Abstract
Conventional reporter gene technology and histological methods cannot routinely be used to track the in vivo behavior of embryonic stem (ES) cells longitudinally after cellular transplantation. Here we describe a protocol for monitoring the in vivo survival, proliferation, and migration of ES cells following surgical administration without necessitating animal sacrifice. Stable ES cell lines containing double fusion (DF; enhanced green fluorescent protein and firefly luciferase) or triple fusion (TF; monomeric red fluorescent protein, firefly luciferase, and herpes simplex virus thymidine kinase) reporter genes can be established within 4–6 weeks by lentiviral transduction followed fluorescence activated cell sorting (FACS). The cell fate and behavior of these DF or TF ES cells can subsequently be tracked non-invasively by bioluminescence and microPET imaging for a prolonged period of time.
Keywords: embryonic stem cells, molecular imaging, bioluminescence imaging, positron emission tomography imaging
1. Introduction
Embryonic stem (ES) cells offer exciting promises as therapeutic donor cells for regenerative medicine. Unlike adult stem cells, ES cells can differentiate into any somatic cell of the human body and have the capacity for unlimited self renewal 1. To fully realize the therapeutic potential of ES cells, however, it is important for investigators to understand the in vivo behavior of transplanted cells following administration. At present, a number of hurdles have hindered the effective translation of ES-cell based therapy to the clinic. These issues include teratoma formation, immune rejection, failure of cells to engraft, and cellular migration outside the area of administration. The development of sensitive and accurate methods to track the survival, proliferation, and migration of ES cells or ES cell derivatives will thus be indispensable for future applications of ES cell therapy in human patients.
In this chapter, we describe a novel method of monitoring in vivo ES cell behavior through the use of bioluminescence imaging (BLI) and positron emission tomography (PET) reporter genes. In these types of cellular imaging, a reporter gene coding for the synthesis of a detectable protein is stably introduced into the genome of a target cell or tissue via a lentiviral vector carrying a constitutively active promoter such as ubiquitin that drives reporter gene expression. Subsequent cell mediated synthesis of the reporter protein produces a probe that can interact with an exogenously administered substrate to generate a detectable signal (Figure 1). In the case of BLI, the reporter gene introduced is firefly luciferase (Fluc). Interaction of Fluc with its substrate D-luciferin catalyzes production of the optically active metabolite oxyluciferin which emits low intensity photons that can be imaged with a cool-charged couple device (CCD) camera for cell localization. In PET imaging, the reporter protein herpes simplex virus thymidine kinase (HSVtk) phosphorylates its substrate, the PET reporter probe 9-4-[18F]fluoro-3-(hydroxylmethylbutyl) guanine ([18F]-FHBG), to produce high-energy photons that can be captured by a PET camera. Our group has used both a double fusion (DF) construct containing enhanced green fluorescent protein (eGFP) and Fluc reporter genes, and a triple fusion (TF) construct containing monomeric red fluorescent protein (mRFP), Fluc, and HSVtk reporter genes, to stably transduce ES cells for reporter gene imaging
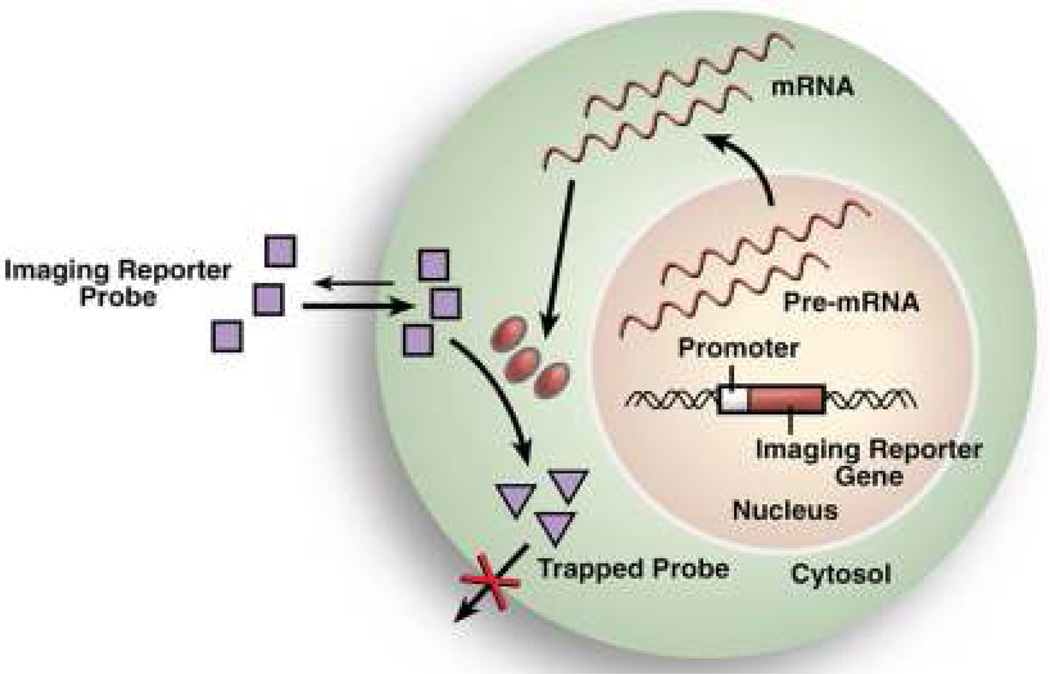
Figure 1.
Conceptual basis of reporter gene imaging. Reporter genes are stably introduced into the genomes of target cells or tissues via lentiviral transduction. Transcription of the reporter gene into mRNA and subsequent translation produces a detectable reporter protein that interacts with an administered reporter probe to generate signal.
As compared to conventional methods of assaying cell fate such as histological analysis and staining for GFP or β-galactosidase (LacZ), reporter gene imaging allows for non-invasive and longitudinal visualization of the spatio-temporal kinetics of cell engraftment and survival in living subjects without requiring animal sacrifice. Because cellular transcription and translation must be intact for synthesis of the reporter proteins, only cells that are alive generate positive signal. In addition, genetic inheritance of the reporter gene from mother to daughter cell permits longitudinal monitoring of cellular proliferation and misbehavior (e.g., teratoma formation). Previous studies have also shown that expression of the reporter genes do not significantly impact ES cell viability, proliferation, and differentiation 2–4. Our laboratory has applied reporter gene imaging to successfully monitor the in vivo survival, migration, and proliferation of transplanted ES cells 4–7 and ES cell derivatives such as cardiomyocytes 8 and endothelial cells 9, 10 over a prolonged period of time.
2. Materials
2.1 Production of DF or TF Lentiviral Transduction Vector
eGFP-Fluc double fusion (DF) construct or mRFP-Fluc-HSVtk triple fusion (TF) construct (available upon request) (Figure 2A).
HEK 293-FT cells (Invitrogen, Carlsbad, CA).
HEK 293-FT cell medium: Dulbecco’s modified Eagle’s medium (DMEM) with high glucose and L-glutamine (Invitrogen, Gibco, Carlsbad, CA), supplemented with 10% fetal bovine serum (FBS, Invitrogen, Gibco, Carlsbad, CA).
psPAX2 packaging plasmid (Addgene, Cambridge, MA).
pMD2G envelope plasmid (Addgene, Cambridge, MA).
Opti-MEM I medium (Invitrogen, Gibco, Carlsbad, CA).
Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
5× PEG-it Lentivirus Concentration Solution (System Biosciences, Mountain View, CA).
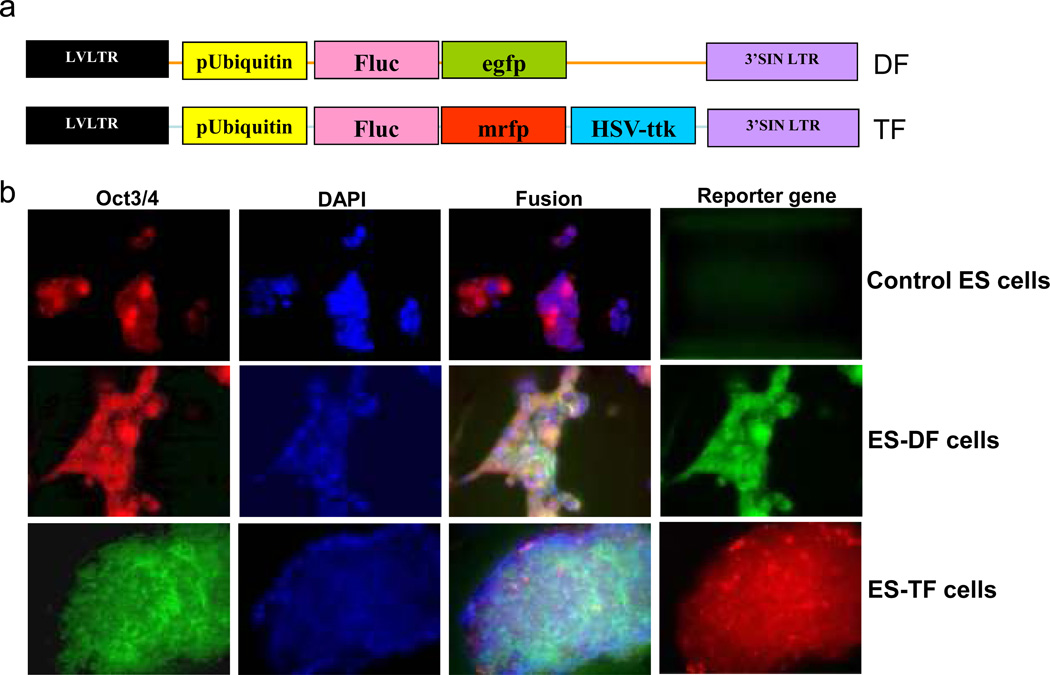
Figure 2.
Characterization of reporter constructs and stably transduced ES cells. (a) Schematic of the DF and TF reporter constructs cloned into a self-inactivating (SIN) lentiviral vector carrying the ubiquitin promoter. (b) Fluorescence microscopy reveals robust expression of mouse ES cell markers (Oct4) in control and stably transduced DF and TF ES cells. Control ES cells are negative for reporter gene expression, whereas DF and TF cells exhibit robust expression of eGFP and mRFP respectively. (Reproduced from reference #7 with permission from Mary Ann Liebert, Inc)
2.2 Lentiviral Transduction of ESCs
Irradiated mouse embryonic fibroblasts (MEFs, ATCC, Manassas, VA).
A mouse ES cell line such as D3 (ATCC, Manassas, MA) or human ES cell line such as H7 or H9 (National Stem Cell Bank, Madison, WI).
MEF medium: DMEM with high glucose with L-glutamine, supplemented with 10% FBS (Gibco) and 5% penicillin/streptomycin (Invitrogen, Gibco, Carlsbad, CA).
Mouse ES cell medium: DMEM with high glucose with L-glutamine, supplemented with 15% FBS (Hyclone, Logan, UT), 2 mM L-glutamine (Invitrogen, Gibco, Carlsbad, CA), 0.1 mM mercaptoethanol (Sigma, St Louis, MI), 0.1 mM nonessential amino acids (Invitrogen, Gibco, Carlsbad, CA), 1000 IU/ml leukemia inhibitory factor (LIF, Invitrogen, Gibco, Carlsbad, CA).
Human ES cell medium: DMEM/F-12 (1:1) (Invitrogen, Gibco, Carlsbad, CA), supplemented with 20% Knockout Serum Replacer (Invitrogen, Gibco, Carlsbad, CA), 1% nonessential amino acids, 2 mM L-glutamine, 0.1 mM mercaptoethanol, and 4 ng/ml basic fibroblast growth factor (bFGF, Invitrogen, Gibco, Carlsbad, CA).
Gelatin (Sigma, St Louis, MI): Prepare as a 0.1% solution and autoclave prior to use.
Polybrene (Sigma, St Louis, MI).
2.3 Derivation of Stable DF or TF ES Cell Lines
Irradiated mouse embryonic fibroblasts.
MEF medium.
Phosphate Buffer Solution (PBS) without Ca2+ and Mg2+ (PBS, Invitrogen, Gibco, Carlsbad, CA).
Cell dissociation buffer (Invitrogen, Gibco, Carlsbad, CA).
70 µm cell strainer (BD Biosciences, San Jose, CA).
Propidium iodide buffer (Invitrogen, Biosource, Carlsbad, CA).
Human ES cell medium + antibiotics/mycotics: add 1% penicillin/streptomycin, 0.25 µg/ml fungizone (Invitrogen, Gibco, Carlsbad, CA) to normal ES cell medium.
Mouse ES cell medium + antibiotics/mycotics: add 1% penicillin/streptomycin, 0.25 µg/ml fungizone to normal ES cell medium.
Collagenase IV (BD Biosciences, San Jose, CA).
Cell scraper (Thomas Scientific, Swedesboro, NJ).
2.4 Verification of Transduction Through In Vitro Imaging
Cell dissociation buffer.
Human or mouse ES cell medium.
D-luciferin (Biosynth, Itasca, IL): Prepare working solution at 45 mg/mlby dissolving 1 gram of D-luciferin in 22 ml PBS and separating into 1–1.5 ml aliquots. Aliquots can be frozen at −20°C for up to six months.
Xenogen In Vivo Imaging System (IVIS, Xenogen Corporation, Alameda, CA).
2.5 Transplantation of DF or TF ES Cells into Animal Models
Cell dissociation buffer (Invitrogen, Gibco, Carlsbad, CA).
PBS.
Growth factor reduced, LDEV-free ES cell compatible Matrigel (BD, Franklin Lakes, NJ).
Portable anesthesia system (Molecular Imaging Products, Bend, OR).
28.5 gauge insulin syringe (BD, Franklin Lakes, NJ).
2.6 Longitudinal Imaging of Transplanted Cells Using BLI
Xenogen IVIS machine with portable anesthesia system.
D-luciferin prepared as above (1 gram D-luciferin dissolved in 22 ml PBS).
28.5 gauge insulin syringe.
BLI acquisition and analysis software package such as Living Image 3.1 (Caliper Life Sciences, Hopkington, MA).
2.7 Longitudinal Imaging of Transplanted Cells Using PET
[18F]FHBG: A cyclotron is needed to produce [18F]FHBG. If your institution does not have a cyclotron facility, order from a cyclotron facility with expertise in [18F]FHBG synthesis.
Dose calibrator (Capintec, Ramsey, NJ).
Portable anesthesia machine.
A MicroPET scanner such as the MicroPET rodent R4 (Concorde Microsystems, Knoxville, TN).
A MicroPET acquisition and analysis software package such as ASI Pro (Concorde Micorystems, Knoxville, TN).
3. Methods
The following steps describe the derivation of murine and human ES cell lines that stably express DF (eGFP-Fluc) and TF (mRFP-Fluc-HSVtk) reporter genes, as well as imaging protocols to monitor the survival, migration, and proliferation of these cells. Because the emphasis of this chapter is on reporter gene transduction and imaging, we have not included instructions on how to make the reporter gene plasmid constructs used in Section 3.1 below. For a description of the PCR and standard molecular cloning techniques used to make these constructs please refer to De et. al 11, Ray et. al 12, and Cao et. al 4.
3.1 Production of DF or TF Lentiviral Transduction Vector
Plate approximately 5×106 HEK 293-FT cells onto a 100 mm culture dish using 10 ml of HEK 293-FT culture medium. Incubate at 37°C overnight.
After a minimum of 12 hours, combine 60 µl of Lipofectamine 2000 with 1.5 ml Opti-MEM I medium in a 15 ml conical centrifuge tube. Incubate at room temperature for 5 minutes.
While the diluted Lipofectamine is incubating, mix 12 µg of the DF or TF plasmid construct, 8 µg of the psPAX2 packaging plasmid, and 4 ug of the pMD2G envelope plasmid with 1.5 ml of Opti-MEM I medium in a separate 15 ml conical centrifuge tube.
Combine the diluted Lipofectamine solution with the plasmid mixture and pipet gently to homogenize. Incubate the solution at room temperature for 20 minutes.
Remove the 100 mm HEK 293-FT dish from the incubator and add the plasmid:Lipofectamine 2000 mixture into the culture medium dropwise. Incubate the plate at 37 °C for 6 hours.
After 6 hours, aspirate the transduction medium and add 10 ml of fresh HEK 293-FT growth medium. Incubate the cells at 37 °C for 24 hours.
The next day, the culture medium will be saturated with virus released by the HEK 293-FT cells. Collect the transduction medium and place it in a 50 ml conical centrifuge tube for storage at 4 °C. Add 10 ml fresh HEK 293-FT growth medium to the cells. Incubate the cells at 37 °C again for 24 hours.
After 24 hours, collect the culture medium again from the plate and combine it with the transduction medium isolated from the previous day. The total volume should be close to 20 ml. Centrifuge the combined transduction medium at 3000× g for 15 min at 4 °C.
Pass the supernatant through a 0.45 µm filter into a separate 50 ml conical centrifuge tube and mix with 5 ml 5× PEG-it Lentivirus Concentration Solution. Incubate at 4 °C for at least 12 hours. This mixture can be stored at 4 °C for up to 2 weeks.
When ready to make the final lentivirus, centrifuge the viral particle:PEG-it mixture at 1500× g for 30 min at 4 °C. The viral particles will form a pellet. Aspirate the supernatant and centrifuge a second time at 1500× g for 5 minutes at 4 °C. Remove any remaining supernatant. Resuspend the pellet in 200 µl of Opti-MEM I medium. Once the lentiviral vector is created, it may be used immediately or stored at −80 °C for up to 3 months.
3.2 Lentiviral Transduction of ES Cells
Prepare a 6 well plate with a feeder layer of inactivated mouse embryonic fibroblasts (MEFs) by coating all 6 wells of a 6-well plate with 2 ml of autoclaved 0.1% gelatin. Incubate the plate at room temperature for at least 15 minutes.
Plate ~3 × 10° inactivated mouse embryonic fibroblasts (MEFs) in 2 ml of MEF growth medium in each well of the 6-well plate. Incubate at 37 °C for 24 hours to allow for the feeder cells to firmly attach to the bottom of the dish.
After 24 hours, verify the feeder layer has firmly attached by looking under a light microscope and aspirate the MEF medium. Plate ~6 × 10° human or mouse ES cells in each well suspended in 2 ml of the appropriate ES cell medium. Incubate at 37 °C for 24 hours.
The next day, replace the culture medium with fresh ES cell medium. Look at the cells under a light microscope to measure confluence. If the cells have are close to confluence, proceed to step 5. If not, replace the existing medium with 2 ml of fresh ES cell medium and check on the cells again the next day. Mouse ES cells will typically reach confluence 2–3 days after plating. Human ES cells may take up to 4–5 days. For optimal transduction, plates should be at least at 80% confluent and colonies should be no larger than 200–400 cells per colony.
Once the cells have reached confluence, prepare the lentivirus transduction medium in a 15 ml conical centrifuge tube. Thaw the viral stock made in Part 3.1 above and add it at a multiplicity of infection (MOI) of 10 to 5 µL of polybrene and 2 ml of fresh ES cell medium for each well of the 6-well plate to be transduced.
Aspirate the ES cell medium from each well of the 6-well plate and replace it with 2 ml of the transduction medium. Incubate at 37 °C for 24 h.
At 24 hours, aspirate the transduction medium and replace it with 2 ml of fresh ES cell medium per well. Continue to incubate the cells at 37 °C for a second day, and change the cell medium again.
2 days after transduction, look at the cells under an inverted epifluorescence microscope to verify successful uptake and expression of the reporter gene. Estimate the percentage of cells that are GFP or RFP positive. Typically, at least 30–40% of total cells must be GFP or RFP positive for successful cell cytometry sorting (Figure 2B).
3.3 Derivation of Stable DF or TF ES Cell Lines
The day before cell sorting, prepare a 12-well plate with a MEF feeder layer by incubating 1 mL of 0.1% autoclaved gelatin in each well at room temperature for a minimum of 15 minutes. Aspirate the gelatin and seed 3 × 10° MEFs per well in 1 mL of MEF medium. Incubate at 37 °C for 24 hours.
The next day, remove the transduced ES cells from the incubator and aspirate the culture medium. Wash the cells with 1 ml of PBS per well and aspirate. Add 300 µl of cell dissociation buffer to each well and incubate at 37 °C for 5–10 minutes.
After 5–10 minutes, begin monitoring the cells under a light microscope. Once the cells begin to detach from one another, add 1 ml of ES cell medium to each well to dilute the dissociation buffer. Use a 2 ml serological pipet or cell lifter to gently break the ES cell colonies into a single cell solution and and transfer the cells into a 15 ml conical centrifuge tube.
Pass the single cell suspension from each well through a 70 µm cell strainer and collect it in a 50 ml conical centrifuge tube. Centrifuge the cell suspension at 500× g for 2 minutes at 4 °C. Aspirate the supernatant and add 5 ml PBS into the centrifuge tube to wash the cells a second time. Centrifuge the cells again at 500× g for 2 minutes at 4 °C. Aspirate the supernatant and resuspend the cells in 1 ml PBS. Estimate the number of cells in suspension using a hemocytometer and add 10 µl of propium iodide buffer to the suspension for every 3 × 105 cells in the mixture. Transfer this solution to a FACS tube and place on ice.
Isolate the GFP or RFP positive cells with a sterile fluorescence activated cell sorter. Plate these cells on the 12-well plate prepared in Step 1 of this section. For optimal cell survival, plate at least 50,000 cells per well. Incubate the cells at 37 °C for 48 hours. Use ES cell medium supplemented with 1% penicillin/streptomycin and 0.25 µg/ml fungizone for cell culture to prevent contamination. It will take close to a week to expand these cells to a sizeable number for a second sort.
After 48 hours, aspirate the ES cell medium, replace with 1 ml of fresh medium, and incubate the cells again at 37 °C. Continue to culture cells, changing media every day and monitoring the cells once a day under a microscope. After 6–9 days, the sorted ES cells should reach a colony size of 300–500 cells per colony. When colonies of this size are reached, prepare a second 12-well plate with a MEF feeder layer as in Step 1 of this section and proceed to the next step.
Place the 12-well dish under a fluorescence microscope and using permanent marker, clearly circle the location of all GFP/RFP positive colonies on the cover of the dish. Use a sterilized glass pipet tip or cell lifter to dislodge the GFP/RFP positive colonies and transfer them to a new MEF coated 12-well plates using a p200 Gibson pipet. Incubate the cells at at 37 °C for 24 hours.
Replace the ES cell medium every 24 hours and continue to culture the cells until they are confluent (5–7 days). When cells near confluency, prepare another 12-well plate with a MEF feeder layer.
When the cells reach confluency, some of the colonies will be more differentiated than others and exhibit less of a stereotypical ES cell morphology. Examine the dish under a microscope and use a permanent marker to clearly mark all differentiated colonies. As in Step 7, use a sterilized glass pipet tip or cell lifter to dislodge the differentiated colonies. Aspirate the culture medium and floating colonies, eliminating them from the dish and leaving only colonies with stereotypical ES cell morphology attached to the plate.
Replace the culture medium with 500 µl of collagenase IV per well prepared at a concentration of 1mg/ml DMEM:F12. Incubate the cells at 37 °C for 5 minutes. Then use a cell lifter to split these colonies onto a new 12-well plate with MEF feeder layer. Culture these plates until cells are confluent.
11. After the split cells reach confluency, prepare for a second round of cell sorting by plating another 6-well culture dish with a MEF feeder layer. Harvest ES cells using the methodologies listed in Steps 3–5 of this section. Sort the cells with a sterile fluorescent activated cell sorter and plate them onto the new MEF coated dish as outlined in Step 6 of this section. These double sorted GFP/RFP positive cells should grow stably, express the reporter genes, and can be used for in vivo transplantation. Cells can be frozen at −80°C for future use.
3.4 Verification of Transduction through In Vitro Imaging
To confirm successful transduction of ES cells, it is useful to take cell plate images of bioluminescence signals and correlate to cell numbers. To image luciferase positive cells, you will need to have a working solution of the reporter probe D-luciferin already prepared. Dissolve a stock of D-luciferin at a concentration of 45 mg/ml PBS (1 gram of d-luciferin in ~22 ml PBS). Store the prepared D-luciferin in 1–1.5 ml aliquots under tin foil at −20 °C for future use.
Aspirate the ES cell medium of 1–2 wells of a 6-well plate and replace it with 1 ml of cell dissociation buffer per well. Incubate the plate at 37 °C for 10 minutes and scrape the well with a cell scraper. Transfer the cells into a 15 ml conical centrifuge tube. Centrifuge the cells at 500× g and aspirate the supernatant. Resuspend the cells in 1 ml of PBS and transfer the cells to a 1.5 ml eppendorf tube on ice. Count the cells with a hemacytometer.
In a 24 well plate, seed a serially diluted series of cells (for example, 5 × 105 cells, 1 × 106 cells, 2 × 106 cells, etc) in a 5 or more wells (Figure 3A).
Thaw a 1 ml aliquot of working solution of D-luciferin and dilute it in 99 ml of PBS. Add 1 ml of diluted D-luciferin to each well of the serially diluted series of cells. BLI signal should be immediately detectable.
Quickly image the cells using an IVIS Xenogen machine. Begin with acquisition intervals of 10 seconds. If the signal is saturated, reduce the acquisition interval. If signal appears to be weak, increase the acquisition interval as necessary.
Acquire cell plate images of bioluminescence signals repeatedly over a 5–10 minute period of time.
Use the BLI acquisition and analysis program Living Image to draw regions of interest (ROIs) over the wells and record the BLI signal in units of maximum photons per second per centimeter square per steradian (photons/s/cm2/sr).
Use a graphing software program such as Microsoft Excel to plot the bioluminescence data as maximum photons per second per centimeter square per steradian (photons/s/cm2/sr) against cell number. Cell number should correlate with bioluminescence signal at an R2 value of >0.90 (Figure 3B).
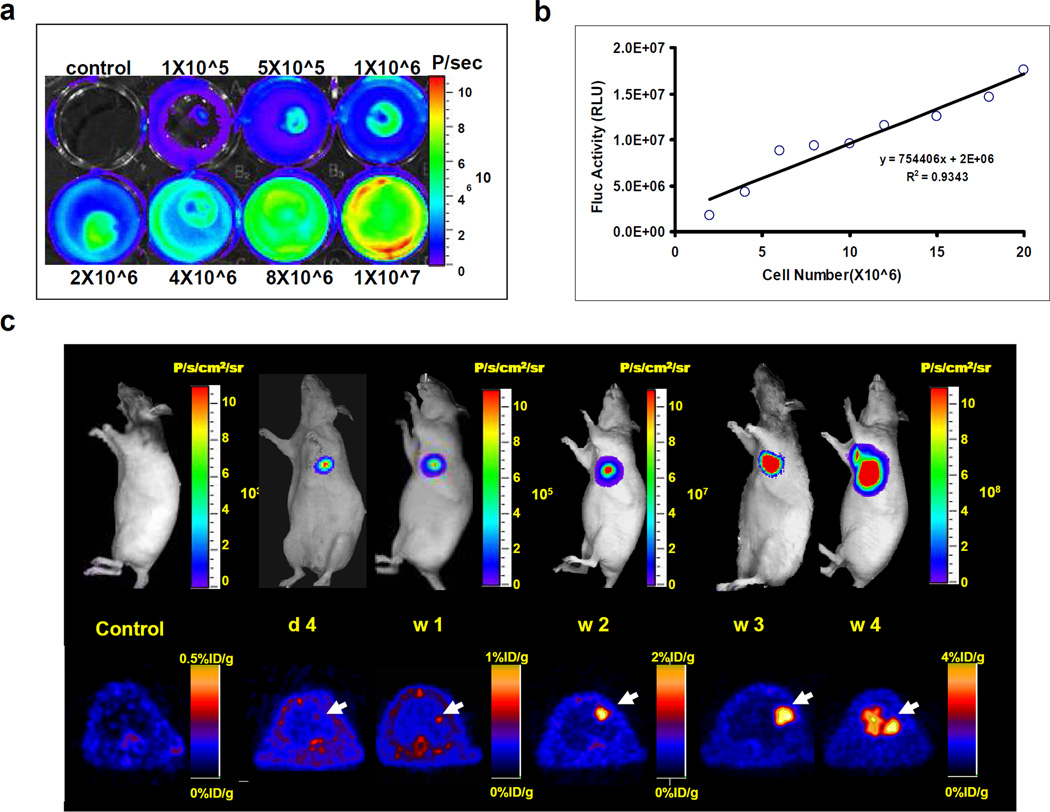
Figure 3.
Reporter gene imaging of ES cells in living animals. (a) of Fluc positive ES cells. (a) Cell plate imaging of TF positive ES cells. 1×105, 5×105, 1×106, 2×106, 4×106, 8×106, and 1×107 mouse ES cells were plated in a 6-well plate. (b) Linear correlation of cell number and BLI signal (photons/second/cm2/steradian) reveals a strong correlation (R2=0.93). (c) Longitudinal imaging of transplanted ES cells in an immunodeficient nude rat. Transplantation of 1×107 ES cells into the myocardium leads to teratoma growth by week 4 as monitored by progressive increases in BLI and PET signals. For PET images, the animal was injected via tail vein with close to 100 μCi of the PET reporter probe [18F]-FHBG. Images were acquired 1 hour after radiotracer administration to allow for adequate biodistribution. (Reproduced from reference #4 with permission from Lippincott Williams & Wilkins)
3.5 Transplantation of DF or TF ES Cells into Animals
Expand DF/TF ES cells to a sufficient number for transplantation. For ES cell injections, our group will typically inject at least 10 thousand cells. Injecting higher cell numbers (e.g., 1 million cells) will yield higher engraftment and faster teratoma formation.
For each well of a 6-well plate, aspirate the ES cell media and wash cells with PBS. Aspirate the PBS and incubate ES cells in 1 ml of cell dissociation buffer at 37 °C for 10 minutes. Dilute the cell dissociation buffer with 2 ml of PBS per well and use a cell scraper to dislodge the cells. Transfer the solution to a conical centrifuge tube and spin the cells down at 800× g for 2 minutes at room temperature. Aspirate the supernatant and resuspend the cells in as low volume as possible of PBS (start with ~100 µl and increase volume as necessary). Homogenize the solution by pipetting gently. Calculate cell concentration using a hemocytometer.
Suspend the desired number of cells in a 1:1 mixture of PBS and human ES qualified Matrigel. Limit the volume of injection to less than 50 µl. Place this mixture on ice.
Using a portable anesthesia machine, anesthetize the animal designated for transplantation following the approved animal study protocol of your institution. In our laboratory, we have used 2% (mice) or 3% (rat) isoflurane to knock down animals. Shave the animal at the site of injection if the animal is not nude.
Use a 28.5 insulin syringe to administer the ES cells to the desired anatomical location. The animal can be imaged immediately after cell transplantation or alternatively hours to days later to minimize prolonged exposure to anesthesia.
3.6 Longitudinal Monitoring of Transplanted ES Cells Using BLI
To determine background bioluminescence levels, anesthetize a control animal that has not received cell transplantation. Image the animal with a Xenogen IVIS machine and record BLI signal in photons/s/cm2/sr as the background signal.
Knock down the experimental animal with 2% isoflurane. Administer 375 mg/kg body weight of D-luciferin working solution (45 mg/ml) by intraperitoneal injection with a 28.5 gauge insulin syringe. Wait for 10 min while keeping the animal anesthetized before imaging to allow for systemic absorption.
Place the animal in the imaging chamber of a Xenogen IVIS machine. Image the animal with 1 second to 5 minute acquisition intervals for 30 minutes to catch the peak BLI signals in photons/s/cm2/sr. Fluc signals will generally peak 20–30 minutes after administration 2.
Image the same animals at set time points over a defined period. We normally image the animals at days 0, 2, 4, 7, 10 and weekly thereafter (Figure 3C).
Analyze the images using the BLI analysis package Living Image.
3.7 Longitudinal Monitoring of Transplanted ES Cells Using PET
For cells transduced with the HSVtk reporter gene, PET imaging can be used to produce high intensity photons for cell localization. Prepare or order a sufficient amount of [18F]FHBG to meet imaging needs. Typically you will want approximately 100 µCi [18F]FHBG per animal. If your institution does not have a cyclotron facility to produce [18F]FHBG, order this radiotracer from an experienced cyclotron facility.
Image a control animal as in Step 1 of Part 3.6 to determine background signal.
Draw approximately 100 µCi [18F]FHBG into a 28.5 gauge insulin syringe. Record the exact activity within the syringe at time of injection using a dose calibrator. Administer the entire syringe content of [18F]FHBG into the animal via tail vein injection and use the dose calibrator after administration to record the remaining activity within the syringe. Record the time of measurement and time of injection. Wait for 55–60 minutes for the PET tracer to biodistribute before proceeding to imaging (Figure 3C).
Prior to imaging, knock the animal down using 2% isoflurane. Secure the animal onto the bed of the mircoPET scanner and image the animal as per manufacturer’s instructions. Record the time of imaging. Typical MicroPET scanners will have an acquisition and analysis software package such as ASI PRO.
Reconstruct the images with a software program provided by the MicroPET manufacturer such as ASI PRO. We have typically used filtered back projection algorithms to reconstruct the images.
Use a software package such as ASI PRO or A Medical Imaging Data Examiner (AMIDE) to analyze the reconstructed images.
Image the animal at set time points over a defined period. Because PET imaging is considerably more expensive than BLI and [18F]FHBG may be hard to acquire, we usually image at a weekly or monthly intervals.
4. Notes
Human ES cells can be transduced on MEF feeder layers or in feeder free conditions. While the original transductions performed in our laboratory were performed on feeder layers, feeder free conditions using growth factor reduced, LDEV-free ES cell compatible Matrigel (BD, Franklin Lakes, NJ), and mTeSR-1 (Stem Cell Technologies, Vancouver, Canada) will maximize transduction efficiency by eliminating MEF uptake of the reporter gene. In our experience, continuous culture in feeder free conditions leads to higher levels of ES cell differentiation as compared to culture on feeder layers. To transduce human ES cells in feeder free conditions, use Matrigel coated 6-well plates in lieu of MEF feeder layers. mTeSR-1 should be used in place of human ES cell medium. All other steps are the same. Once DF or TF ES cell lines have been established, we recommend continuous culture on MEF feeder layers. We have found this to be more effective in keeping cells in an undifferentiated state as compared to feeder free conditions. Mouse ES cells can be transduced directly on MEFs.
To calculate multiplicity of infection (MOI), please refer to Tiscornia et. al 13.
Our laboratory uses standard WiCell public protocols for ES cell culture and maintenance. For guidance with efficacious ES cell culture please consult the WiCell website at: http://www.wicell.org
When dissociating or splitting cells using cell dissociation buffer or collagenase IV, monitor cells under a light microscope after 5–10 minutes to monitor for overdigestion. Overdigestion of cells by dissociation buffers may compromise cell quality.
Prior to transplantation of human DF or TF ES cells, we recommend splitting to feeder free conditions for 1–2 passages. This will increase purity of transplanted ES cells by eliminating presence of MEFs.
For intramyocardial injections, suspension of ES cells in Matrigel may lead to higher levels of mortality due to formation of clots and emboli. We recommend suspending DF or TF ES cells in PBS alone for cardiovascular injection.
While the maximum volume we recommend cells to be suspended in for murine injection is 50 µl, the lower the total volume of suspended cells, the lower likelihood cells will be dispersed following transplantation. Dispersal of ES cells following transplantation leads to reduced rates of engraftment and cell survival.
Following cell transplantation BLI will generally reveal acute cell death (signal decline) followed by cell proliferation (signal gain). To monitor the phenomenon of cell death followed by proliferation we typically acquire images at days 0, 2, 4, 7, 10 and 14. After day 14, cellular growth is monitored weekly.
When recording BLI signal, acquire images serially for a 30 minute period following injection. BLI signal should peak 20–30 minutes after intraperitoneal administration of D-luciferin. Record and average the top three signal intensities as the peak value.
Acknowledgements
This work was supported by Howard Hughes Medical Institute research fellowship (AL), R21 HL091453 (JCW), and R21/R33 HL089027 (JCW).
References
- 1.Thomson JA et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Wu JC et al. Proteomic analysis of reporter genes for molecular imaging of transplanted embryonic stem cells. Proteomics. 2006;6:6234–6249. doi: 10.1002/pmic.200600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JC et al. Transcriptional profiling of reporter genes used for molecular imaging of embryonic stem cell transplantation. Physiol Genomics. 2006;25:29–38. doi: 10.1152/physiolgenomics.00254.2005. [DOI] [PubMed] [Google Scholar]
- 4.Cao F et al. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005–1014. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swijnenburg RJ et al. In vivo imaging of embryonic stem cells reveals patterns of survival and immune rejection following transplantation. Stem Cells Dev. 2008;17:1023–1029. doi: 10.1089/scd.2008.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swijnenburg RJ et al. Immunosuppressive therapy mitigates immunological rejection of human embryonic stem cell xenografts. Proc Natl Acad Sci U S A. 2008;105:12991–12996. doi: 10.1073/pnas.0805802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao F et al. Molecular imaging of embryonic stem cell misbehavior and suicide gene ablation. Cloning Stem Cells. 2007;9:107–117. doi: 10.1089/clo.2006.0E16. [DOI] [PubMed] [Google Scholar]
- 8.Cao F et al. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS ONE. 2008;3:e3474. doi: 10.1371/journal.pone.0003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z et al. Comparison of reporter gene and iron particle labeling for tracking fate of human embryonic stem cells and differentiated endothelial cells in living subjects. Stem cells (Dayton, Ohio) 2008;26:864–873. doi: 10.1634/stemcells.2007-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z et al. Differentiation, survival, and function of embryonic stem cell derived endothelial cells for ischemic heart disease. Circulation. 2007;116:I46–I54. doi: 10.1161/CIRCULATIONAHA.106.680561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De A, Lewis XZ, Gambhir SS. Noninvasive imaging of lentiviral-mediated reporter gene expression in living mice. Mol Ther. 2003;7:681–691. doi: 10.1016/s1525-0016(03)00070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray P, De A, Min JJ, Tsien RY, Gambhir SS. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res. 2004;64:1323–1330. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]