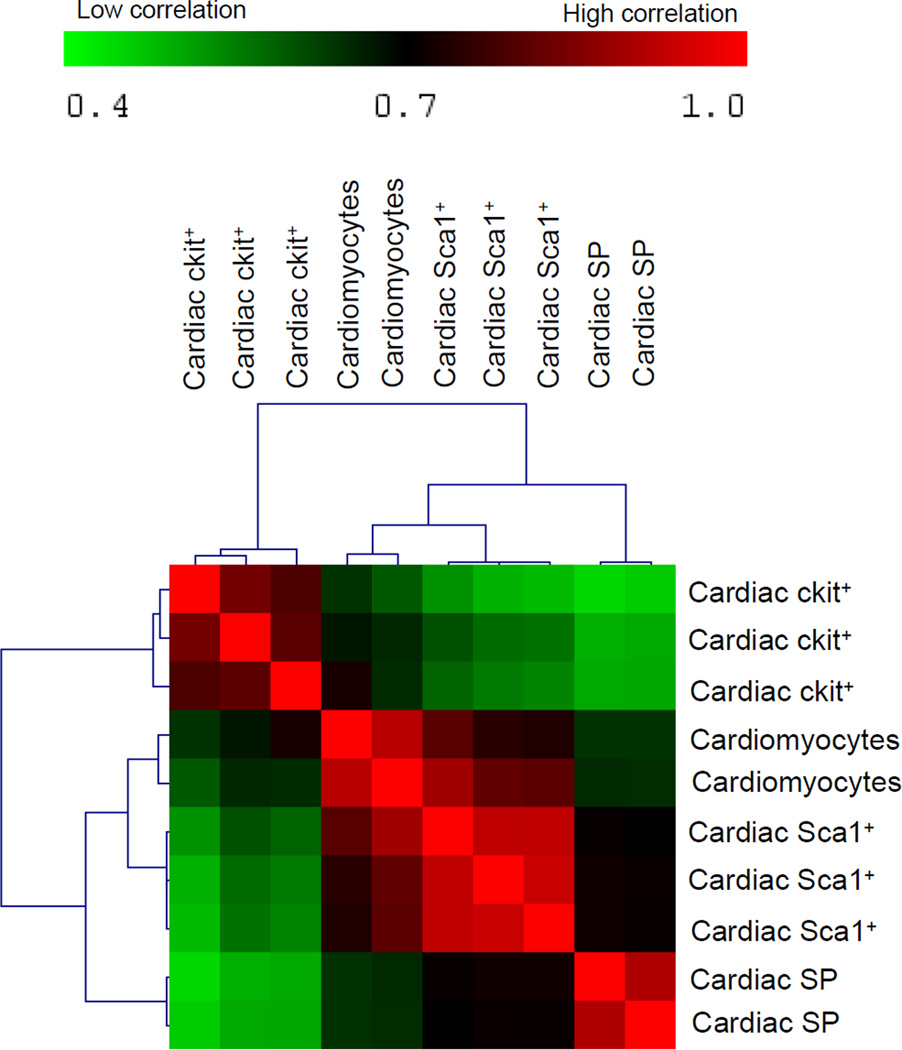
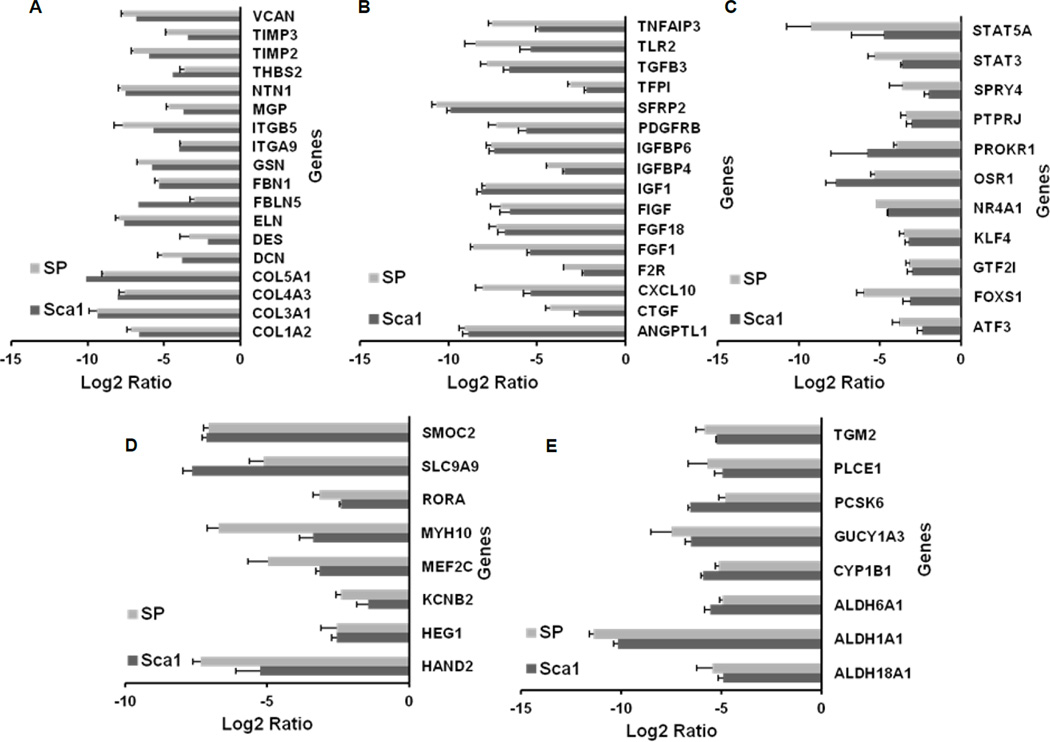
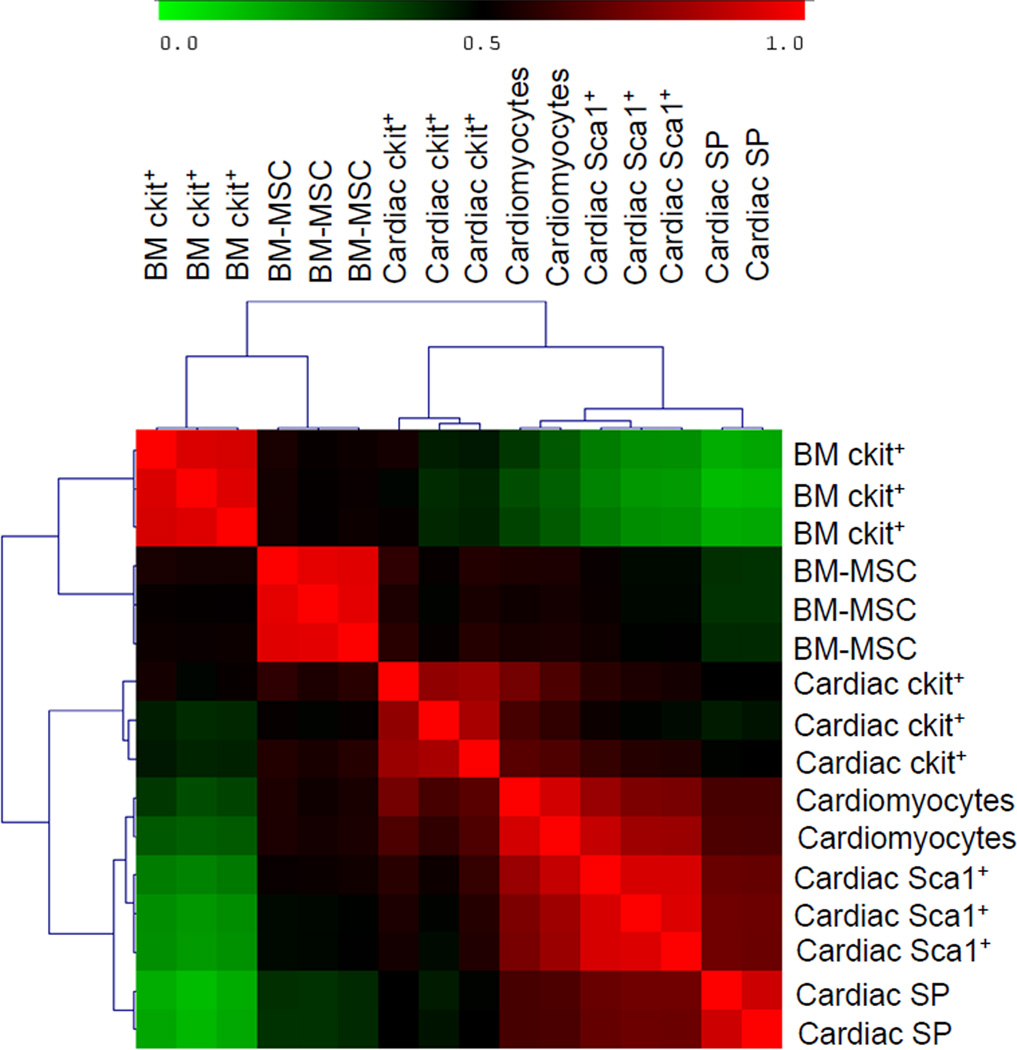
Our first set of analyses involved comparison of the cardiac-derived cells (ckit+, Sca1+, SP, and cardiomyocytes). By applying analysis of variance and including only genes that showed a significant (p<0.05) expression difference of at least two folds among the different classes of CPCs and between CPCs and cardiomyocytes, 1,438 genes were identified. Upregulated and downregulated gene lists were grouped into functional categories based on gene ontology classifications and analyzed using Ingenuity Pathway Analysis (IPA) software. This independent approach highlighted the presence of functional gene cores, differentially represented in ckit+, Sca1+, and SP (Figures 2A–B), which allows the identification of molecular pathways showing different state of activation or repression in the three CPC classes. These pathways are known to play an intrinsic role in the biological processes of cell survival, proliferation, and tissue-specific functions. Cardiomyocytes provided a positive control for expression of lineage-specific genes. Cardiac myosin heavy chain (MYH6) was restricted to cardiomyocytes and was absent in CPCs; this finding was validated by qRT-PCR (Figure 2C–D). Although the overall expression of genes characteristic of mature, terminally differentiated myocytes was significantly downregulated in CPCs with respect to cardiomyocytes, the degree of commitment to myocyte lineage differed in the three classes of CPCs. By comparative analysis and hierarchical clustering, Sca1+ showed the highest correlation with cardiomyocytes, followed by SP CPCs (Figure 3 and Online Figure V). ckit+ appeared to be a distinct cell pool, having the lowest correlation with the other two CPC classes and cardiomyocytes. A subset of myocyte-specific transcription factors, contractile proteins, ion channels, and calcium-binding proteins was upregulated in Sca1+ and SP with respect to ckit+ cells (Supplemental Table I and Supplementary Results).
Figure 2. Relationship among the cardiac-derived cells.
Heatmaps represent hierarchical clustering for different functions among cardiac-derived cells. (A) Cell signaling genes. (B) Cell survival genes (right panel shows an enlarged view of the area within the indicated region). (C) Cardiac and/or muscle-specific genes. (D) Real-time PCR validation of cardiomyocyte specific myosin heavy chain (MYH6). Fold changes in gene expression was calculated with respect to cardiomyocytes. Expression level of this gene was undetectable for SP cells. Each bar represents mean fold change of three replicates per cell population and three technical repeats for each experiment. Error bar represents standard deviation.
Figure 3. Hierarchical clustering among cardiac-derived cells.
Correlation among the three cardiac-derived CPCs and cardiomyocytes represented as a hierarchical cluster matrix, based on Pearson’s correlation of significant (p<0.05) differentially expressed genes (≥ 2-fold) among all samples. Red represents high correlation; green represents low correlation.
Multiple developmentally related, mesoderm-specific genes and stem cell signaling molecules were found to be upregulated in cardiac ckit+ cells
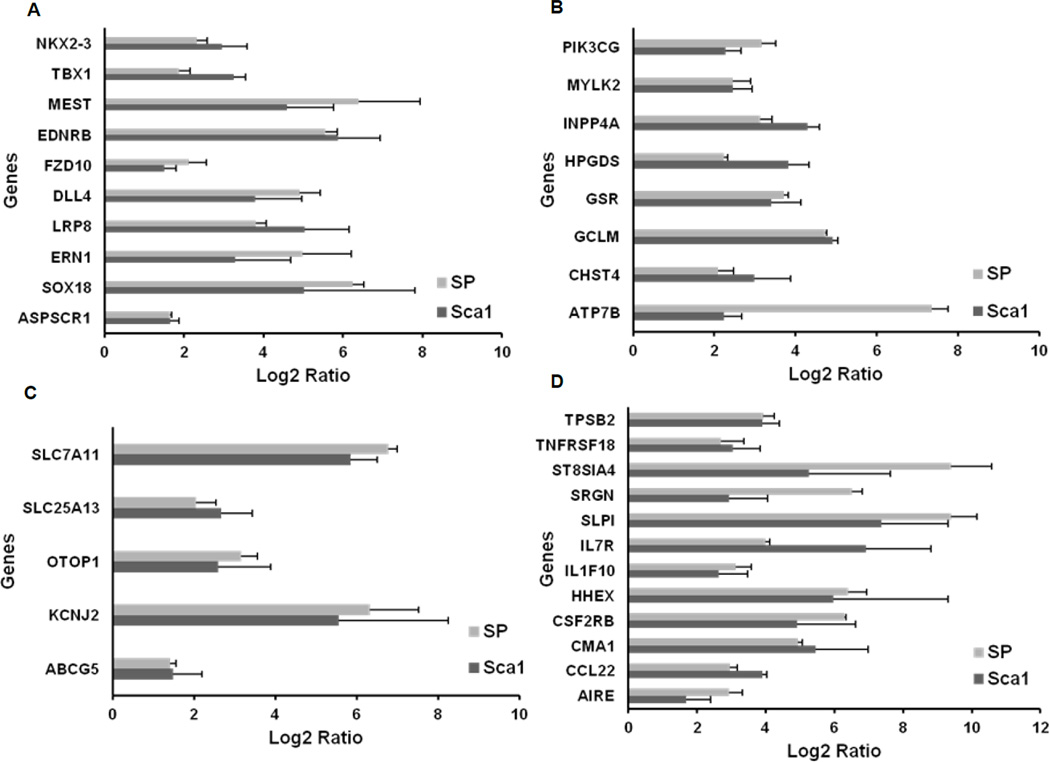
When the expression profile of ckit+ cells was individually compared with Sca1+ and SP cells, a set of 36 common genes emerged, which could be classified into four broad categories: (i) developmental, mesodermal-specific, and stem cell related genes; (ii) genes encoding enzymes in metabolic pathways; (iii) transporters and ion channels; and (iv) hematopoietic specific genes (Figure 4 and Supplementary Tables II–V). Multiple developmental genes, especially those expressed in early mesoderm such as MEST, NKX2.3, and TBX1, as well as stem and progenitor cell specific genes like members of Notch and canonical Wnt signaling pathways (DLL4, FZD10, and LRP8), were found to be exclusively upregulated in cardiac ckit+ cells (Figure 4A). Key enzymes mediating cell signaling and metabolism, such as phosphoinositide 3-kinase (PI3K), phosphatase (INPP4A), and dehydrogenases (ADH7) showed enriched expression in cardiac ckit+ cells compared to both the CPCs (Figure 4B). A third category of upregulated molecules included different isoforms of surface transporters like the ATP-binding cassette transporter, ABCG5, and anionic transporters of the solute carrier (SLC) family (Figure 4C). ckit+ cells also showed enriched expression of a set of genes encoding chemokines, interleukin receptors (CCL22, TNFRSF18, IL7R, and CSF2RB/CD131) and genes expressed in mast cells among other cell types (CMA1, TPSB2, and SRGN) (Figure 4D).
Figure 4. Functional categorization of upregulated genes in cardiac-derived ckit+ cells when compared to Sca1+ and SP cells.
(A) Developmental, mesodermal-specific, and stem cell related genes. (B) Genes encoding components of metabolic pathways. (C) Ion channels and transporter genes. (D) Hematopoeitic cell specific genes.
In order to validate these results, total ckit+ cells and ckit+CD45− cells were sorted by flow cytometry, and were subjected to quantitative real-time PCR for CMA1, TPSB2, and SRGN. The expression of these genes was significantly lower in the ckit+CD45− fraction, compared to the total ckit+ population (Online Figure VI). These data indicate that a clear separation exists between ckit+ CPCs and other hematopoietic cells, including mast cells. Importantly, the culture medium employed for the expansion of ckit+ CPCs resulted in apoptosis of CD45+ cells. Ingenuity Pathway Analysis (IPA) of the factors promoting cardiogenesis in ckit+ cells also showed differences between ckit+ progenitors and the other two CPC populations (Online Figure VII). Interestingly, multiple genes encoding microRNAs appeared to be highly upregulated in ckit+ cells only with respect to Sca1+ cells (miRNA 8, 17, 24, 25, 181, 145, 196, 338, 362, and let-7), but not when compared to SP cells.
Genes involved in cell-cell and cell-ECM interactions are downregulated in cardiac ckit+ cells
As part of the detailed analysis of molecular differences among cardiac ckit+ CPCs with respect to Sca1+ and SP cells, the most significantly downregulated functional gene core that came up in ckit+ cells consisted of genes encoding for extracellular matrix (ECM) proteins, integrins, matrix metalloproteases, and gap junctions (Figure 5A, Supplementary Table VI). IPA also indicated an overall downregulation of the actin cytoskeletal pathway in ckit+ cells (Online Figure VIII), though the number of molecular markers downregulated with respect to Sca1+ cells was more than the number when compared to SP cells. The next category was comprised of genes encoding for growth factors and transcription factors specifically expressed in vascular endothelial cells, fibroblasts, and those involved in connective tissue formation, function, or remodeling (Figures 5B–5C, Supplemental Table VII-VIII). As detailed earlier, a set of cardiac-specific genes was found downregulated in ckit+ cells compared to the other two CPC populations (Figure 5D). Interestingly, multiple isoforms of the enzyme aldehyde dehydrogenase (ALDH) were seen to be significantly downregulated in cardiac ckit+ cells (Figure 5E, Supplemental Table IX).
Figure 5. Functional categorization of genes downregulated in cardiac-derived ckit+ cells when compared to Sca1+ and SP cells.
(A) Genes encoding extracellular matrix (ECM) proteins, matrix metalloproteases (MMPs), and junction proteins. (B) Genes encoding growth factors. (C) Transcription factors. (D) Cardiac-specific genes. (E) Genes encoding enzymes.
Overall, our comparison of cardiac-derived progenitors indicated that the ckit+ population is most distinct and least correlated with Sca1+ and SP progenitors. Cardiac derived ckit+ cells were unique in their enrichment of transcripts involved in early mesoderm development, stem cell signaling pathways, growth factors, cytokines and downregulation of genes encoding for ECM and gap junction proteins. Correlation studies and IPA also indicated that Sca1+ CPCs were closest to cardiomyocytes, followed by the SP.
Comparative analysis of the transcription profile between cardiac-derived and bone marrow-derived progenitor cells
We next sought to compare the cardiac-derived progenitors with the two most widely used extracardiac progenitors, BM-derived ckit+ cells and MSCs. All three replicates of the BM-derived progenitors showed high correlations in Pearson’s correlation test. For the first analysis, difference in gene expression was calculated between the average intensity of expression of all three cardiac-derived CPCs and average of BM-derived progenitor cells. Hierarchical clustering analysis indicated that the BM-derived group was distinct from the cardiac-derived progenitors (Figure 6 and Online Figure IX). Approximately 4,531 differentially expressed genes were identified among the six samples. Functional categorization of these genes demonstrated significant enrichment of mRNAs within distinct functional categories in cardiac-derived progenitors, namely; ECM, cytoskeletal elements, gap junction, integrin signaling and cardiac-specific (downregulated in BM-derived progenitors) (Online Figure X & XI; Supplemental Tables X–XVIII). The surface markers CD40 and CDH13 were exclusively upregulated in the cardiac group, whereas CD52 and CD48 were exclusively upregulated in the BM group. A panel of surface markers was unique to BM-MSCs (CD28, CD300LB, and CD101), whereas PECAM1 was found to be exclusively downregulated in BM-derived MSCs. On the other hand, a large gene set comprising of genes involved in DNA replication, repair, and cell cycle regulation was downregulated in the cardiac group (highly upregulated in BM group) (Online Figure XII & XIII; Supplemental Tables XIX–XXIV).
Figure 6. Hierarchical clustering among cardiac-derived and bone marrow-derived cells.
Correlation among the three cardiac-derived CPCs, cardiomyocytes, and BM-derived cells, represented as a hierarchical cluster matrix, based on Pearson’s correlation test of significant (p<0.05) differentially expressed genes (≥ 2-fold) among all samples. Red represents high correlation; green represents poor correlation.
BM-derived ckit+ population is the most distinct progenitor cell type
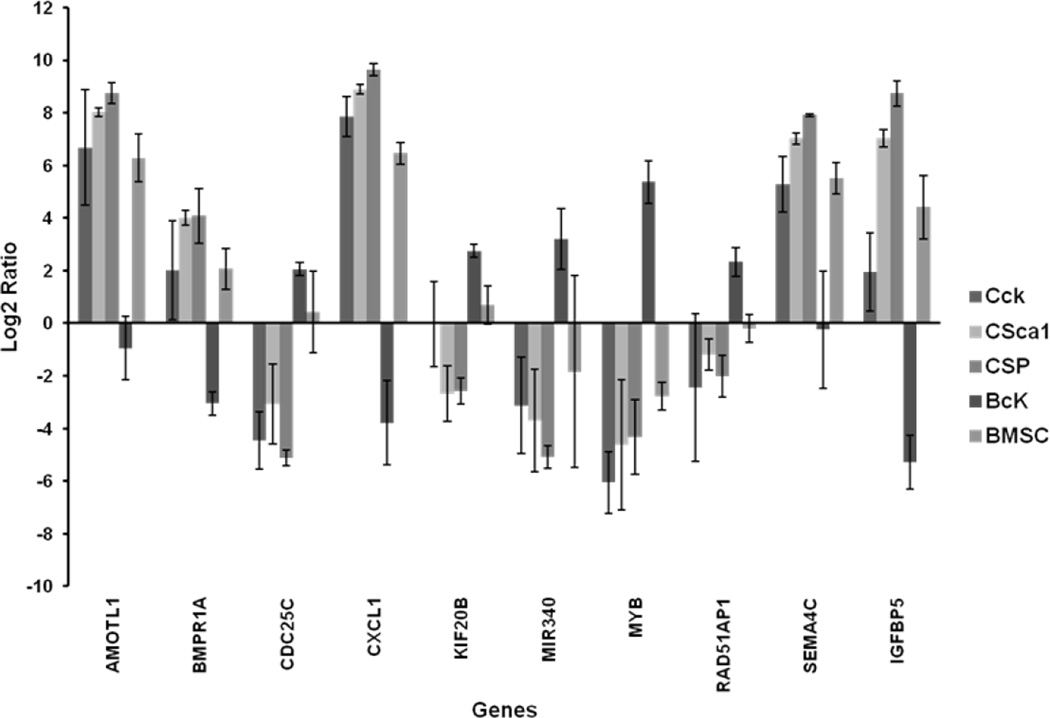
On analyzing the transcript profile of each cardiac-derived and BM-derived progenitors individually, we observed a significant number of genes (50–100 top differentially expressed genes) whose expression in BM-derived ckit+ cells was inversely correlated to all other progenitor cells, including the BM-derived MSCs (Figure 7). This group consisted of genes encoding for growth factors, their receptors, cytokines, ion channels, gap junctions, and DNA replication. Interestingly, most of the genes involved in DNA replication, repair, and cell cycle regulation showed highest upregulation in BM-derived ckit+ cells.
Figure 7. Comparison of expression profiles between bone marrow-derived ckit+ cells and all other progenitors.
(log)2 ratio among the cardiac- and BM-derived progenitor cells for a representative set of genes. (CcK: cardiac ckit+ cells; CSca1: cardiac Sca1+ cells; CSP: Cardiac SP; BcK: BM ckit+ cells; BMSC: BM mesenchymal stem cells)
BM-derived ckit+ cells are distinct from BM-derived MSCs in upregulation of neutrophil specific and DNA replication, repair genes
Considering that both ckit+ cells and MSCs are derived from the same BM source, we sought to compare the differences between these two groups. We analyzed the top 100–130 upregulated and downregulated genes (with highest fold differences) in BM-derived ckit+ cells compared to BM-derived MSCs (Online Figure XIV). Close to 30% of highly upregulated genes in BM-derived ckit+ cells were neutrophil- and macrophage-specific (ELANE, NGP, CEBPE, NKG7, and GZMB). The next largest group was the lectin family (carbohydrate-binding proteins and sphingosine transporters), followed by genes involved in DNA repair and replication (Online Figure XV). In the top 100 downregulated genes in BM-derived ckit+ cells (those upregulated in MSCs), the largest category included gap junctions and integrins, MMP coding genes, and growth factor signaling (Online Figure XVI). Genes encoding growth factor binding proteins came up as the second largest group, and the third largest group was composed of enzymes involved in redox reactions.
Cardiac-derived ckit+ population differs from BM-derived ckit+ cells by having increased expression of genes encoding ECM, gap junction proteins, and cytoskeletal elements
The receptor tyrosine kinase CD117, or ckit, is a well-known stem cell marker in multiple tissues. In our study, we had two ckit+ populations, derived from the heart and the bone marrow. To understand the molecular differences between these two cell types, we analyzed the top 100 upregulated and top 100 downregulated genes between them, based on highest fold differences, as described before. Following this, these genes were categorized into different functional groups. Genes encoding for ECM and gap junction proteins (31 genes) appeared predominant among the most highly enriched mRNAs in cardiac-derived ckit+ cells, followed by transcripts for cytokines, growth factors (CXCL1, CXCL2, NGF, EREG, ADM, etc.) and multiple transcripts involved in cytoskeletal structure and rearrangements (MYL2, FHL1, Caldesmon, Nckap1, Mtap, among others). The list also included a few endothelial cell specific genes or those involved in angiogenesis (ESM1, Cyr61, Procr, and Wt1) (Online Figure XVIIA). Downregulated gene list in cardiac-derived ckit+ cells (upregulated in BM-derived ckit+ cells) pinpoints DNA repair and replication as the functional category having highest number of genes (33) (Online Figure XVIIB and XX). Several hematopoietic specific genes (19) also came up in the downregulated gene list. Multiple genes encoding for enzymes of ATPase and dehydrogenase family and lectins (Clec4b2, MRC1, and FCNB) were found to be downregulated in cardiac-derived ckit+ cells. IPA indicated a predominant upregulation of tight junction components, actin cytoskeletal, PDGF, and TGFβ signaling in cardiac-derived ckit+ cells (Online Figure XVIII), with significant downregulation of genes involved in immune cell pathways and DNA repair/replication with respect to BM-derived ckit+ cells (Online Figure XIX). Our overall analysis indicates that BM-derived ckit+ cells might be closely related to a primitive granulocyte lineage, and is distinct from the other progenitors, including BM-derived MSCs and cardiac-derived ckit+ population.
‘Cardiosphere-derived cells’ (CDCs) are closely related to BM-derived MSCs
Cardiospheres are spontaneously aggregating structures in vitro, derived from a mixture of cells, and ‘cardiosphere derived cells’ (CDCs) come from in vitro culture of cardiospheres. All cell types in our study (except the BM-derived MSCs), were freshly isolated and had not been cultured/expanded in vitro. Hence, although cardiospheres and CDCs make up a distinct cardiac-derived population implicated in cardiac regeneration, we did not include these cells as part of this study due to their in vitro derivation and mixed identity16. However, considering that these cells are being tested in clinic (CADUCEUS), we also compared publicly available microarray database for cardiospheres from Cho et al.17 with our data shown in Supplementary Results & Online Figure XXI–XXIII.
DISCUSSION
For a long time the adult mammalian heart has been considered to be a post-mitotic organ, with no regenerative potential. The first indication of existence of stem-like cells in the adult heart dated from a decade ago in a study of patients with myocardial infarction19. This study showed the presence of dividing cardiomyocytes in the infarct border zone, which theoretically could have originated from a cardiac resident or circulating pool of stem cells. A subsequent study identified this population as clonogenic, multipotent, and self-renewing cardiac resident lineage-negative, ckit-positive cells4, henceforth referred to as ‘cardiac ckit+ cells’. Shortly after this, two groups reported the isolation of two additional types of adult cardiac progenitor cells based either on expression of the surface marker Sca15 or by a functional Hoechst 33342 efflux assay, representing the cardiac ‘side population’ (SP)20. Sca1+ as well as SP cells were successfully shown to ‘home’ in on the myocardium after an ischemia/reperfusion injury, and to differentiate into functional cardiomyocytes21. The original report on Sca1+ cells demonstrated that >93% of cardiac SP cells were Sca1+, and neither of these populations expressed the ckit antigen. In addition to resident CPCs, BM-derived ckit+ cells and MSCs have also been shown to improve heart function post MI15, 22. However, all these reports have utilized different animal models, strains, lineage marker cocktails, and isolation/culture methods, thereby making it very difficult to compare the progenitors on the same platform to understand their molecular relationships.
To better compare the progenitors under more consistent conditions, the cardiac- and BM-derived progenitor cells were isolated from age- and sex-matched C57BL/6 mice. Our study design therefore eliminated the variability confounding the other studies, enabling a detailed molecular comparison between cardiac and BM-derived progenitors at the transcript level. Comparisons among the three cardiac progenitor cells in our study indicated that the transcriptional landscape of cardiac ckit+ cells was farthest away and most distinct from Sca1+ and SP cells. Upregulation of myocyte-specific transcription factors, contractile proteins, ion channels, and calcium binding proteins in Sca1+ and SP cells with respect to ckit+ CPCs suggested that early lineage-specific genes are poised for expression in Sca1+ and SP cells, because these cardiac-related genes are typically present in embryonic myocytes and not widely distributed in the adult myocardium. MEF2C is an essential component of the core network of transcription factors that control heart morphogenesis23. Together with Nkx2.5, TBX5, and GATA4, MEF2C regulates myocyte formation and differentiation in the embryo. The absence of MEF2C does not affect myocyte lineage specification but interferes with the differentiation of contractile cells. MEF2C physically interacts with HAND2, a protein that is expressed throughout development, becoming gradually enriched in the right ventricle. In the adult heart, MEF2C and HAND2 have been detected in newly formed cells derived from the activation and commitment of resident stem cells following injury23. Transcripts of the Kv2.2 isoform were found highly enriched in Sca1+ and SP cells in our study with respect to ckit+ CPCs. Importantly, Kv2.2 transcripts have been found in embryonic myocytes at very early stages of development24, suggesting that this potassium channel subunit constitutes the immature counterpart of the adult protein. Similarly, the higher levels of non-muscle myosin heavy chain (MYH10) in Sca1+ and SP cells with respect to ckit+ CPCs is intriguing because deletion of this gene is coupled with early embryonic lethality dictated by severe abnormalities in the formation of the heart and brain25. Interestingly, the absence of MYH10 is coupled with a defect in cytokinesis, leading to a decreased number of embryonic cardiac myocytes. Expression of the skeletal and smooth muscle genes TNNT3 and ACTA2 in Sca1+ and SP cells may also be coupled with early stages of cardiomyocyte lineage specification.
Collectively, the expression of immature cardiac-related genes (e.g., MEF2C, HAND2, Kv2.2, Myh10, TNNT3, and ACTA2) in Sca1+ and SP cells is consistent with the view that cardiomyogenesis in the adult heart recapitulates heart development. These two populations may have properties similar to the early committed precursors identified in the embryonic organ. The relatively lower expression of myocardial genes in ckit+ CPCs is dictated by the highly undifferentiated phenotype of this cell class, characterized by the repression of lineage-related genes and activation of stemness-related genes (e.g., NKX2.3, MEST, and SOX). This molecular adaptation is essential for preservation of multipotency in ckit+ CPCs. Upregulation of the hematopoietic transcription factor HHEX, receptors for interleukins, CSF2RB, cytokines, IL7R, CMA1, TPSB2, AIRE, and SRGN in cardiac ckit+ cells with respect to both Sca1+ and SP cells, was most likely dictated by presence of a small subset of cells expressing the pan-leukocyte marker CD45 together with the c-kit receptor26. (See Supplemental Discussion)
While cardiac ckit+ cells appear to be the most ‘undifferentiated’, the Sca1+ cells appear to be the most ‘committed’ to differentiation, given their transcriptional similarity with cardiomyocytes. By comparison, cardiac ‘side population’ (SP) appears to be intermediate between the cardiac ckit+ and Sca1+ cells. Importantly, SP shares Sca1 antigen, whereas neither of these two populations share the ckit surface marker, indicating a closer relationship between SP and Sca1+ cells. Hence, based on our study, we believe that the cardiac ckit+, Sca1+ and SP represent three distinct cell populations functioning at different levels of commitment to differentiation. Our study generated useful insights about the relative level of “stemness” and “early commitment” among the three cardiac progenitors. It must be noted that in order to firmly establish whether the cell of origin for cardiac Sca1+ and SP is the ckit+ cells, in vivo lineage tracing studies must be performed.
Transcriptional profile of Sca1+ cells is closest to cardiomyocytes and very distinct from BM-derived progenitors. This has been independently observed in two earlier studies27, 28. A few reports have stated that Sca1+ CPCs express markers of mesenchymal cells29. In our comparison of Sca1+ cells and BM-derived MSCs, we observed a similar expression of growth factor and their receptors, but there were also significant differences in expression of genes encoding for ECM related proteins, membrane transporters (highly upregulated in Sca1+ cells), and DNA replication/repair molecules (upregulated in MSCs) (data not shown).
Our analysis indicated the upregulation of a large gene network involved in DNA replication, repair, and cell cycle regulation in BM-derived cells when compared with cardiac cells. Stem/progenitor cells in the BM retain a high proliferative capacity during the lifetime of an animal30. In adult mice, long-term repopulating HSCs have been shown to be in a constant slow cycling state31. Natural mobilization of HSCs starts with mitotic expansion of the stem cell progeny, followed by their release in blood in G1 phase to seed secondary tissues32. Cardiac progenitors are characterized by a relatively quiescent state in the non-injured organ33, 34. However, a large fraction of cardiac progenitors enter the cell cycle following injury in an attempt to reconstitute the lost muscle mass. Based on our data and existing literature, the level of expression of genes involved in DNA replication, repair, and cell cycle regulation may distinguish progenitor cells of cardiac from BM origin in the organ in steady state.
An interesting observation in our analysis was the distinct profile of BM-derived ckit+ cells when compared to all the cardiac-derived progenitor cells and BM-derived MSCs individually. The stem cell identity of ‘mesenchymal stem cells’ has been challenged by several studies35, with reports demonstrating that MSCs might be an ‘intermediate’ state of differentiation of HSCs, which finally give rise to mature mesenchymal cells36. It has also been suggested that adult CPCs might undergo EMT, giving rise to precursors that may undergo a reversible commitment to either mesenchymal or cardiac lineage depending on the niche37. Unlike BM-derived hematopoietic progenitors, MSCs are not known to undergo extensive cell cycling; they are instead involved in differentiation into tissue-specific mesenchymal cells38 with immunomodulatory activity39, both functions being subject to the level and combination of local cytokines and growth factors. Given these facts, BM-derived MSCs appear to be distinct from other marrow hematopoietic progenitors or cardiac cells. Finally, a comparison of our transcriptome data with that of the cardiac-derived in vitro population, ‘cardiosphere derived cells’ (CDCs) indicates their similarity with MSCs, as noted previously40. For details on CDCs vis-à-vis the other cardiogenic cells, please refer to Supplemental Discussion.
Significant enrichment of transcripts encoding granulocyte progenitor-specific genes, lectins and cell cycle related genes was observed in BM-derived ckit+ cells, compared to both BM-derived MSCs and cardiac-derived ckit+ cells. Multiple reports have demonstrated a strong expression of lectins exclusively in immature neutrophils of the bone marrow41, 42. In one of these studies, lectin expression was not detected either in more primitive myeloblasts or in mature immune cells. Analysis of our data in conjunction with existing literature indicates that BM-derived ckit+ cells might represent an immature neutrophil precursor population or a subset of immature granulocytes.
In summary, our study indicates that in the adult rodent heart, there are progenitor populations at distinct stages of commitment, with the cardiac-derived ckit+ cells representing the most primitive cell type, as reflected by significant enrichment of early developmental and stem cell-specific genes in this population. These are followed by the more committed ‘side population’, which express few early myocyte-specific genes, and finally the Sca1+ cells, whose transcript profile is closest to cardiomyocytes. The mRNA profile of the extracardiac BM-derived MSCs is intermediate between cardiac progenitors and BM-derived ckit+ cells. Finally, BM-derived ckit+ population is the most distinct from all other cardiogenic progenitors in its enrichment of transcripts involved in cell cycle regulation, DNA replication and repair, with significant downregulation of mRNAs encoding for cell-cell and cell-matrix interactions.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
1. What is known?
The past decade in cardiovascular medicine has witnessed a surge in ‘stem cell based therapies’ for heart diseases.
The discovery of stem cells resident in the adult cardiac heart resulted in the surge. Interestingly, multiple populations of ‘cardiac stem cells’ were reported, unlike other organs, where a single population predominates as ‘stem cells’.
An intriguing observation has been that when resident stem cells in the bone marrow were harvested and injected in an infarcted heart, they could generate functional cardiomyocytes. This resulted in explosion of ‘bone marrow stem cell based therapies’ in patients post myocardial infarction.
2. What new information does this article contribute?
Our study focused on understanding the complex relationship among all the cardiac- and bone marrow-derived cardiogenic progenitors by analyzing their global gene expression profile on the same platform, thereby identifying one of the populations among the cardiac resident progenitors as being distinct and most primitive compared to the others.
Genes involved in cell-cell and cell-extracellular matrix adhesion were highly enriched in the cardiac populations, but significantly lower in the bone marrow-derived cells.
The bone marrow-derived cells had high level of expression of cell division and DNA repair genes, which was negligible in cardiac-derived cells.
SUMMARY.
Cardiovascular medicine has seen a surge in stem cell based therapies with the discovery of resident stem cells in the adult heart and demonstration of the cardiogenic potential of bone marrow-derived stem cells. Despite the multiple cell types used in preclinical studies and clinical trials, a rigorous comparison among them is still lacking. This would require critical understanding of the molecular relationship among these cells and the mechanistic role being played by them during cardiac regeneration. Here we analyzed the global molecular profile of all the stem cell populations in a mouse model. Among the cardiac resident progenitors, one specific population was found to be the most primitive. Genes involved in cell-cell and cell-matrix adhesions were highly enriched in cardiac populations while bone marrow-derived progenitors had high levels of cell division and DNA repair genes. Unlike previous studies, each of which have focused on individual stem cell populations, we have for the first time isolated cells from age- and sex-matched mice of the same strain, and analyzed them on an identical platform. We envisage that this study will serve as a basis to help identify an ideal combination of cardiogenic progenitors for robust cardiac repair in the future.
ACKNOWLEDGEMENTS
We would like to acknowledge Dr. Joseph Gold at Stanford Cardiovascular Institute for his feedback on the manuscript and Grigoriy Losyev at the FACS Core of Brigham and Women's Hospital for his help with cell sorting. We also thank funding support from the NIH R01 EB009689, NIH R01 HL113006, NIH P01 GM099130, and Burroughs Wellcome Foundation (JCW).
List of Abbreviations
- ABCG
ATP Binding Cassette Group
- ADM
Adrenomedullin
- AIRE
Autoimmune Regulator
- ALDH
Aldehyde Dehydrogenase
- ANOVA
Analysis of Variance
- ATP
Adenosine Triphosphate
- BM MSC
Bone Marrow Mesenchymal Stem Cells
- CCL
Chemokine (C−C motif) ligand
- CEBPE
CCAAT/Enhancer Binding Protein, epsilon
- CMA
Chymase
- CPC
Cardiac Progenitor Cells
- CSF2RB
Colony Stimulating Factor Receptor
- CXCL
Chemokine (C−X−C motif) Ligand
- Cyr
Cysteine rich angiogenic inducer
- DNA
Deoxyribonucleic Acid
- ECM
Extracellular Matrix
- ELANE
Elastase, Neutrophil expressed
- EMT
Epithelial Mesenchymal Transition
- EREG
Epiregulin
- ESM
Endothelial Cell Specific Molecule
- F2R
Coagulation Factor 2
- FCNB
Ficolin B
- FHL
Four and a half LIM domains
- GZMB
Granzyme B
- HAND
Heart and Neural Crest expressed derivative
- HHEX
Hematopoietically Expressed Homeobox
- HSC
Hematopoietic Stem Cell
- IL
Interleukin
- IPA
Ingenuity Pathway Analysis
- MEST
Mesoderm Specific Transcript
- MI
Myocardial infarction
- Mir
MicroRNA
- MMP
Matrix Metalloprotease
- MRC
Mannose Receptor C
- MSC
Mesenchymal Stem Cells
- Mtap
Methylthioadenosine phosphorylase
- MYH
Myosin Heavy Chain
- MYL
Myosin Light Chain
- Nckap
NCK Associated Protein
- NGF
Nerve Growth Factor
- NGP
Neutrophilic Granule Protein
- NKG
Natural Killer Cell Group
- OSR
Odd Skipped Related
- PDGF
Platelet Derived Growth Factor
- PI3K
Phosphoinositide-3 Kinase
- Procr
Protein C Receptor, endothelial
- RNA
Ribonucleic Acid
- RT-PCR
Reverse Transcriptase Polymerase Chain Reaction
- Sca
Stem Cell Antigen
- SCF
Stem Cell Factor
- SLC
Solute Carrier
- SP
Side Population
- SRGN
Serglycin
- TBX1
T-Box 1
- TGFβ
Transforming Growth Factor, beta
- TNFR
Tumor Necrosis Factor Receptor
- TPSB
Tryptase beta
- Wt1
Wilms Tumor 1
Footnotes
DISCLOSURES
None
REFERENCES
- 1.Quaini F, Urbanek K, Beltrami AP, Finato N, Beltrami CA, Nadal-Ginard B, Kajstura J, Leri A, Anversa P. Chimerism of the transplanted heart. N Engl J Med. 2002;346:5–15. doi: 10.1056/NEJMoa012081. [DOI] [PubMed] [Google Scholar]
- 2.Glaser R, Lu MM, Narula N, Epstein JA. Smooth muscle cells, but not myocytes, of host origin in transplanted human hearts. Circulation. 2002;106:17–19. doi: 10.1161/01.cir.0000021923.58307.8f. [DOI] [PubMed] [Google Scholar]
- 3.Laflamme MA, Myerson D, Saffitz JE, Murry CE. Evidence for cardiomyocyte repopulation by extracardiac progenitors in transplanted human hearts. Circ Res. 2002;90:634–640. doi: 10.1161/01.res.0000014822.62629.eb. [DOI] [PubMed] [Google Scholar]
- 4.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 5.Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, Garry DJ, Entman ML, Schneider MD. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci U S A. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin CM, Meeson AP, Robertson SM, Hawke TJ, Richardson JA, Bates S, Goetsch SC, Gallardo TD, Garry DJ. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004;265:262–275. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 7.Scharenberg CW, Harkey MA, Torok-Storb B. The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood. 2002;99:507–512. doi: 10.1182/blood.v99.2.507. [DOI] [PubMed] [Google Scholar]
- 8.Bolli R, Chugh AR, D'Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Solankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, Anversa P. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Taylor DA, Atkins BZ, Hungspreugs P, Jones TR, Reedy MC, Hutcheson KA, Glower DD, Kraus WE. Regenerating functional myocardium: improved performance after skeletal myoblast transplantation. Nat Med. 1998;4:929–933. doi: 10.1038/nm0898-929. [DOI] [PubMed] [Google Scholar]
- 10.Strauer BE, Brehm M, Zeus T, Kostering M, Hernandez A, Sorg RV, Kogler G, Wernet P. Repair of infarcted myocardium by autologous intracoronary mononuclear bone marrow cell transplantation in humans. Circulation. 2002;106:1913–1918. doi: 10.1161/01.cir.0000034046.87607.1c. [DOI] [PubMed] [Google Scholar]
- 11.Orlic D, Kajstura J, Chimenti S, Limana F, Jakoniuk I, Quaini F, Nadal-Ginard B, Bodine DM, Leri A, Anversa P. Mobilized bone marrow cells repair the infarcted heart, improving function and survival. Proc Natl Acad Sci U S A. 2001;98:10344–10349. doi: 10.1073/pnas.181177898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei HM, Wong P, Hsu LF, Shim W. Human bone marrow-derived adult stem cells for post-myocardial infarction cardiac repair: current status and future directions. Singapore Med J. 2009;50:935–942. [PubMed] [Google Scholar]
- 13.Wang Y, Haider H, Ahmad N, Zhang D, Ashraf M. Evidence for ischemia induced host-derived bone marrow cell mobilization into cardiac allografts. J Mol Cell Cardiol. 2006;41:478–487. doi: 10.1016/j.yjmcc.2006.06.074. [DOI] [PubMed] [Google Scholar]
- 14.Fazel S, Cimini M, Chen L, Li S, Angoulvant D, Fedak P, Verma S, Weisel RD, Keating A, Li RK. Cardioprotective c-kit+ cells are from the bone marrow and regulate the myocardial balance of angiogenic cytokines. J Clin Invest. 2006;116:1865–1877. doi: 10.1172/JCI27019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbash IM, Chouraqui P, Baron J, Feinberg MS, Etzion S, Tessone A, Miller L, Guetta E, Zipori D, Kedes LH, Kloner RA, Leor J. Systemic delivery of bone marrow-derived mesenchymal stem cells to the infarcted myocardium: feasibility, cell migration, and body distribution. Circulation. 2003;108:863–868. doi: 10.1161/01.CIR.0000084828.50310.6A. [DOI] [PubMed] [Google Scholar]
- 16.Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, Berman D, Czer LS, Marban L, Mendizabal A, Johnston PV, Russell SD, Schuleri KH, Lardo AC, Gerstenblith G, Marban E. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 379:895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho HJ, Lee HJ, Youn SW, Koh SJ, Won JY, Chung YJ, Cho HJ, Yoon CH, Lee SW, Lee EJ, Kwon YW, Lee HY, Lee SH, Ho WK, Park YB, Kim HS. Secondary sphere formation enhances the functionality of cardiac progenitor cells. Mol Ther. 20:1750–1766. doi: 10.1038/mt.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, Fiordaliso F, Salio M, Battaglia M, Latronico MV, Coletta M, Vivarelli E, Frati L, Cossu G, Giacomello A. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 19.Beltrami AP, Urbanek K, Kajstura J, Yan SM, Finato N, Bussani R, Nadal-Ginard B, Silvestri F, Leri A, Beltrami CA, Anversa P. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- 20.Jackson KA, Majka SM, Wang H, Pocius J, Hartley CJ, Majesky MW, Entman ML, Michael LH, Hirschi KK, Goodell MA. Regeneration of ischemic cardiac muscle and vascular endothelium by adult stem cells. J Clin Invest. 2001;107:1395–1402. doi: 10.1172/JCI12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oyama T, Nagai T, Wada H, Naito AT, Matsuura K, Iwanaga K, Takahashi T, Goto M, Mikami Y, Yasuda N, Akazawa H, Uezumi A, Takeda S, Komuro I. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. J Cell Biol. 2007;176:329–341. doi: 10.1083/jcb.200603014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer GP, Wollert KC, Lotz J, Steffens J, Lippolt P, Fichtner S, Hecker H, Schaefer A, Arseniev L, Hertenstein B, Ganser A, Drexler H. Intracoronary bone marrow cell transfer after myocardial infarction: eighteen months' follow-up data from the randomized, controlled BOOST (BOne marrOw transfer to enhance ST-elevation infarct regeneration) trial. Circulation. 2006;113:1287–1294. doi: 10.1161/CIRCULATIONAHA.105.575118. [DOI] [PubMed] [Google Scholar]
- 23.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 24.Burger C, Ribera AB. Xenopus spinal neurons express Kv2 potassium channel transcripts during embryonic development. J Neurosci. 1996;16:1412–1421. doi: 10.1523/JNEUROSCI.16-04-01412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang A, Ma X, Conti MA, Liu C, Kawamoto S, Adelstein RS. Nonmuscle myosin II isoform and domain specificity during early mouse development. Proc Natl Acad Sci U S A. 2010;107:14645–14650. doi: 10.1073/pnas.1004023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metcalfe DD, Baram D, Mekori YA. Mast cells. Physiol Rev. 1997;77:1033–1079. doi: 10.1152/physrev.1997.77.4.1033. [DOI] [PubMed] [Google Scholar]
- 27.Matsuura K, Nagai T, Nishigaki N, Oyama T, Nishi J, Wada H, Sano M, Toko H, Akazawa H, Sato T, Nakaya H, Kasanuki H, Komuro I. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J Biol Chem. 2004;279:11384–11391. doi: 10.1074/jbc.M310822200. [DOI] [PubMed] [Google Scholar]
- 28.Oh H, Chi X, Bradfute SB, Mishina Y, Pocius J, Michael LH, Behringer RR, Schwartz RJ, Entman ML, Schneider MD. Cardiac muscle plasticity in adult and embryo by heart-derived progenitor cells. Ann N Y Acad Sci. 2004;1015:182–189. doi: 10.1196/annals.1302.015. [DOI] [PubMed] [Google Scholar]
- 29.da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 30.Weissman IL. Translating stem and progenitor cell biology to the clinic: barriers and opportunities. Science. 2000;287:1442–1446. doi: 10.1126/science.287.5457.1442. [DOI] [PubMed] [Google Scholar]
- 31.Bradford GB, Williams B, Rossi R, Bertoncello I. Quiescence, cycling, and turnover in the primitive hematopoietic stem cell compartment. Exp Hematol. 1997;25:445–453. [PubMed] [Google Scholar]
- 32.Morrison SJ, Wright DE, Weissman IL. Cyclophosphamide/granulocyte colony-stimulating factor induces hematopoietic stem cells to proliferate prior to mobilization. Proc Natl Acad Sci U S A. 1997;94:1908–1913. doi: 10.1073/pnas.94.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urbanek K, Cesselli D, Rota M, Nascimbene A, De Angelis A, Hosoda T, Bearzi C, Boni A, Bolli R, Kajstura J, Anversa P, Leri A. Stem cell niches in the adult mouse heart. Proc Natl Acad Sci U S A. 2006;103:9226–9231. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meinhardt A, Spicher A, Roehrich ME, Glauche I, Vogt P, Vassalli G. Immunohistochemical and flow cytometric analysis of long-term label-retaining cells in the adult heart. Stem Cells Dev. 2011;20:211–222. doi: 10.1089/scd.2009.0203. [DOI] [PubMed] [Google Scholar]
- 35.Ogawa M, Larue AC, Watson PM, Watson DK. Hematopoietic stem cell origin of mesenchymal cells: opportunity for novel therapeutic approaches. Int J Hematol. 2010;91:353–359. doi: 10.1007/s12185-010-0554-4. [DOI] [PubMed] [Google Scholar]
- 36.Ebihara Y, Masuya M, Larue AC, Fleming PA, Visconti RP, Minamiguchi H, Drake CJ, Ogawa M. Hematopoietic origins of fibroblasts: II. In vitro studies of fibroblasts, CFU-F, and fibrocytes. Exp Hematol. 2006;34:219–229. doi: 10.1016/j.exphem.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Wessels A, Perez-Pomares JM. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat Rec A Discov Mol Cell Evol Biol. 2004;276:43–57. doi: 10.1002/ar.a.10129. [DOI] [PubMed] [Google Scholar]
- 38.Friedenstein AJ, Gorskaja JF, Kulagina NN. Fibroblast precursors in normal and irradiated mouse hematopoietic organs. Exp Hematol. 1976;4:267–274. [PubMed] [Google Scholar]
- 39.Ryan JM, Barry FP, Murphy JM, Mahon BP. Mesenchymal stem cells avoid allogeneic rejection. J Inflamm (Lond) 2005;2:8. doi: 10.1186/1476-9255-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marban E. Heart to heart: cardiospheres for myocardial regeneration. Heart Rhythm. 9:1727–1731. doi: 10.1016/j.hrthm.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Perrin C, Bayle J, Bannwarth S, Michiels JF, Heudier P, Lefebvre JC, Giordanengo V. Expression of LSLCL, a new C-type lectin, is closely restricted, in bone marrow, to immature neutrophils. C R Acad Sci III. 2001;324:1125–1132. doi: 10.1016/s0764-4469(01)01392-0. [DOI] [PubMed] [Google Scholar]
- 42.Denda-Nagai K, Kubota N, Tsuiji M, Kamata M, Irimura T. Macrophage C-type lectin on bone marrow-derived immature dendritic cells is involved in the internalization of glycosylated antigens. Glycobiology. 2002;12:443–450. doi: 10.1093/glycob/cwf061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.