Abstract
Nanoparticles conjugated with D-maltoheptaose (G7) showed a striking increase in the internalization by Escherichia coli. This applies to strains with and without the maltodextrin transport channel and particles ranging from a few to a hundred nanometers.
A plethora of nanomaterials has been developed and has shown high potentials as vessels carrying therapeutic and/or diagnostic agents to treat human diseases.1 Because of their small size and large specific surface area, nanomaterials can accommodate high density of ligands to target specific disease locations such as cancer cells or pathogenic microorganism. In theranosis, efficient transport and uptake of nanomaterials by the target biological species is of paramount importance. Thus, surface modification of nanomaterials that can specifically target and gain entry into pathological bacteria or cancerous cells can greatly facilitate the diagnosis of disease states and the development of effective therapeutics.2 A number of studies have been conducted to understand the mechanism of translocation and nanoparticle passage to cells. These studies conclude a receptor-mediated endocytosis to account for the uptake of nanoparticles by mammalian cells.3
For bacterial cells, however, the conventional wisdom does not support the mechanism of endocytosis, pinocytosis, or exocytosis.4 Nonetheless, nanoparticles, for example, Ag nanoparticles, and ceramic powders of ZnO and MgO have shown remarkable antibacterial activities.5 In these cases, the damage of bacterial cell membranes by released metal ions frequently follows the internalization of nanoparticles, and the cellular response correlates strongly with the nanoparticle uptake.5a,6 It can therefore be envisioned that surface ligands that can improve the uptake of nanomaterials to bacterial cells should provide a powerful means of targeting a payload delivery to a potential disease causing strain. In this article, we investigate the impact of surface-conjugated D-maltoheptaose (G7) on the internalization of nanoparticles into cell-wall containing enteric bacteria Escherichia coli (E. coli), a rod-shaped gram-negative prokaryotic organism. Protecting the intracellular matrix of the E. coli cell is a capsule with a triple layer peptidoglycan cell wall. The outer membrane of E. coli is rendered as a molecular sieve in which the porins allow passage for nutrients, and plays a major role in cell-cell interaction, cell recognition and cell to solid phase attachment.7 Maltodextrin, a major source of glucose for metabolic activity, is a preferred nutrient in E. coli metabolism. Among maltodextrins, G7 is the largest carbon source that can be transported into cytoplasm and utilized by E. coli.8 The trimeric LamB porins on the outer membrane are responsible for the guided diffusion of maltodextrin from the extracellular medium to the periplasmic space, where it binds to maltodextrin-binding protein for active transport towards the intracellular matrix.9 Larger maltodextrins, although not transported into the cytoplasm, can nevertheless bind the LamB porin receptor and the periplasmic maltose binding protein with good affinity.8
In this study, we conjugate G7 on nanoparticles (NPs), and investigate the uptake of G7-NP conjugates by E. coli. In particular, we aim at testing whether the immobilized G7 would promote the internalization of NPs. Studies were conducted using silica nanoparticles (SNPs), magnetic nanoparticles (MNPs), silica-coated magnetic nanoparticles (SMNPs) and silica-coated quantum dots (SQDs). The G7 conjugates of these nanoparticles were then treated with four different strains of E. coli and the uptake of the nanoparticles by the bacteria cells were analyzed.
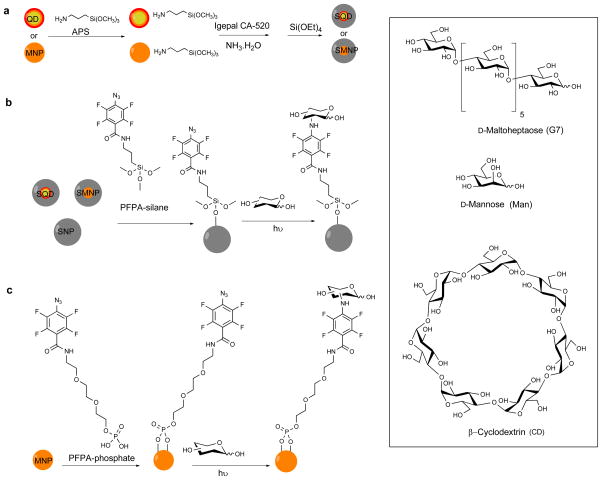
MNPs were prepared by heating iron acetylacetonate, 1,2-hexadecanediol and oleic acid in benzyl ether at 200 °C for 2 h under Ar followed by 300 °C for 1 h (Scheme 1a).10 Silica coating was introduced to MNPs using the reverse micelle method by treating MNPs with APS followed by IGEPAL, NH4OH and TEOS (see ESI† for experimental details).11 TEM image showed successful encapsulation of MNPs in silica where individual MNPs (5.6 ± 0.7 nm, Figure S1a, ESI†) were evenly coated with a silica layer (25.2 ± 2.1 nm, Figure S1b). Core-shell QDs (CdSe/CdS/ZnS) were prepared following the procedure of Peng et al.12 Silica coating was subsequently introduced following the reverse micelle method to give SQDs (Scheme 1a, see ESI† for experimental details). The particle size increased from 8.3 ± 0.9 nm to 25.4 ± 2.2 nm after silica encapsulation (Figures S1e and S1f), and the fluorescence emission remained unchanged at 622 nm (Figure S3b). SNPs were synthesized using the classic Stöber protocol via NH4OH-mediated hydrolysis and condensation of TEOS13 to give particles of 81.2 ± 7.3 nm in size by TEM (Figure S1g) and 92.5 ± 8.7 nm by DLS (Figure S2). SMNPs, SQDs and SNPs were functionalized with PFPA by treating the particles with PFPA-silane (Scheme 1b).14 MNPs without the silica coating (5.6 ± 0.7 nm, Figure S1a) were treated with PFPA-phosphate having the ethylene oxide spacer to enhance the solubility of the resulting PFPA-functionalized MNPs (Scheme 1c).15 Carbohydrate immobilization was carried out using the photocoupling method developed previously in our laboratory by irradiating the nanoparticles in the presence of the carbohydrate followed by dialysis (Scheme 1b, 1c, see ESI† for experimental details).15–16 The density of carbohydrates on SNPs was determined from a colorimetric assay using anthrone/sulfuric acid16a,17 to yield the surface coverage of 51 ± 9%, 48 ± 13% and 52% ± 6%, for Man, G7 and CD, respectively (Table S1).
Scheme 1.
(a) Synthesis of SMNP and SQD. (b) Conjugation of carbohydrates to SQD, SMNP, SNP. (c) Conjugation of carbohydrates to MNP.
Four strains of E. coli were used in this study: ATCC 33456, JW3392-1, ORN 178 and ORN 208. The outer membrane of ATCC 33456 is reported to have LamB porins that present binding sites for maltodextrin.8,9b E. coli strain JW3392-1 is a LamB mutant, which lacks the maltodextrin transporter channel. ORN 178 possesses FimH, a Man-binding protein, on its pili, whereas FimH is absent in the mutant ORN 208.18 When Man-conjugated nanoparticles (Man-SMNP, Man-SQD, Man-SNP) were treated with ORN 178, all three NPs were observed on the pili of the bacteria cells (Figures S4a–c). On ORN 208, however, very little nanoparticles were seen due to the absence of FimH (Figures S4d–f). These results were similar to what we observed previously,14a,15 confirming the successful conjugation of Man to these nanoparticles.
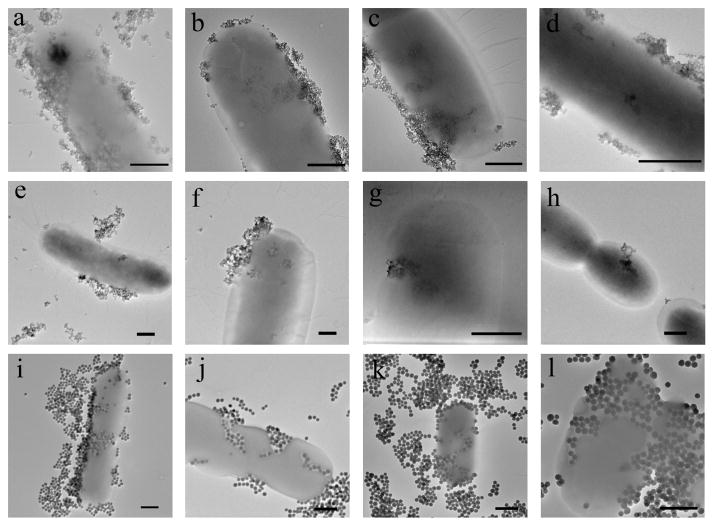
G7, a member of the maltodextrin family that contains seven glucose units via the α1→4 linkage, was used to test the hypothesis that maltodextrin would increase the uptake and internalization of nanoparticles. Thus, G7-conjugated NPs (G7-SMNP, G7-SQD, G7-SNP) were incubated at 37 °C for 2 h with E. coli strain ATCC 33456 that was harvested at 0.5 OD600. After excess nanoparticles were removed from the medium, the sample was examined by TEM. Unlike Man-conjugated NPs which were attached to the pili of ORN 178, NPs functionalized with G7 adhered to the surface of ATCC 33456 and gained entry into the cells (Figures 1a, 1e, 1i). Because ATCC 33456 possesses the maltodextrin transporter, one possibility of the uptake would be a pathway facilitated by the maltodextrin transport channel. In the work of Murthy and coworkers,19 maltohexaose was labeled with a fluorescent dye, and the dye-maltohexaose conjugate was then used as an imaging probe to detect bacteria cells. Their results demonstrated that the dye-maltohexaose conjugate was internalized by ATCC 33456 through the maltodextrin transporter pathway. The conclusion was further supported by the finding that E. coli strain JW3392-1, which lacks the maltodextrin transport channel, showed much lower cellular uptake. To test the selectivity of the nanoparticle internalization in our case, G7-NPs were treated with JW3392-1. Surface adhesion and subsequent cell wall crossing and internalization were observed for all G7-functionalized nanoparticles (Figures 1b, 1f, 1j). The experiment was then repeated on ORN 178 and ORN 208. Similar to ATCC 33456 and JW3392-1, G7-NPs were uptaken by ORN 178 (Figures 1c, 1g, 1k) as well as ORN 208 (Figures 1d, 1h, 1l). These results failed to support a mechanism involving the maltodextrin transporter.
Figure 1.
TEM images of G7-SMNP incubated with E. coli strain (a) ATCC 33456, (b) JW3392-1, (c) ORN 178, (d) ORN 208; G7-SQD incubated with E. coli strain (e) ATCC 33456, (f) JW3392-1, (g) ORN 178, (h) ORN 208; G7-SNP incubated with E. coli strain (i) ATCC 33456, (j) JW3392-1, (k) ORN 178, (l) ORN 208. Scale bars: 500 nm.
To further confirm that the nanoparticles were inside the bacteria cells, thin section samples of ATCC 33456 treated with G7-MNP were prepared. Results showed the presence of G7-MNPs inside the cytoplasm as well as throughout the cell walls (Figure 2a). A higher amount of nanoparticles were seen at locations close to the inner membrane of the bacteria cells, which may perhaps be the major barrier for the translocation of nanoparticles into the cytoplasm. In the case of G7-SNP, the nanoparticles pressed upon the bacterial cell walls and created tunnels within the cells (Figures 1i–l). Interestingly, even under the massive invasion of G7-SNPs, the bacterial cells did not seem to lose cell contents. Confocal laser scanning image also confirmed that the cells were intact (Figure 3).
Figure 2.
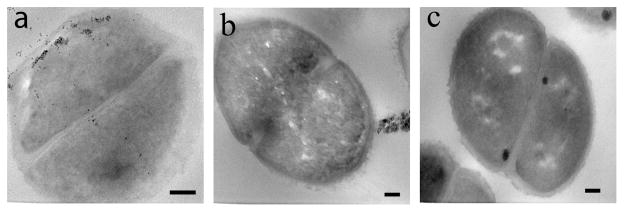
TEM thin section images of ATCC 33456 after treating with (a) G7-MNP and (b) CD-MNP. (c) TEM thin section image of ATCC 33456. Scale bars: 100 nm.
Figure 3.
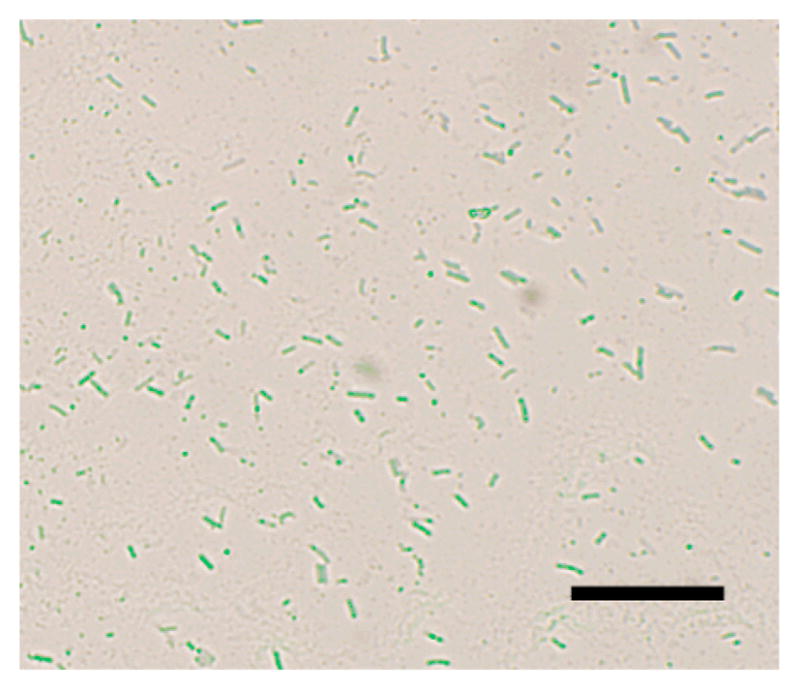
Overlay of confocal laser scanning microscopy image and bright field image of E. coli ATCC 33456 treated with G7-SNP. SNPs were doped with fluorescein.17 Scale bar: 10 μm.
The viability of the bacteria after treating with NPs was then tested. E. coli ATCC 33456 incubated with different NPs was subjected to resazurin-based cytotoxicity assay. High cell viability of 94%, 99% and 88% was obtained for G7-SMNP, G7-SQD and G7-SNP, respectively (Table S2). As a comparison, the viability of cells after treating with unfunctionalized nanoparticles was 97%, 98 and 98% for SMNP, SQD and SNP, respectively (Table S2).
Control experiments were carried out using unfunctionalized NPs and NPs functionalized with β-cyclodextrin (CD). CD is a cyclic member of the maltodextrin family, and like G7, it contains seven glucose units via the α1→4 linkage. The nanoparticles were treated with the four strains of E. coli under the same conditions as G7-conjugated NPs. NPs functionalized with CD (CD-SMNP, CD-SQD, CD-SNP) showed very little surface adherence to all four E. coli strains (Figures S5). Among unfunctionalized nanoparticles, SMNP and SQD had minimal surface attachment, and no visible internalization to the bacteria cells was observed (Figures S6a–h). SNP showed some surface adhesion, likely due to the non-specific adsorption of SNPs on the bacterial cells (Figures S6i–l). The uptake and internalization observed in the G7-functionalized NPs was also absent in these samples. Detailed examination from the thin section sample of ATCC 33456 treated with CD-MNP further supported the lack of internalization of nanoparticles conjugated with CD (Figure 2b). These data strongly support that the internalization of G7-NPs by bacteria cells was the result of the specific functionalization of G7 on the nanoparticles.
In conclusion we have demonstrated that the conjugation of a maltodextrin, G7, on nanoparticles resulted in significantly increased surface binding and internalization of nanoparticles to E. coli. This applies to silica nanoparticles, iron oxide nanoparticles, silica-coated quantum dots and iron oxide nanoparticles ranging from several to about a hundred nanometers in particle diameter, and to different E. coli strains with or without the maltodextrin transport channel. TEM images including the thin section samples revealed the uptake of nanoparticles in cell walls and inside the cytoplasm. Unfunctionalized nanoparticles and nanoparticles functionalized with CD, a cyclic analog of G7, showed little or no binding to the E. coli cell surface, and no obvious internalization of the nanoparticles was observed. Man-functionalized nanoparticles bound to the pili of ORN 178 through the well-known Man-binding lectin (FimH) rather than cell internalization. The preferential uptake of nanoparticles is clearly driven by surface G7, however, the uptake mechanism and whether the particle translocation is simply a wrapping process or involves pore creation in the membrane warrant further investigation. The inherent non-toxicity and non-immunogenicity of maltodextrin makes it an attractive promoter facilitating the translocation of nanomaterials into bacteria cells. Furthermore, maltodextrin-NP conjugates can be readily prepared using the versatile photocoupling chemistry which applies to underivatized maltodextrin structures as well as technologically important nanomaterials. This general strategy of using maltodextrin to facilitate the internalization of nanoparticles into the bacteria cells may lead to a wide range of applications in theranosis and treatment of bacterial infection.
Supplementary Material
Acknowledgments
This work was supported by NIH (R01GM080295 and 2R15GM066279), and a startup fund from University of Massachusetts Lowell. The authors thank Professor Paul Orndorff of North Carolina State University for his kind donation of E. coli strains ORN 178 and ORN 208, and Christopher Santeufemio for his assistance in the preparation of TEM thin section samples.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental details on the synthesis of nanoparticles, TEM and DLS characterization of nanoparticles, carbohydrate density measurement, TEM sample preparation and additional TEM images. See DOI: 10.1039/b000000x/
Notes and references
- 1.(a) Elsabahy M, Wooley KL. Chem Soc Rev. 2012;41:2545–2561. doi: 10.1039/c2cs15327k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang AZ, Langer R, Farokhzad OC. Annu Rev Med. 2012;63:185–198. doi: 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]; (c) Ng KK, Lovell JF, Zheng G. Acc Chem Res. 2011;44:1105–1113. doi: 10.1021/ar200017e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Prow TW, Grice JE, Lin LL, Faye R, Butler M, Becker W, Wurm EMT, Yoong C, Robertson TA, Soyer HP, Roberts MS. Adv Drug Delivery Rev. 2011;63:470–491. doi: 10.1016/j.addr.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 2.(a) Ding L, Ju H. J Mater Chem. 2011;21:18154–18173. [Google Scholar]; (b) Saha K, Bajaj A, Duncan B, Rotello VM. Small. 2011;7:1903–1918. doi: 10.1002/smll.201100478. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Verma A, Stellacci F. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]; (d) Hild W, Pollinger K, Caporale A, Cabrele C, Keller M, Pluym N, Buschauer A, Rachel R, Tessmar J, Breunig M, Goepferich A. Proc Natl Acad Sci U S A. 2010;107:10667–10672. doi: 10.1073/pnas.0912782107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Song M, Wang X. 2010 [Google Scholar]
- 3.(a) Canton I, Battaglia G. Chem Soc Rev. 2012;41:2718–2739. doi: 10.1039/c2cs15309b. [DOI] [PubMed] [Google Scholar]; (b) Cardoso MM, Peca IN, Roque ACA. Curr Med Chem. 2012;19:3103–3127. doi: 10.2174/092986712800784667. [DOI] [PubMed] [Google Scholar]; (c) Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. J Am Chem Soc. 2010;132:5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]; (d) Wang M, Thanou M. Pharmacol Res. 2010;62:90–99. doi: 10.1016/j.phrs.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Lonhienne TGA, Sagulenko E, Webb RI, Lee KC, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA. Proc Natl Acad Sci USA. 2010;107:12883–12888. doi: 10.1073/pnas.1001085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Li Y, Zhang W, Niu J, Chen Y. ACS Nano. 2012;6:5164–5173. doi: 10.1021/nn300934k. [DOI] [PubMed] [Google Scholar]; (b) Chaloupka K, Malam Y, Seifalian AM. Trends Biotechnol. 2010;28:580–588. doi: 10.1016/j.tibtech.2010.07.006. [DOI] [PubMed] [Google Scholar]; (c) Rai M, Yadav A, Gade A. Biotechnol Adv. 2009;27:76–83. doi: 10.1016/j.biotechadv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 6.(a) Yan D, Yin G, Huang Z, Li L, Liao X, Chen X, Yao Y, Hao B. Langmuir. 2011;27:13206–13211. doi: 10.1021/la2008107. [DOI] [PubMed] [Google Scholar]; (b) Kumar A, Pandey AK, Singh SS, Shanker R, Dhawan A. Cytometry Part A. 2011;79A:707–712. doi: 10.1002/cyto.a.21085. [DOI] [PubMed] [Google Scholar]; (c) Prasad TNVKV, Kambala VSR, Naidu R. Curr Nanosci. 2011;7:531–544. [Google Scholar]; (d) Allahverdiyev AM, Abamor ES, Bagirova M, Rafailovich M. Future Microbiol. 2011;6:933–940. doi: 10.2217/fmb.11.78. [DOI] [PubMed] [Google Scholar]
- 7.Nikaido H, Rosenberg EY. J Bacteriol. 1983;153:241–252. doi: 10.1128/jb.153.1.241-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferenci T. Eur J Biochem. 1980;108:631–636. doi: 10.1111/j.1432-1033.1980.tb04758.x. [DOI] [PubMed] [Google Scholar]
- 9.(a) Orlik F, Andersen C, Benz R. Biophys J. 2002;82:2466–2475. doi: 10.1016/S0006-3495(02)75589-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Duan X, Hall JA, Nikaido H, Quiocho FA. J Mol Biol. 2001;306:1115–1126. doi: 10.1006/jmbi.2001.4456. [DOI] [PubMed] [Google Scholar]
- 10.Sun S, Zeng H, Robinson DB, Raoux S, Rice PM, Wang SX, Li G. J Am Chem Soc. 2003;126:273–279. doi: 10.1021/ja0380852. [DOI] [PubMed] [Google Scholar]
- 11.Selvan ST, Tan TT, Ying JY. Adv Mater. 2005;17:1620–1625. [Google Scholar]
- 12.(a) Jing P, Zheng J, Ikezawa M, Liu X, Lv S, Kong X, Zhao J, Masumoto Y. J Phys Chem C. 2009;113:13545–13550. [Google Scholar]; (b) Li JJ, Wang YA, Guo W, Keay JC, Mishima TD, Johnson MB, Peng X. J Am Chem Soc. 2003;125:12567–12575. doi: 10.1021/ja0363563. [DOI] [PubMed] [Google Scholar]
- 13.Stoeber W, Fink A, Bohn E. J Colloid Interface Sci. 1968;26:62–69. [Google Scholar]
- 14.(a) Wang X, Ramstrom O, Yan M. Chem Commun. 2011;47:4261–4263. doi: 10.1039/c0cc05299j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gann JP, Yan M. Langmuir. 2008;24:5319–5323. doi: 10.1021/la7029592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu LH, Dietsch H, Schurtenberger P, Yan M. Bioconjugate Chem. 2009;20:1349–1355. doi: 10.1021/bc900110x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Wang X, Ramström O, Yan M. Anal Chem. 2010;82:9082–9089. doi: 10.1021/ac102114z. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang X, Ramstrom O, Yan M. J Mater Chem. 2009;19:8944–8949. doi: 10.1039/B917900C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong Q, Wang X, Wang H, Kubo T, Yan M. Anal Chem. 2012;84:3049–3052. doi: 10.1021/ac203455b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris SL, Spears PA, Havell EA, Hamrick TS, Horton JR, Orndorff PE. J Bacteriol. 2001;183:4099–4102. doi: 10.1128/JB.183.13.4099-4102.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ning X, Lee S, Wang Z, Kim D, Stubblefield B, Gilbert E, Murthy N. Nat Mater. 2011;10:602–607. doi: 10.1038/nmat3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.