Abstract
We performed global gene expression analyses in mouse hearts with progressive respiratory chain deficiency and found a metabolic switch at an early disease stage. The tissue-specific mitochondrial transcription factor A (Tfam) knockout mice of this study displayed a progressive heart phenotype with depletion of mtDNA and an accompanying severe decline of respiratory chain enzyme activities along with a decreased mitochondrial ATP production rate. These characteristics were observed after 2 weeks of age and became gradually more severe until the terminal stage occurred at 10-12 weeks of age. Global gene expression analyses with microarrays showed that a metabolic switch occurred early in the progression of cardiac mitochondrial dysfunction. A large number of genes encoding critical enzymes in fatty acid oxidation showed decreased expression whereas several genes encoding glycolytic enzymes showed increased expression. These alterations are consistent with activation of a fetal gene expression program, a well-documented phenomenon in cardiac disease. An increase in mitochondrial mass was not observed until the disease had reached an advanced stage. In contrast to what we have earlier observed in respiratory chain-deficient skeletal muscle, the increased mitochondrial biogenesis in respiratory chain-deficient heart muscle did not increase the overall mitochondrial ATP production rate. The observed switch in metabolism is unlikely to benefit energy homeostasis in the respiratory chain-deficient hearts and therefore likely aggravates the disease. It can thus be concluded that at least some of the secondary gene expression alterations in mitochondrial cardiomyopathy do not compensate but rather directly contribute to heart failure progression.
Respiratory chain dysfunction can generate symptoms in almost any organ with varying ages of onset (1, 2). Many distinct genetic mitochondrial disease syndromes caused by mutations in nuclear genes or mtDNA have been extensively studied (3). Furthermore, mitochondrial dysfunction is also implicated in common age-associated diseases as well as in the normally occurring aging process (4, 5). Respiratory chain dysfunction thus plays an important role in human pathology, and it has generally been assumed that the pathogenesis is directly related to deficient mitochondrial ATP production rate (MAPR). However, there is very little direct experimental support for this hypothesis, and other mechanisms may have been overlooked. Gene expression studies of respiratory chain-deficient budding yeast cells have demonstrated a variety of responses, including major reconfiguration of the metabolic pathways and induction of peroxisomal biogenesis (6). We have generated mouse models for studies of the pathogenesis of respiratory chain deficiency in vivo by using a conditional knockout strategy to disrupt the mitochondrial transcription factor A (Tfam) gene in selected cell types (7-12). Increased cell death is a common consequence of prolonged respiratory chain deficiency, with the mechanism for cell death induction differing between affected tissues (8, 9, 13, 14). We have found evidence for tissue-specific secondary alterations of metabolism as exemplified by increased levels of transcripts encoding the glycolytic enzyme GAPDH in respiratory chain-deficient heart muscle (14) and normal GAPDH transcript levels in respiratory chain-deficient cerebral cortex (9). Our studies have also demonstrated that the mitochondrial mass increases in respiratory chain-deficient embryos and differentiated mouse tissues, suggesting that increased mitochondrial biogenesis represents a cellular compensatory mechanism. Measurement of the MAPR has demonstrated that an increase of mitochondrial mass increases overall ATP production to near normal levels in mitochondrial myopathy mice (11). MAPR may thus not be as critical for the pathophysiology of respiratory chain deficiency as previously thought, and other mechanisms, such as changes in reduction/oxidation status and secondary metabolic alterations, may be important. In this article, we have characterized temporal changes in gene expression patterns and induction of mitochondrial biogenesis in Tfam knockout mouse hearts with progressive respiratory chain deficiency. Surprisingly, we found that an early switch in metabolism may worsen the situation and contribute to the disease progression in respiratory chain-deficient hearts.
Materials and Methods
Mating and Genotyping of Transgenic Animals. TfamloxP/TfamloxP mice of mixed genetic background (10, 12) were mated to heterozygous transgenic mice (+/Ckmm-NLS-cre) (15). Double heterozygotes (+/TfamloxP,+/Ckmm-NLS-cre) were obtained and backcrossed to the TfamloxP/TfamloxP strain to generate tissue-specific knockouts (TfamloxP/TfamloxP,+/Ckmm-NLS-cre). We analyzed a total of 416 offspring from this cross and observed Mendelian distribution of the four obtained genotypes: TfamloxP/TfamloxP,+/Ckmm-NLS-cre (n = 93), TfamloxP/TfamloxP (n = 105), +/TfamloxP (n = 116), and +/TfamloxP,+/Ckmm-NLS-cre (n = 102).
RNA Isolation, Target Preparation, and Microarray Hybridization. Total RNA was isolated by using the TRIzol Reagent (GIBCO/BRL) and equal amounts of RNA from at least four animals were pooled for each experiment. Target preparation and hybridization to Affymetrix Mu11K Probe Arrays were performed according to the protocol supplied by Affymetrix (Santa Clara, CA).
Microarray Data Analysis. Raw data were analyzed with the Affymetrix software microarray suite 4.0. We used global scaling to correct for differences in hybridization performance between the arrays. Only values scored as “Present” by the software were used for analysis. Genes with no “Present” values for any of the experiments were eliminated, resulting in a data set of 7,706 genes. The intensities for each probe set, designated average differences (AvgDiff) by the software, were averaged for all experiments and used as the denominator to calculate a fold change ratio for each gene and experiment. The ratios were log transformed, and the data were analyzed by the cluster program (16). We filtered the data to keep genes with data available for at least 90% of the samples. We then performed hierarchical clustering of the arrays, by using average linkage clustering. The results were displayed with the treeview program (16). Each branch of the resulting array tree contained a number of experiments that were used as a group for further analyses. The mean intensity (mean AvgDiff) for each gene and group was calculated and compared with the other groups to obtain the fold change of their respective intensities. The Student t test was used to compare the different groups. The signal intensities for a gene represented by two or more probe sets were normalized to each other and then used to calculate the fold change and the P value. Annotations and gene ontology information were obtained from the NetAffx Analysis Center, available on the Affymetrix website (17). We further categorized the genes manually according to function as defined by public databases and literature.
Southern and Northern Blot Analysis. Hearts were collected (n = 3) and prepared for isolation of DNA and RNA as described (10). RNA and DNA levels were quantified with a Fuji BAS 2000 phosphorimager and the image gauge 3.3 software.
Transmission Electron Microscopy. Small pieces from the left myocardium (n = 3-5) were fixed in 2% glutaraldehyde, 0.5% paraformaldehyde, 0.1 M sodiumcacodylate, 0.1 M sucrose, and 3 mM CaCl2 (pH 7.4) at room temperature for 30 min, followed by 24 h at 4°C. Specimens were rinsed in a buffer containing 0.15 M sodium cacodylate and 3 mM CaCl2 (pH 7.4), postfixed in 2% osmiumtetroxide, 0.07 M sodium cacodylate, 1.5 mM CaCl2 (pH 7.4) at 4°C for 2 h, dehydrated in ethanol followed by acetone and embedded in LX-112 (Ladd, Burlington, VT) (18). Ultra-thin sections (≈40-50 nm) from longitudinal parts were cut and contrasted with uranyl acetate followed by lead citrate and examined in a Tecnai 10 transmission electron microscope (Fei, Eindhoven, The Netherlands) at 80 kV.
Volume Density Measurements. Digital images at ×6200 magnification were collected randomly on myofibrils from longitudinal sections of the myocardium. Printed digital images were used for point counting by using a 2-cm square lattice to determine the mitochondrial volume density (Vv) as described (19). A pilot study was performed to determine the number of blocks and pictures needed for an appropriate sample by using a cumulative mean plot for evaluation (19). Thus, two different blocks from one animal were sectioned, and 10 random pictures were collected from each block. The point counting was done twice, and the mean value from each picture was used.
Biochemical Evaluation of Respiratory Chain Function and Glycolysis. Heart tissue samples from the left ventricle of knockout (n = 4-5) and control mice (n = 5-6) were collected at 2, 4, and 8 weeks of age, and mitochondria were isolated. The respiratory chain enzyme activities and MAPR were measured blindly as described (11, 20). For determination of glycolytic enzyme activities, a frozen sample of heart tissue (≈10 mg) was thawed and homogenized (1:50 wt/vol) in a Potter Elvehjem homogenizer containing ice-cold Tris(hydroxymethyl)aminomethane 50 mmol/liter, EDTA 1 mmol/liter, and MgSO4 5 mmol/liter (pH 8.2) buffer. This pH was chosen to avoid acid inactivation of phosphofructokinase (PFK) (21), and it had no or very minor effects on the other enzyme activities. The homogenate was stored at -80°C for subsequent analyses of the following enzyme activities: hexokinase (HXK, EC 2.7.1.1) (22), glucose-6-phosphate isomerase (GPI, EC 5.3.1.9) (23), PFK (EC 2.7.1.11) (24), and phosphoglycerate kinase (PGK, EC 2.7.2.3) (25). All enzyme activities were determined spectrophotometrically at 35°C.
Statistical Analyses. A two-way ANOVA was used to analyze the statistical significance of observed difference in enzyme activities and MAPR.
Results
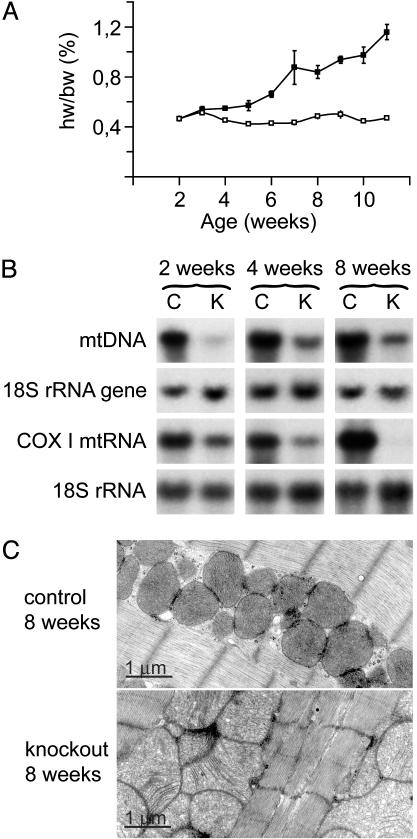
Progressive Cardiomyopathy and Increased Mitochondrial Mass in Knockout Mice. We have previously characterized a mouse strain with tissue-specific disruption of Tfam in the heart and skeletal muscle (TfamloxP/TfamloxP,+/Ckmm-cre) causing dilated cardiomyopathy, atrioventricular heart conduction blocks, and death at 2-4 weeks of age (10). This strain does not develop a muscle phenotype during their limited life span (10), consistent with our recent observation that it takes ≈4 months before TFAM protein depletion in skeletal muscle causes severely reduced MAPR (11). The usefulness of this knockout strain was limited because of short postnatal survival, and the yield of knockout mice was low due to linkage between the transgene chromosomal insertion site and the Tfam locus (10). We have now circumvented these problems by developing a novel knockout strain (TfamloxP/TfamloxP, +/Ckmm-NLS-cre). These knockouts have a longer life span of ≈10-12 weeks and show a progressive increase of heart size from ≈3 weeks of age (Fig. 1A). The new strain enabled us to follow temporal events during the progression of heart failure. The relative levels of mtDNA in knockouts in comparison with controls were 25.1 ± 4.2% (mean ± SEM) at 2 weeks, 23.8 ± 6.0% at 4 weeks, and 16.6 ± 2.9% at 8 weeks of age (Fig. 1B). The relative levels of Cox1 transcripts in knockouts in comparison with controls were 48.0 ± 5.4% at 2 weeks, 29.5 ± 5.2% at 4 weeks, and 3.4 ± 0.4% at 8 weeks of age (Fig. 1B). This result is consistent with our previous observation of different turnover rates for mtDNA and mitochondrial transcripts in Tfam knockouts (11). The mitochondria in knockout hearts had a pathological appearance on electron microscopy with abnormally shaped tubular cristae from 4 weeks of age onwards (Fig. 1C). The mitochondrial volume density was 54.2 ± 2.7% (mean ± SEM), 50.8 ± 2.0% and 45.2 ± 1.3% in control hearts at 2, 4, and 8 weeks of age whereas it was 55.5 ± 5.4%, 50.5 ± 0.1%, and 56.6 ± 0.1% in knockout hearts at 2, 4, and 8 weeks of age. This result corresponds to a ratio of mitochondrial volume density in knockouts in comparison with controls of 103 ± 9% (mean ± SEM), 100 ± 2%, and 126 ± 4% at 2, 4, and 8 weeks of age. There was thus no difference in mitochondrial volume density in knockout hearts at 2 and 4 weeks of age whereas the 8 week-old knockouts had an ≈25% increase in mitochondrial volume density.
Fig. 1.
Hearts of Tfam knockouts (TfamloxP/TfamloxP,+/Ckmm-NLS-cre) show increased size, accumulation of mitochondria, and reduced amounts of mtDNA and mtDNA-encoded cytochrome c oxidase (COX) I mtRNA. (A) Heart weight/body weight (hw/bw) ratio in knockout and control (TfamloxP/TfamloxP) mice at different ages. (B) Southern and Northern blot analysis showing levels of mtDNA and mtRNA in knockout (K) and control (C) mouse hearts at 2, 4, and 8 weeks of age. Membranes were first hybridized with a Cox1 DNA probe and then reprobed with an 18S rRNA DNA probe as a loading control. (C) Transmission electron microscopy performed on heart sections from knockout and control mice at 8 weeks of age.
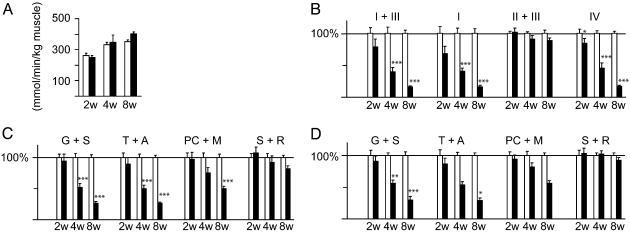
Impaired Respiratory Chain Function in Knockout Animals. We found a tendency toward increased citrate synthase activity in 8-week-old knockouts (Fig. 2A) consistent with the increase in mitochondrial mass documented by volume density measurements. The progressive reduction in mtDNA expression (Fig. 1B) correlated with a dramatic progressive respiratory chain dysfunction in knockout mice (Fig. 2B). All of the respiratory chain complexes containing mtDNA-encoded subunits were affected whereas the succinate cytochrome c reductase (complex II and III) activity, mainly dependent on the function of the nucleus-encoded complex II, was preserved. The observed MAPR was in good agreement with the enzyme activity measurements (Fig. 2 C and D). MAPR was reduced when substrates entering at the level of complex I and II (glutamate plus succinate), complex IV (N, N, N′, N′-tetramethyl-p-phenylenediamine dihydrochloride plus ascorbate), or through fatty acid oxidation (palmitoyl-l-carnitine plus malate) were added. In contrast, MAPR was essentially normal when substrates entering at the level of complex II (succinate plus rotenone) were used.
Fig. 2.
Biochemical analysis of respiratory chain function in knockout (filled bars) and control (open bars) mouse hearts at 2 (2w), 4 (4w), and 8 (8w) weeks of age. Bars present mean levels ± SEM. Asterisks indicate level of statistical significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (A) Citrate synthase (CS) activity in heart muscle. (B) Relative enzyme activities of respiratory chain enzymes. NADH cytochrome c reductase (NCR), corresponding to complex I + III; NADH coenzyme Q reductase (NQR), complex I; succinate:cytochrome c reductase (SCR), complex II + III; cytochrome c oxidase (COX), complex IV. The relative enzyme activities presented as 100% in the figure correspond to the following absolute ratios of enzyme activity per unit of CS activity at 2, 4, and 8 weeks of age, respectively: NCR, 1.32, 0.74 and 0.75; NQR, 0.35, 0.22, and 0.23; SCR, 0.77, 0.66, and 0.78; COX, 2.88, 2.45, and 2.36. (C) Measurements of MAPR, per unit of CS activity, by using substrates that enter the respiratory chain at different points. The relative MAPR/CS presented as 100% in the figure corresponds to the following absolute ratios of MAPR/unit of CS activity at 2, 4, and 8 weeks of age, respectively: glutamate plus succinate (G + S), 0.27, 0.26, and 0.27; TMPD plus ascorbate (T + A), 0.25, 0.26, and 0.26; palmitoyl-l-carnitine plus malate (PC + M), 0.22, 0.15, and 0.14; succinate plus rotenone (S + R), 0.05, 0.05 and 0.05. (D) Measurements of MAPR per kg of muscle. The relative MAPR/kg presented as 100% in the figure corresponds to the following absolute ratios of MAPR/kg of heart muscle (mmol/ATP/min/kg heart muscle) at 2, 4, and 8 weeks of age, respectively: G + S, 71, 86, and 97; T + A, 66, 86, and 94; PC + M, 57, 50, and 52; S + R, 13, 17, and 19.
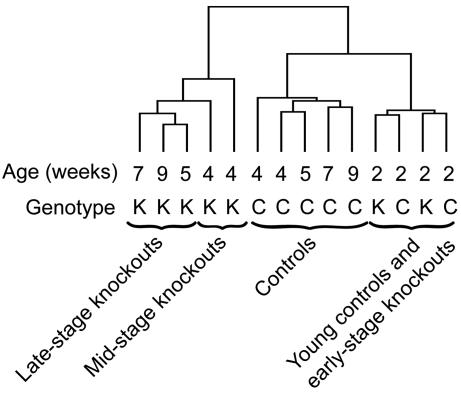
Major Alterations in Gene Expression Occur Concomitant with Onset of Respiratory Chain Deficiency. We observed no major changes in gene expression in 2-week-old knockouts, hereafter denoted “early-stage (ES) knockouts,” as their gene expression patterns clustered closely together with the patterns of the 2-week-old controls (Fig. 3). The gene expression patterns of 4- to 9-week-old knockouts clustered together and formed a separate branch on the tree distinct from the cluster formed by the age-matched controls (Fig. 3). The 5- to 9-week-old knockouts, hereafter denoted “late-stage (LS) knockouts,” formed their own subbranch of the tree separate from the 4-week-old knockouts, hereafter denoted “mid-stage knockouts (MS).” We proceeded to compare the MS and LS knockouts with the 4 to 9-week-old control group, hereafter denoted “controls” (C), and we calculated the fold change of the mean intensity for each gene. We chose to group the genes with altered expression according to their biological function and found that a substantial fraction of the genes were involved in metabolism (Tables 1 and 2). We found a profound reduction of Tfam transcripts (Table 2) confirming efficient disruption of the Tfam gene in knockout hearts. We found reduced expression of sarcoplasmic reticulum Ca2+ ATPase (Serca2) and increased expression of atrial natriuretic factor (Anf) mRNA (data not shown) consistent with our previous results (14) and observations in other heart failure models (26-28). Several changes in gene expression were compatible with compensation of impaired mitochondrial function, e.g., genes involved in import, synthesis, and turnover of mitochondrial proteins showed increased expression in MS and LS knockouts (Table 2). The majority of genes encoding respiratory chain enzyme subunits showed no change in mRNA expression in MS knockouts and reduced expression in LS knockouts (Table 2). The LON protease gene (involved in degradation of mitochondrial matrix proteins) was highly induced in MS and LS knockouts (Table 2).
Fig. 3.
Dendrogram showing results from cluster analysis of gene expression patterns. Pools of RNA from knockout (K) and control (C) mouse hearts at 2, 4, 5, 7, and 9 weeks of age were hybridized to gene microarrays. Some samples were performed in duplicate. The RNA pools with the most similar gene expression profile will cluster together within the same branch of the tree.
Table 1. Expression of glucose and fatty acid metabolism genes.
| Fold change |
||
|---|---|---|
| Gene | MS/C | LS/C |
| Glucose metabolism | ||
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase | 0.76 | 0.51** |
| Aldolase | 1.03 | 0.71* |
| Glucose transporter 4 | 0.97 | 0.66** |
| Hexokinase 1 | 1.04 | 1.26* |
| Hexokinase 2 | 1.00 | 0.74** |
| Phosphofructokinase, platelet | 2.36** | 2.12** |
| Phosphofructokinase, muscle | 0.89 | 0.59*** |
| Phosphoglycerate kinase 1 | 1.38** | 1.54* |
| Phosphoglycerate mutase 1 | 1.51*** | 2.63*** |
| Pyruvate kinase | 0.94 | 0.87* |
| Fatty acid metabolism | ||
| Acyl-CoA dehydrogenase, long-chain | 0.96 | 0.88* |
| Acyl-CoA dehydrogenase, medium-chain | 0.81* | 0.67*** |
| Acyl-CoA dehydrogenase, short-chain | 0.75* | 0.88 |
| Acyl-CoA oxidase 1, palmitoyl | 0.94 | 0.70** |
| Acyl-CoA synthetase, long-chain | 0.89 | 0.56* |
| AU RNA-binding protein/enoyl-CoA hydratase | 0.80* | 0.61** |
| Carnitine palmitoyltransferase 2 | 0.70** | 0.45 |
| Enoyl-CoA hydratase 1, peroxisomal | 0.63* | 0.39** |
| Enoyl-CoA hydratase, short-chain, 1, mitochondrial | 0.89 | 0.65** |
| Fatty acid-binding protein 3, muscle and heart | 1.55* | 0.74 |
| Fatty acid-binding protein 4, adipocyte | 0.78** | 0.79** |
| Fatty acid-binding protein 5, epidermal | 1.04 | 0.81* |
| Mitochondrial carnitine/acylcarnitine translocase | 0.82 | 0.73** |
| Peroxisome proliferator-activated receptor α | 0.44 | 0.46** |
Fold changes between MS knockouts and controls (C), and LS knockouts and controls. Asterisks represent level of significance: *, P<0.05; **, P<0.01; ***, P<0.001.
Table 2. Expression of nuclear genes encoding mitochondrial proteins.
| Fold change |
Fold change |
||||
|---|---|---|---|---|---|
| Gene | MS/C | LS/C | Gene | MS/C | LS/C |
| Respiratory chain subunits | |||||
| ATP synthase a, isoform 1 | 0.98 | 0.74*** | Cox, subunit VIIb | 0.80** | 0.79* |
| ATP synthase b | 1.04 | 0.73* | Cox, subunit VIIc | 0.99 | 0.59* |
| ATP synthase, subunit c, isoform 3 | 1.04 | 0.69* | Cox, subunit VIIIb | 0.80 | 0.71* |
| ATP synthase, subunit F | 0.95 | 0.75* | SDH, subunit A | 0.94 | 0.73* |
| ATP synthase, subunit F, isoform 2 | 0.89 | 0.79* | SDH, subunit C | 1.01 | 0.61** |
| ATP synthase, O subunit | 0.98 | 0.81* | Ubiquinol-Cyt c reductase, core protein 1 | 0.85 | 0.67* |
| Cox, subunit VIa, polypeptide 1 | 1.44 | 1.91* | Ubiquinol-Cyt c reductase, subunit VII | 0.90 | 0.74* |
| Cox, subunit VIIa, polypeptide 2-like | 0.99 | 1.29*** | |||
| Mitochondrial protein import, synthesis, and degradation | |||||
| ANT1 | 0.98 | 0.74** | Ribosomal protein L35 | 1.99*** | 2.04* |
| ANT2 | 1.41** | 1.56*** | Ribosomal protein L39 | 0.76* | 0.80* |
| Citrate carrier | 0.94 | 0.77* | Ribosomal protein S24 | 0.74** | 0.84* |
| Creatine kinase, mt 2 | 0.85 | 0.71* | Ribosomal protein S31 | 1.25** | 1.05 |
| Deoxynucleotide carrier | 0.92 | 0.61* | Ribosomal protein S34 | 1.13* | 1.70*** |
| Dicarboxylate carrier | 0.87 | 0.65** | SOD 2, mitochondrial | 0.97 | 0.77* |
| Hsp 1 (chaperonin 10) | 1.22* | 1.92 | Tfam | 0.27*** | 0.15*** |
| Hsp, A (homologous to stress-70 protein precursor) | 1.66** | 1.89** | Tim 8 homolog a (yeast) | 1.39 | 2.31* |
| Lon protease homolog precursor | 3.09*** | 4.66*** | Tim 10 homolog (yeast) | 2.00*** | 2.19*** |
| Polymerase γ | 0.84* | 0.85 | Tim 13 homolog a (yeast) | 1.07 | 1.50*** |
| Ribosomal protein L2 | 1.01 | 1.41* | Tim a | 1.45** | 1.20 |
| Ribosomal protein L3 | 1.14 | 1.76** | Tim 44 | 1.41** | 1.56*** |
| Ribosomal protein L9 | 1.39** | 1.19 | Vdac 1 | 0.94 | 0.84* |
| Ribosomal protein L15 | 1.31* | 1.29* | Vdac 2 | 1.17** | 1.34** |
| Ribosomal protein L20 | 1.48 | 2.00** | Vdac 3 | 1.44** | 1.44** |
Fold changes between MS knockouts and controls (C), and LS knockouts and controls. Asterisks represent level of significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001. ANT, adenine nucleotide translocator; SDH, succinate dehydrogenase; Tim, translocase of inner mitochondrial membrane; Vdac, voltage-dependent anion channel.
Coordinated Reduction in the Expression of Fatty Acid Oxidation Enzyme Transcripts. Transcripts encoding citric acid cycle (data not shown) and fatty acid oxidation enzymes (Table 1) were coordinately down-regulated in the knockouts. Peroxisome proliferator activated receptor α (PPARα) is a major regulator of the fatty acid oxidation pathway, and its expression was reduced in knockouts from 2 weeks of age (Table 1 and data not shown). Many PPARα target genes showed reduced mRNA expression from 4 weeks onwards, e.g., fatty acyl-CoA oxidase, acyl-CoA synthetase, carnitine palmitoyl transferase 2 (CPT-2), the acyl-CoA dehydrogenases, and the fatty acid binding proteins (Table 1). Strikingly, there was a coordinately reduced expression of genes regulating many different steps of fatty acid metabolism, e.g., fatty acid binding proteins (transport of fatty acids in cytoplasm), CPT-2, and carnitine/acylcarnitine translocase (transport of long-chain fatty acids from cytoplasm to mitochondrial matrix), acyl-CoA dehydrogenases, and enoyl-CoA hydratase (mitochondrial β-oxidation), and fatty acyl-CoA oxidase (peroxisomal fatty acid oxidation).
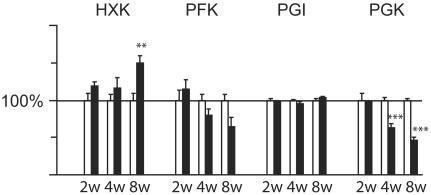
Altered Expression and Activity Levels of Glycolytic Enzymes. Several genes encoding glycolytic enzymes, e.g., hexokinase I (Hk1), PGK, and phosphoglycerate mutase, had increased expression at the RNA level in the knockouts (Table 1). The enzymatic activity of hexokinase was increased in knockouts (Fig. 4) consistent with the increased mRNA levels (Table 1). There was no change in phosphoglucose isomerase (PGI) activity (Fig. 4) in good agreement with the finding that the mRNA expression was not significantly changed (data not shown). We found a tendency of decreased PFK activity in knockouts (Fig. 4). The PFK platelet isoform mRNA was increased whereas the muscle isoform mRNA was decreased (Table 1), suggesting that the PFK muscle isoform is the main determinant of PFK activity in heart muscle. The PGK activity was significantly reduced (Fig. 4) despite the up-regulated mRNA levels (Table 1).
Fig. 4.
Biochemical analysis of glycolytic enzymatic activities. Relative enzymatic activities assayed from heart of knockout and control mice at 2, 4, and 8 weeks of age. Bars present the mean ± SEM. Asterisks indicate level of statistical significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001. The relative enzymatic activities presented as 100% in the figure correspond to the following absolute enzymatic activities (mmol/min per kg of heart tissue) at 2, 4, and 8 weeks of age, respectively: hexokinase (HXK) 6.6, 8.1, and 9.3; PFK 41.6, 63.9, and 50.3; phosphoglucose isomerase (PGI) 229, 267, and 279; PGK 121, 171, and 190.
Discussion
We have performed global gene expression analyses in mouse hearts with progressive respiratory chain deficiency, leading to cardiac dysfunction, and have revealed that there is an early switch in metabolism. Several genes encoding glycolytic enzymes show increased expression whereas genes encoding enzymes in fatty acid oxidation show decreased expression. These alterations are consistent with activation of a fetal gene expression program in the heart, a well documented phenomenon in cardiac disease (29). The supply of oxygen is low during mammalian fetal development, and the heart therefore depends heavily on glycolysis for energy production. The preferential use of glycolysis avoids the consumption of large amounts of oxygen associated with energy production by fatty acid oxidation. Shortly after birth, the heart switches from glucose to rely on fatty acids as its principal substrate for energy production (29), leading to a dramatic induction in the expression of genes encoding fatty acid oxidation enzymes (30). Reports of patients and animal models with heart failure have consistently shown that the myocardium switches to increased dependency on glucose as the substrate during cardiac hypertrophy and failure (29). This metabolic shift coincides with a down-regulation of expression of fatty acid oxidation enzymes and has been interpreted as a reactivation of the fetal gene expression program (31). The knockout mice of this study display an early onset of reduced expression of PPARα transcripts, encoding the major regulator of the fatty acid oxidation pathway. In addition, many of the PPARα target genes show simultaneously reduced expression. Furthermore, reduced transcript levels of peroxisomal enoyl-CoA hydratase 1 and acyl-CoA oxidase indicate that there is also a down-regulation of fatty acid oxidation in the peroxisomes. These findings contrast with observations in budding yeast cells where respiratory chain deficiency is followed by induced expression of genes that encode peroxisomal enzymes (6).
In a previous study it was found that patients with different mtDNA mutations and rearrangements have an increased expression of genes encoding glycolytic enzymes in affected tissues (32). We found that the glycolytic enzyme activities do not directly correlate with the altered levels of gene expression, suggesting that there are posttranscriptional regulatory events involved. The increased transcript levels of Hk1 and decreased transcript levels of Hk2 in knockout hearts are consistent with reintroduction of a fetal expression pattern in the heart because Hk1 is predominant in the heart during fetal development whereas Hk2 is more common in the adult heart.
The knockout mice of this study display a progressive phenotype with increasing heart size, depletion of mtDNA, and severe decline of respiratory chain function in heart. We first observed abnormal mitochondrial morphology in 4-week-old knockouts whereas increased mitochondrial mass is first documented in 8-week-old knockouts. In parallel with the progression of respiratory chain dysfunction, there is an increase in the expression of genes responsible for mitochondrial protein import and mitochondrial protein synthesis, probably representing an attempt to increase the amount of respiratory chain subunits in mitochondria. The LON protease, with the role of degrading misfolded, unassembled, or oxidatively damaged proteins (33, 34), is very highly induced in the knockouts. This finding suggests that there is an increased protein turnover, possibly due to an imbalance between the availability of mtDNA- and nucleus-encoded respiratory chain subunits. The expression of nucleus-encoded respiratory chain subunits was reduced in LS knockout mouse hearts, which suggests a negative feedback mechanism to prevent bombarding mitochondria with respiratory chain subunits that cannot be assembled. The adenine nucleotide translocator 2 (ANT2) had higher expression in late-stage knockouts consistent with the suggestion that ANT2 may allow cytosolic ATP to enter mitochondria if the respiratory chain function is deficient (32, 35).
We have previously shown that respiratory chain deficiency can be partly compensated by an increase in mitochondrial mass in mitochondrial myopathy mice with skeletal muscle-specific disruption of Tfam (11). However, the situation in the heart knockouts seems to be different as the MAPR is similarly reduced when compared with either citrate synthase activity (a measure of mitochondrial mass) or total muscle mass (Fig. 2 C and D). It is possible that the high volume density of mitochondria in the heart (≈45-55%) does not permit a sufficiently large increase of mitochondrial mass to partly compensate for the deficient respiratory chain function. It is also possible that the down-regulation of the fatty acid oxidation pathway in respiratory chain-deficient hearts further aggravates the problem. It is interesting to note that we observed an increased oxidation of palmitoyl-l-carnitine in respiratory chain-deficient skeletal muscle (11) whereas it was decreased in respiratory chain-deficient heart muscle (Fig. 2 C and D).
Taken together, the differences in gene expression patterns between Tfam knockout and control hearts suggest a reactivation of the fetal gene expression program during the development of mitochondrial cardiomyopathy. Interestingly, these events occur at an early stage and precede the observed increase of mitochondrial mass. It seems unlikely that this metabolic switch would be beneficial to the myocardium in sustaining energy production. Our findings suggest that adaptive metabolic changes in fact may be part of the problem and may aggravate the consequences of cardiac respiratory chain deficiency. There is currently no efficient treatment for respiratory chain diseases and increased knowledge of the pathophysiology may reveal secondary metabolic changes that are more accessible to therapeutic intervention than the respiratory chain dysfunction per se.
Acknowledgments
We thank Greg Barsh, Stanford University, for helpful discussions. We thank Maud Åkerberg and Zekiye Cansu for excellent technical assistance. N.-G.L. is supported by the Swedish Research Council, Funds of Karolinska Institutet, Torsten and Ragnar Söderbergs Stiftelse, the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine, the Swedish Heart and Lung Foundation, and the Swedish Foundation for Strategic Research (Functional Genomics). R.W. is supported by Karolinska Institutet Fonder and Freemasons in Stockholm Foundation for Children's Welfare. N.H. holds a European Commission Marie Curie Individual Postdoctoral Fellowship.
Abbreviations: Tfam, mitochondrial transcription factor A; MAPR, mitochondrial ATP production rate; PPARα, peroxisome proliferator activated receptor α; CS, citrate synthase; PFK, phosphofructokinase; LS, late-stage; MS, mid-stage; Hk1, hexokinase I; PGK, phosphoglycerate kinase.
References
- 1.Larsson, N.-G. & Clayton, D. A. (1995) Annu. Rev. Genet. 29 151-178. [DOI] [PubMed] [Google Scholar]
- 2.Larsson, N.-G. & Luft, R. (1999) FEBS Lett. 455 199-202. [DOI] [PubMed] [Google Scholar]
- 3.Graff, C., Bui, T. H. & Larsson, N.-G. (2002) Best Pract. Res. Clin. Obstet. Gynaecol. 16 715-728. [DOI] [PubMed] [Google Scholar]
- 4.Wallace, D. C. (1992) Science 256 628-632. [DOI] [PubMed] [Google Scholar]
- 5.Wallace, D. C. (1999) Science 283 1482-1488. [DOI] [PubMed] [Google Scholar]
- 6.Epstein, C. B., Waddle, J. A., Hale, W. t., Dave, V., Thornton, J., Macatee, T. L., Garner, H. R. & Butow, R. A. (2001) Mol. Biol. Cell 12 297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li, H., Wang, J., Wilhelmsson, H., Hansson, A., Thoren, P., Duffy, J., Rustin, P. & Larsson, N.-G. (2000) Proc. Natl. Acad. Sci. USA 97 3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva, J. P., Kohler, M., Graff, C., Oldfors, A., Magnuson, M. A., Berggren, P. O. & Larsson, N.-G. (2000) Nat. Genet. 26 336-340. [DOI] [PubMed] [Google Scholar]
- 9.Sorensen, L., Ekstrand, M., Silva, J. P., Lindqvist, E., Xu, B., Rustin, P., Olson, L. & Larsson, N. G. (2001) J. Neurosci. 21 8082-8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, J., Wilhelmsson, H., Graff, C., Li, H., Oldfors, A., Rustin, P., Bruning, J. C., Kahn, C. R., Clayton, D. A., Barsh, G. S., Thoren, P. & Larsson, N.-G. (1999) Nat. Genet. 21 133-137. [DOI] [PubMed] [Google Scholar]
- 11.Wredenberg, A., Wibom, R., Wilhelmsson, H., Graff, C., Wiener, H. H., Burden, S. J., Oldfors, A., Westerblad, H. & Larsson, N.-G. (2002) Proc. Natl. Acad. Sci. USA 99 15066-15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsson, N.-G., Wang, J., Wilhelmsson, H., Oldfors, A., Rustin, P., Lewandoski, M., Barsh, G. S. & Clayton, D. A. (1998) Nat. Genet. 18 231-236. [DOI] [PubMed] [Google Scholar]
- 13.Silva, J. P. & Larsson, N.-G. (2002) Biochim. Biophys. Acta 1555 106-110. [DOI] [PubMed] [Google Scholar]
- 14.Wang, J., Silva, J. P., Gustafsson, C. M., Rustin, P. & Larsson, N.-G. (2001) Proc. Natl. Acad. Sci. USA 98 4038-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruning, J. C., Michael, M. D., Winnay, J. N., Hayashi, T., Horsch, D., Accili, D., Goodyear, L. J. & Kahn, C. R. (1998) Mol. Cell 2 559-569. [DOI] [PubMed] [Google Scholar]
- 16.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, G., Loraine, A. E., Shigeta, R., Cline, M., Cheng, J., Valmeekam, V., Sun, S., Kulp, D. & Siani-Rose, M. A. (2003) Nucleic Acids Res. 31 82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Förster, C., Mäkela, S., Wärri, A., Kietz, S., Becker, D., Hultenby, K., Warner, M. & Gustafsson, J. Å. (2002) Proc. Natl. Acad. Sci. USA 99 15578-15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weibel, E. (1979) Stereological Methods: Practical Methods for Biological Morphometry (Academic, London), Vol. 1.
- 20.Wibom, R., Hagenfeldt, L. & von Dobeln, U. (2002) Anal. Biochem. 311 139-151. [DOI] [PubMed] [Google Scholar]
- 21.Mansour, T. E. (1965) J. Biol. Chem. 240 2165-2172. [PubMed] [Google Scholar]
- 22.Bass, A., Brdiczka, D., Eyer, P., Hofer, S. & Pette, D. (1969) Eur. J. Biochem. 10 198-206. [DOI] [PubMed] [Google Scholar]
- 23.Beutler, E., Blume, K. G., Kaplan, J. C., Lohr, G. W., Ramot, B. & Valentine, W. N. (1977) Br. J. Haematol. 35 331-340. [DOI] [PubMed] [Google Scholar]
- 24.Opie, L. H. & Newsholme, E. A. (1967) Biochem. J. 103 391-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiMauro, S., Dalakas, M. & Miranda, A. F. (1983) Ann. Neurol. 13 11-19. [DOI] [PubMed] [Google Scholar]
- 26.Arai, M., Matsui, H. & Periasamy, M. (1994) Circ. Res. 74 555-564. [DOI] [PubMed] [Google Scholar]
- 27.Razeghi, P., Young, M. E., Alcorn, J. L., Moravec, C. S., Frazier, O. H. & Taegtmeyer, H. (2001) Circulation 104 2923-2931. [DOI] [PubMed] [Google Scholar]
- 28.Wankerl, M. & Schwartz, K. (1995) J. Mol. Med. 73 487-496. [DOI] [PubMed] [Google Scholar]
- 29.Lehman, J. J. & Kelly, D. P. (2002) Clin. Exp. Pharmacol. Physiol. 29 339-345. [DOI] [PubMed] [Google Scholar]
- 30.Kelly, D. P., Gordon, J. I., Alpers, R. & Strauss, A. W. (1989) J. Biol. Chem. 264 18921-18925. [PubMed] [Google Scholar]
- 31.Sack, M. N., Rader, T. A., Park, S., Bastin, J., McCune, S. A. & Kelly, D. P. (1996) Circulation 94 2837-2842. [DOI] [PubMed] [Google Scholar]
- 32.Heddi, A., Stepien, G., Benke, P. J. & Wallace, D. C. (1999) J. Biol. Chem. 274 22968-22976. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, C. K., Suda, K., Wang, N. & Schatz, G. (1994) Science 264 273-276; and [DOI] [PubMed] [Google Scholar]; erratum (1994) 264 891. [Google Scholar]
- 34.Bota, D. A. & Davies, K. J. (2002) Nat. Cell. Biol. 4 674-680. [DOI] [PubMed] [Google Scholar]
- 35.Giraud, S., Bonod-Bidaud, C., Wesolowski-Louvel, M. & Stepien, G. (1998) J. Mol. Biol. 281 409-418. [DOI] [PubMed] [Google Scholar]