Abstract
Nucleotide metabolic pathways provide numerous successful targets for antiparasitic chemotherapy, but the human pathogen Cryptosporidium parvum thus far has proved extraordinarily refractory to classical treatments. Given the importance of this protist as an opportunistic pathogen afflicting immunosuppressed individuals, effective treatments are urgently needed. The genome sequence of C. parvum is approaching completion, and we have used this resource to critically assess nucleotide biosynthesis as a target in C. parvum. Genomic analysis indicates that this parasite is entirely dependent on salvage from the host for its purines and pyrimidines. Metabolic pathway reconstruction and experimental validation in the laboratory further suggest that the loss of pyrimidine de novo synthesis is compensated for by possession of three salvage enzymes. Two of these, uridine kinase-uracil phosphoribosyltransferase and thymidine kinase, are unique to C. parvum within the phylum Apicomplexa. Phylogenetic analysis suggests horizontal gene transfer of thymidine kinase from a proteobacterium. We further show that the purine metabolism in C. parvum follows a highly streamlined pathway. Salvage of adenosine provides C. parvum's sole source of purines. This renders the parasite susceptible to inhibition of inosine monophosphate dehydrogenase, the rate-limiting enzyme in the multistep conversion of AMP to GMP. The inosine 5′ monophosphate dehydrogenase inhibitors ribavirin and mycophenolic acid, which are already in clinical use, show pronounced anticryptosporidial activity. Taken together, these data help to explain why widely used drugs fail in the treatment of cryptosporidiosis and suggest more promising targets.
Keywords: Cryptosporidium parvum, horizontal gene transfer, drug target, thymidine kinase, IMPDH
The protozoan parasite Cryptosporidium parvum is widely distributed. The largest documented U.S. outbreak, in Milwaukee 1993, was responsible for an estimated 403,000 cases of symptomatic gastrointestinal disease (1). Epidemics are usually linked to contaminated drinking or recreational water. The infective oocyst stage is highly resistant to standard water treatment, which has heightened biodefense concerns. Although cryptosporidiosis is usually an acute and self-limiting infection, immunocompromized patients develop protracted and life-threatening illness (2, 3). Chronic severe diarrhea due to C. parvum is a common complication in AIDS patients and contributes to AIDS wasting syndrome and significantly shortens life expectancy. No effective drug is currently available to treat cryptosporidiosis in these patients (4). Progress in the identification of drug targets has been thwarted by the inability to culture C. parvum continuously in vitro. In light of this limitation, the complete genome sequence of C. parvum provides a critical resource.
Purine and pyrimidine nucleotides are the basic building blocks of DNA and RNA as well as crucial components of other metabolic processes and the nucleotide biosynthetic pathways are a rich source of therapeutic targets. Here we have used bioinformatic and experimental analyses to delineate this important pathway in C. parvum. We show the loss of pyrimidine de novo synthesis in this pathogen as well as the acquisition of several nucleotide salvage enzymes most likely by means of gene transfer. Pharmacological data further suggest that these divergent pathways might be exploited to develop antiparasitic drugs.
Materials and Methods
Parasites and Host Cells. C. parvum oocysts from the type 2 IOWA isolate were obtained from Michael Arrowood (Centers for Disease Control, Atlanta) and Charles Sterling (University of Arizona, Tucson). A total of 105 oocysts were added to confluent Mardin-Darby canine kidney cell (MDCK) coverslip cultures, and medium was replaced 3 h after infection to remove residual oocysts. To score C. parvum development parasites were cultured for 48 h, processed for immunofluorescence (5) and labeled with the parasite-specific monoclonal antibody c3c3 (6), an IgG3 antibody directed against meront and gamont cytoplasmic antigens, and either the DNA dye 6′-diamidino-2-phenylinidole (DAPI) or propidium iodide (PI; only PI staining is compatible with the DNA denaturation procedure used in the thymidine kinase (TK) assay described below). The number of type 1 meronts (which are unambiguously recognizable by their six to eight nuclei) was recorded for 25 microscopic fields per coverslip. Each data point represents the average of three independent coverslip cultures, the bar indicates the respective standard deviation. Toxoplasma gondii RH strain and transgenic lines derived thereof were cultured in human foreskin fibroblasts, transfected, and selected for stable plasmid integration as described (7). T. gondii growth was measured by using the fluorescence assay (8) or the monolayer disruption assay (9). Host cell growth was measured by counting nuclei per 25 fields after DAPI labeling. All drugs were obtained from Sigma, radiochemicals were obtained from Movarek (Brea, CA), and Alexa-conjugated antibodies and fluorescent dyes were obtained from Molecular Probes.
Data Mining. A custom blast searchable database of all available apicomplexan genomic, GSS, and EST sequences was constructed. This database, (ApiDB), contains a total of 443,562,576 nucleotides and the complete or nearly complete genomic sequences for Plasmodium falciparum and Plasmodium yoelii (5×) obtained from http://PlasmoDB.org, T. gondii (10×), Eimeria tenella (5×), and Theileria annulata (8×) (see below for sources) and C. parvum (7×) obtained from http://CryptoDB.org. Additional genomic, GSS, and EST sequence were incorporated from: Plasmodium chabaudi, Plasmodium vivax, Plasmodium knowlesi, T. gondii, E. tenella, and Babesia bovis, obtained from ftp://ftp.sanger.ac.uk/pub/pathogens and www.cbil.upenn.edu/paradbs-servlet/index.html. Sarcocystis neurona and Neospora caninum were obtained from GenBank. dbEST and EST cluster consensus sequences were used whenever possible. The results of tblastn (wu-blast 2.0; ref. 10) searches to identify putative homologs of the enzymes discussed in the manuscript are presented in Table 1. All blast searches used the same database, so P values are comparable. Query sequences from the same organism were not always possible; the query sequence used for each of the searches is as indicated in Table 1. Preliminary (T. gondii) genomic and/or cDNA sequence data were accessed via http://ToxoDB.org and/or www.tigr.org/tdb/t_gondii. Genomic data were obtained from The Institute for Genomic Research and by the Sanger Institute. EST sequences were generated by Washington University. These sequence data (Theileria annulata, T. gondii, P. berghei, P. chabaudi, P. reichenowi, P. vivax, E. tenella, B. bovis, Entamoeba histolytica, and Dictyostelium discoideum) were produced by the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens and www.sanger.ac.uk/Projects/D_discoideum. The sequence encoding uridine kinase-uracil phosphoribosyltransferase (UK-UPRT) in Chlamydomonas reinhardtii was kindly provided by the Chlamydomonas Resource Center, www.biology.duke.edu/chlamy_genome.
Table 1. Comparative genomic analysis of nucleotide biosynthesis in Apicomplexa.
| Gene/pathway | C. parvum | T. gondii | Theileria annulata | P. falciparum | Query |
|---|---|---|---|---|---|
| De novo pyrimidine | |||||
| Carbamoyl phosphate synthetase II | Absent | Present | Present | Present | Tg |
| P = 0.003 | P = 0.0 | P = 2.1e-307 | P = 0.0 | ||
| Aspartate carbamoyl-transferase | Absent | Present | Present | Present | Pf |
| P = 0.99 | P = 7.7e-46 | P = 3.1e-58 | P = 7.1e-184 | ||
| Dihydroorotase | Absent | Present | Present | Present | Pf |
| No hits | P = 6.2e-67 | P = 9.1e-46 | P = 7e-197 | ||
| Dihydroorotate dehydrogenase | Absent | Present | Present | Present | Pf |
| P = 0.013 | P = 3.9e-77 | P = 1.8e-31 | P = 5.3e-309 | ||
| Orototate-PRT | Absent | Present | Present | Present | Pf |
| P = 0.0014 | P = 3.7e-12 | P = 2.3e-7 | P = 3.1e-146 | ||
| Oritidine monophosphate decarboxylase | Absent | Present | Present | Present | Pf |
| P = 0.97 | P = 1.1e-5* | P = 3.23-27 | P = 3.3e-174 | ||
| Pyrimidine salvage | |||||
| UPRT | Eukaryotic | Eukaryotic | Absent | Absent | Tg |
| P = 4.1e-50 | P = 8.9e-113 | No hits | No hits | ||
| UK-UPRT | Eukaryotic | Absent | Absent | Absent | Cp |
| P = 2.1e-243 | P = 1.2e-35† | No hits | No hits | ||
| TK | Bacterial | Absent | Absent | Absent | Cp |
| P = 1.6e-101 | No hits | No hits | No hits | ||
| Dihydrofolate reductase-thymidylate synthase | Eukaryotic | Eukaryotic | Eukaryotic | Eukaryotic | Tg |
| P = 4.8e-114 | P = 1.5e-284 | P = 1.5e-66 | P = 6.2e-115 | ||
| Purine salvage | |||||
| Hypoxanthine-xanthine-guanine- PRT | Absent | Present | Absent | Present | Tg |
| No hits | P = 1.3e-149 | No hits | P = 1.9e-53 | ||
| AK | Present | Present | Absent | Absent | Tg |
| P = 1.8e-22 | P = 4.6e-192 | No hits | No hits | ||
| Inosine monophosphate dehydrogenase | Bacterial | Eukaryotic | Eukaryotic | Eukaryotic | Pf |
| P = 1.5e-27 | P = 4.6e-84 | P = 8.6e-77 | P = 7.9e-269 | ||
| Adenine-PRT | Absent | Absent | Absent | Absent | Ld |
| No hits | No hits | No hits | No hits |
All publicly available apicomplexan sequence data were combined in a database and searched for the indicated enzyme genes (see Materials and Methods for details). Statistical significance for each hit is provided as P value (note that P values are comparable here because searches were performed in the same database. The respective gene used to query the database was as indicated; Tg, T. gondii; Pf, P. falciparum; Cp, C. parvum; Ld, Leishmania donovani; PRT, phosphoribosyltransferase.
This gene is fragmented by introns. After intron removal, blast queries using the putative coding sequence results in an e-18 hit against P. falciparum OMPD.
This hit represents the unfused T. gondii UPRT (19), matching only the UPRT portion of UK-UPRT.
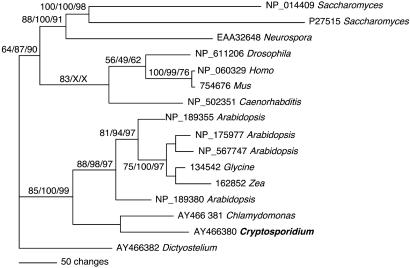
Phylogenetic Analysis. Multiple sequence alignments were created for UK-UPRT (16 taxa) and TK (21 taxa) representing the major domains of life for which sequence information exists. Note that most organisms lack a fused UK-UPRT and many organisms lack TK, thus, the trees appear taxonomically incomplete. The alignments were created with clustal x 1.8 (11) and refined by eye. Only unambiguously aligned positions were included in the analyses (see supporting information, which is published on the PNAS web site). Parsimony analyses were performed with paup* version 4.0b10 (12). A heuristic search using random stepwise addition and tree bisection-reconnection (TBR) was performed including 1,000 bootstrap replicates. A single most parsimonious tree (length = 1,699) was recovered for UK-UPRT, and four equally parsimonious trees for TK (length = 1,357) were obtained. Distance analyses were performed by using the phylip v3.6a3 programs protdist, neighbor, seqboot, and consense. The Jones-Taylor-Thornton (JTT) substitution model was used. One hundred bootstrap replicates were performed (13). Maximum likelihood analyses were performed by using tree-puzzle version 5.1 for Unix (14). Quartet likelihood used exact maximum likelihood computation, 10,000 puzzling steps were used, parameter estimation was exact. A mixed JTT substitution model with (eight γ distributed and one invariable category) was used. Parameters were estimated from the data. Trees were visualized with paup* and drawgram. UK-UPRT accession numbers are as indicated in Fig. 2, accession numbers for TK are provided in the supporting information.
Fig. 2.
C. parvum harbors a UK-UPRT with similarity to plant and algal UK-UPRT. The tree shown is the single most parsimonious tree. Numbers above the branches indicate the bootstrap values for parsimony and neighbor-joining analyses followed by maximum likelihood puzzle frequencies. Only values >50% are shown. If a given method did not provide significant support for the relationship indicated, an “X” has been inserted. The scale is as indicated. GenBank and TIGR EGO accession numbers are as listed. See Materials and Methods and supporting information for details on alignment and analysis.
Molecular Methods. The coding sequence of adenosine kinase (AK) was amplified by PCR (5′-TCGCCTAGGATGAGGGGAAAGAAAATATTTGG, 5′-AGCCTGCAGTTAATTATTTTTAGATGAAGTTAGTTG, restriction sites are underlined) using C. parvum genomic DNA as template and cloned (AvrII/PstI) into T. gondii expression plasmid CAT-GFP (5) replacing GFP. The coding region of TK was amplified by using primers (5′ATCCCTAGGATGGCAAAAATTATACTTTTACTATTC, 5′GATCTGCAGTTAGAAATTGTATTCTTCACAATTAATTATATG) and the resulting DNA molecule was radiolabeled by random priming and used as probe for Southern analysis. Isolation of genomic DNA and Southern blot analysis (5 μg per lane) was performed essentially as described (9).
Enzyme Assays. TK activity was detected in situ by BrdUrd labeling. Infected cultures were incubated with 10 μM BrdUrd for 4 h, fixed (3% paraformaldehyde), permeabilized (0.25% Triton X-100), treated with RNase A, and denatured with 2 M HCl for 2 h. Incorporation was detected by immunofluorescence (5) using a monoclonal antibody against BrdUrd (Sigma, 1:64) and PI as a DNA counter stain. To biochemically measure AK activity, parasites were disrupted by sonication in 5 mM MgCl2/10 mM NaF/2.5 mM ATP (5 × 107 per ml, ref. 15) and incubated at 37°C in the presence of 2.5 μCi [2,8,3H]adenosine (1 Ci = 37 GBq). Five microliters of the reaction was spotted onto DE 81 filters (Whatman) at indicated times, and filters were washed to remove unbound nucleoside and measured by scintillation counting.
Results and Discussion
C. parvum Depends on Salvage for both Purines and Pyrimidines. Nucleotide biosynthesis is essential to parasite growth and has been a rich target for the development of antiparasitic drugs. Parasitic protists generally salvage purine nucleotides from their host, but synthesize pyrimidine nucleotides de novo (16). We have mined the C. parvum genome database (CryptoDB, http://CryptoDB.org, ref. 17) to evaluate nucleotide biosynthesis in C. parvum as a target for antimicrobial therapy. The metabolic model established through this analysis is summarized in Fig. 1. Both the pyrimidine and purine synthesis pathways diverge from those previously characterized in related parasites. Most notably, C. parvum appears to have lost the capacity to perform de novo pyrimidine synthesis because we were unable to identify any of the genes encoding the six enzymes involved in this pathway (Table 1). To ensure that this was not a reflection of poor sequence conservation of these genes among Apicomplexa or caused by other technical issues, we performed a wider comparative genomic analysis. A database was established combining all publicly available apicomplexan sequence data (443,562,576 nucleotides, see Materials and Methods for detail and acknowledgments) and searched by using the blast algorithm. In contrast to C. parvum, we identified all genes for the pyrimidine de novo pathway in the genome data sets for related apicomplexans including P. falciparum, Theileria annulata and T. gondii with significant statistical confidence (see Table 1).
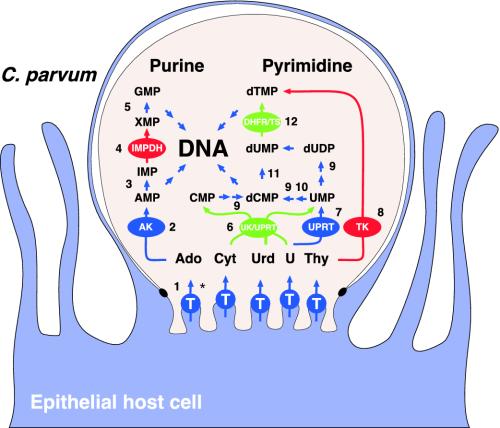
Fig. 1.
The C. parvum nucleotide biosynthetic pathway is a phylogenetic mosaic. Enzymes labeled in red show strong phylogenetic association with eubacteria, those in green show association with plants and algae. Metabolic reconstruction and phylogenetic analyses presented here are based on the C. parvum type 2 IOWA genome [analysis of the type 1 data set (36) yielded identical results]. Supporting comparative genomic analyses are provided in Table 1. Two arrows indicate two or more enzymatic steps. Most nucleoside mono- and diphosphate kinase and phosphorylase steps have been omitted for simplicity. A complete set of these genes is present in the genome. The membrane topology of the feeder organelle (37) has been schematized. *, Transporter: the localization of nucleoside transporters is hypothetical, a localization outside the feeder organelle is equally possible. 1, adenosine transporter; 2, AK; 3, adenosine monophosphate deaminase; 4, IMPDH; 5, guanosine monophosphate synthase; 6, UK-UPRT; 7, uracil phosphoribosylltransferase; 8, TK; 9 ribonucleotide diphosphate reductase; 10, cytosine triphosphate synthetase; 11, deoxycytosine monophosphate deaminase; 12, dihydrofolate reductase-thymidylate synthase.
The loss of the entire pathway was surprising because recent genetic analysis in the related parasite T. gondii demonstrated that functional pyrimidine de novo synthesis is essential for the development, and hence virulence, of this parasite in the host (18). As indicated in Fig. 1, C. parvum appears to compensate for this loss by possessing genes for three pyrimidine salvage enzymes: UPRT, UK-UPRT, and TK. Whereas the presence of UPRT is well established in T. gondii (19), UK-UPRT and TK have not been previously reported in apicomplexan organisms, nor have we detected these genes in the ≈443 million nucleotides of sequence data for Apicomplexa in ApiDB (Table 1).
Gene Transfer as a Mechanism for the Acquisition of Salvage Enzymes. To determine the evolutionary origin of the divergent pyrimidine salvage pathway, we performed detailed phylogenetic analyses of the genes encoding these enzymes. The bifunctional enzyme UK-UPRT shows a strong association with plant and algal sequences whether analyzed with parsimony, distance, or maximum likelihood (Fig. 2). The phylogenetic distribution of fused UK-UPRT genes across the tree of life is spotty and appears to be limited to eukaryotes. We constructed more taxonomically complete data sets for genes encoding individual UK and UPRT enzymes. Phylogenetic analyses of UK and UPRT data sets including artificially split UK-UPRT sequences again demonstrated, albeit with less statistical confidence, a relationship of Cryptosporidium with algal and plant sequences (alignment provided in the supporting information). One scenario consistent with this observation is that the fused UK-UPRT was acquired by means of intracellular gene transfer from an algal endosymbiont. C. parvum, unlike the other apicomplexans examined to date, does not contain an apicoplast organelle (20). However, phylogenetic studies on glyceraldehyde-3-phosphate dehydrogenase (21) have suggested that the acquisition of the algal endosymbiont, which gave rise to the apicoplast, occurred very early in the evolution of the Alveolata (which implies the absence of a plastid in C. parvum to be secondary). Alternatively, the presence of UK-UPRT in plants and C. parvum could be due to their shared ancestry (22). Because the gene was not detected in any other member of the apicomplexa, (Table 1) ciliates, dinoflagellates, diatoms, or kinetoplastids (for which several essentially completed genomes were available to our searches), this model would assume gene loss in all other members of this “super group” with the exception of plants, algae, and C. parvum. Currently, the taxonomic sampling for UK-UPRT genes among protists is not sufficient to convincingly distinguish the singular retention model from the transfer model. Regardless of its mechanism of acquisition, the unique presence of UK-UPRT in C. parvum results in important differences in the pharmacology of nucleotide analogs. Whereas T. gondii is highly resistant to cytosine-arabinoside, a prodrug activated by uridine kinase, C. parvum was found to show a surprising level of susceptibility to this drug (23, 24).
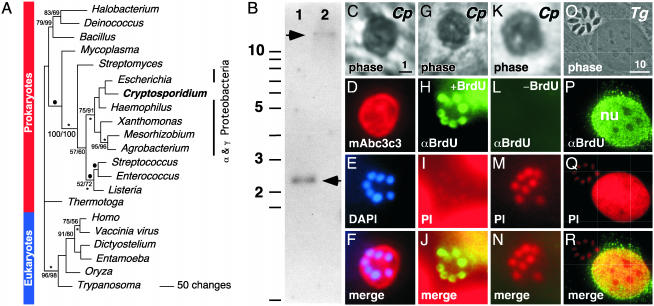
Phylogenetic evidence suggests that the second salvage enzyme, TK, is the result of a horizontal gene transfer from a bacterium (Fig. 3A). Parsimony, distance, and maximum likelihood analyses consistently place C. parvum TK with the α and γ-proteobacteria (TK is absent from several completed genomes of δ- and ε-proteobacteria). The TK gene is found in the bovine and human Cryptosporidium genotype data sets and Southern analysis further confirms this sequence as a genuine C. parvum gene (Fig. 3B). TK is an important target for antiviral therapy (25), we therefore wanted to experimentally validate the activity of this enzyme in the parasite by BrdUrd labeling. BrdUrd is converted to nucleotide in a TK-dependent reaction and incorporated into DNA during S-phase; this incorporation can be detected in situ by using a BrdUrd-specific monoclonal antibody. Twenty-four hours after infection, cultures were incubated with 10 μM BrdUrd for 4 h before fixation and processing for immunofluorescence. As shown in Fig. 3H, incubation with a BrdUrd-specific antibody resulted in labeling of C. parvum nuclei. No antibody staining is observed in the absence of BrdUrd (Fig. 3L) or in T. gondii, which lacks a TK gene (Fig. 3P, note robust incorporation into the host cell nucleus, nu).
Fig. 3.
C. parvum expresses a TK of eubacterial origin. (A) Phylogenetic analysis of TK. The tree shown is one of four most parsimonious trees obtained. Solid bullets indicate branches not found on the other most parsimonious trees. Numbers above the branches (space permitting) indicate the bootstrap values for parsimony and neighbor-joining analyses. Only percentages >50% are shown. Asterisks below branches indicate nodes with ≥50% frequency support from maximum likelihood puzzle analysis. GenBank accession numbers, multiple sequence alignment, and analysis details are available in Materials and Methods and supporting information. Many organisms lack a TK gene including δ- and ε-proteobacteria. (B) Genomic Southern blot of C. parvum type 2 DNA probed with TK coding sequence. Lane 1, EcoRI; lane 2, BamHI. Sizes are as indicated (expected sizes based on genome sequence data are 2.4 and >11.5 kb, respectively). (C) C. parvum type 1 meronts were identified in infected tissue cultures based on their reactivity with the C. parvum-specific mAb c3c3 (D) and the presence of six to eight nuclei (E, I, and M). These nuclei could be detected after BrdUrd labeling with an antibody specific for incorporated BrdUrd (H). No antibody labeling was observed without prior BrdUrd labeling (L) or in T. gondii (P), which lacks a TK gene. DAPI, 6′-diamidino-2-phenylinidole; PI, propidium iodide; Cp, C. parvum; Tg, T. gondii; nu, host cell nucleus. [Scale bar represents 1 (C-N) or 10 μm (O-R), respectively.]
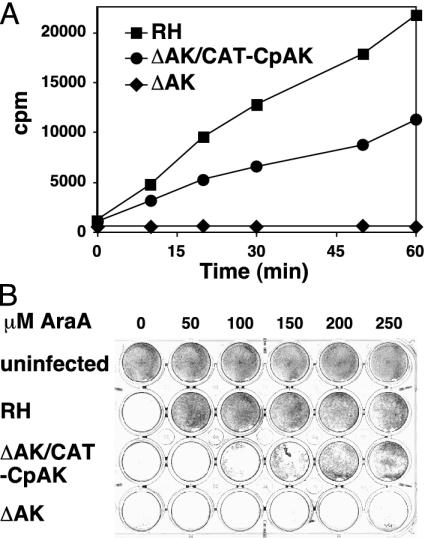
Adenosine Is the Sole Source of Purine for C. parvum. Horizontal gene transfers in C. parvum are not limited to the pyrimidine pathway. Recently inosine 5′ monophosphate dehydrogenase (IMPDH) an enzyme involved in purine salvage (Fig. 1), was shown to have been acquired from an ε-proteobacterium (9). IMPDH is an established drug target and the drugs ribavirin, mycophenolic acid, and mizoribine are in clinical use for antiviral and immunosuppressive chemotherapy (26, 27). Susceptibility to IMPDH inhibition can be limited by guanine salvage, and a previous study has described hypoxanthine-xanthine-guanine phosphoribosyltransferase (HXGPRT) and adenosine phosphoribosyltransferase activity in crude extracts of C. parvum (28). Genomic analyses do not detect the genes for either of these two enzymes in C. parvum (Table 1), but instead predict a streamlined pathway relying entirely upon the salvage of adenosine via AK (Fig. 1). To confirm the presence of a functional AK, we used T. gondii as a closely related transgenic model. The putative C. parvum AK gene was introduced into an expression plasmid and used to stably transfect a T. gondii mutant that lacks endogenous AK activity due to a targeted gene deletion (29). As shown in Fig. 4A, wild-type T. gondii parasites (squares) show time-dependent AK activity absent in the knockout mutant (diamonds, measured by following the conversion of [2,8,3H]adenosine into DE81 retained AMP; ref. 15). Expression of the C. parvum gene resulted in complementation of this mutant's phenotype to approximately half of the wild-type activity (circles). The transgenic parasites also showed renewed susceptibility to adenosine-arabinoside a prodrug activated by AK, which had been used to select the AK mutant (again not to full wild-type level, Fig. 4B). These experiments confirm the presence of AK in C. parvum and demonstrate the facility of T. gondii as a transgenic model for C. parvum.
Fig. 4.
C. parvum encodes an active AK. Transgenic expression of the putative C. parvum AK gene complements enzymatic activity in the T. gondii AK null mutant (measured by conversion of [2,8,3H]adenosine to AMP, DE81 filter retained cpm versus incubation time) (A) and also partially restores the sensitivity of this mutant to the AK activated prodrug adenosine-arabinoside (AraA) (B). Parasite growth under drug was measured by the monolayer disruption assay (9) in 24-well tissue cultures (white wells represent complete host cell lysis due to uninhibited parasite growth, and dark wells indicate parasite inhibition).
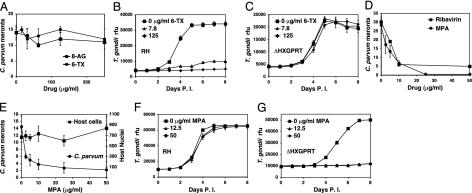
To test for the presence of HXGPRT in C. parvum a pharmacological experiment was performed. 6-thioxanthine (6-TX) is a prodrug that is activated by HXGPRT (30). C. parvum development was not affected by up to 250 μg/ml 6-TX or 8-azaguanine, a HXGPRT activated guanine analog (Fig. 5A). In contrast, T. gondii, which contains HXGPRT, is highly susceptible to 6-TX with an IC50 of <7.8 μg/ml, but becomes resistant when the HXGPRT gene is removed (Fig. 5 B and C; ref. 31). These experiments strongly suggest that C. parvum does not contain HXGPRT.
Fig. 5.
C. parvum lacks HXGPRT, making it susceptible to IMPDH inhibition. (A) C. parvum development in tissue culture is resistant to the HXGPRT-activated prodrugs 6-thioxanthine and 8-azaguanine (6-TX, 8-AG, number of type 1 meronts per 25 microscopic fields, data points represent three independent coverslip cultures with bars indicating the standard deviation) but susceptible to the IMPDH inhibitors ribavirin and mycophenolic acid (D). This pattern is equivalent to the T. gondii HXGPRT null mutant (C and G) growth for T. gondii was measured daily by fluorescence in YFP-YFP expressing lines (8); rfu, relative fluorescence units; four independent wells per data point. Note susceptibility of wild-type RH T. gondii to 6-TX (B). Supplementation of the medium with 100 μM guanine completely rescues growth under mycophenolic acid (MPA) in host cells (E) and wild type T. gondii (F), which can salvage guanine through H(X)GPRT but not in C. parvum (E) or the T. gondii HXGPRT null mutant (G).
Inhibition of IMPDH Inhibits Parasite Growth. The absence of HXGPRT in C. parvum suggests that the parasite should be sensitive to IMPDH inhibition. Indeed, the two IMPDH inhibitors, mycophenolic acid and ribavirin, inhibit C. parvum development in a dose-dependent manner (Fig. 5D; ref. 24). C. parvum is an intracellular parasite living in close dependence of the host cell's metabolism. To ensure that the effect on the parasite is caused by direct inhibition of the parasite enzyme and not the host IMPDH, we repeated the mycophenolic acid experiment in the presence of excess guanine. Mammalian cells are able to overcome an IMPDH block by guanine salvage through HGPRT (32). Although host cell growth is completely rescued by 100 μM guanine, C. parvum development remains inhibited (Fig. 5E). In additional control experiments, wild-type T. gondii is rescued by guanine, but the strain lacking HXGPRT remains sensitive to IMPDH inhibitors. These experiments further confirm that C. parvum lacks HXGPRT and demonstrate that IMPDH is a potential target for anti-cryptosporidial drugs.
Conclusion
C. parvum's nucleotide biosynthesis diverges significantly from other Apicomplexa in that it relies entirely upon salvage from the host for both purines and pyrimidines. The enzymes used represent an evolutionary patchwork stitched together from protozoan, eubacterial, and potentially algal sources. Our analysis of UK-UPRT provides molecular evidence of an algal or “plant-like” gene in the plastid-less genus Cryptosporidium. This evidence, combined with the presence of the bifunctional enzyme dihydrofolate reductase-thymidylate synthase, DHFR, in C. parvum (33) and other apicomplexans provides an additional phylogenetic link between the Apicomplexa and plants. Whether the presence of both fused genes in these lineages indicates monophyly (21, 22) or multiple independent gene transfers (34) remains to be determined. Our discovery of a TK gene of α- or γ-proteobacterial ancestry in C. parvum represents a second, and probably distinct eubacterial horizontal gene transfer because IMPDH was acquired from an ε-proteobacterium (9).
Our genomic and experimental data are consistent with the metabolic model presented in Fig. 1 and have important implications for the development of anticryptosporidial drugs. The discovery of redundant pathways supplying dTMP in this pathogen provides a compelling explanation for the resistance of C. parvum to antifolates (33), a class of drugs widely used against other apicomplexans. Acting primarily on the folate pathway, these drugs exert their main growth-inhibiting effect through the subsequent tetrahydrofolate starvation of the thymidylate synthase reaction. The design of IMPDH inhibitors and TK-activated prodrugs, however, could be a very promising avenue based on C. parvum's unique salvage pathways and the divergent bacterial origin of these enzymes. Large compound collections targeting both enzymes are already available. Transgenic T. gondii expressing C. parvum genes might not only be a tool for target evaluation but also provide an urgently needed vehicle for high-throughput screening using existing fluorescent protein and β-galactosidase assays (8, 35).
Supplementary Material
Acknowledgments
We thank M. Abrahamsen (University of Minnesota, www.cbc.umn.edu/ResearchProjects/AGAC/Cp), G. Buck (Virginia Commonwealth University, www.parvum.mic.vcu.edu), and G. Widmer and S. Tzipori (Tufts University) for access to the C. parvum genome data. Preliminary T. gondii genomic and/or cDNA sequence data were accessed via http://ToxoDB.org and/or www.tigr.org/tdb/t_gondii. Genomic data were provided by The Institute for Genomic Research (supported by National Institutes of Health Grant AI05093) and the Sanger Institute (Wellcome Trust). EST sequences were generated by Washington University (National Institutes of Health Grant 1R01AI045806-01A1). Theileria annulata, E. histolytica, and D. discoidium sequence data were produced by the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/T_annulata or E_histolytica and www.sanger.ac.uk/Projects/D_discoideum. The sequence encoding UK-UPRT in Chlamydomonas reinhardtii was kindly provided by the Chlamydomonas Resource Center, www.biology.duke.edu/chlamy_genome. We thank M. Arrowood (Centers for Disease Control, Atlanta) for critical reagents and advice and R. Tarleton for comments on the manuscript. J.H. and M.-J.G. were supported by postdoctoral fellowships from the American Heart Association. This work was supported by National Institutes of Health grants (to B.S. and L.H.) and grants from the University of Georgia Research Foundation (to J.C.K.) and Merck Research Laboratories (to B.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AK, adenosine kinase; HXGPRT, hypoxanthine-xanthine-guanine phosphoribosyltransferase; IMPDH, inosine 5′ monophosphate dehydrogenase; TK, thymidine kinase; UK-UPRT, uridine kinase-uracil phosphoribosyltransferase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY466379-AY466383 and AY466390-AY466394).
References
- 1.MacKenzie, W. R., Schell, W. L., Blair, K. A., Addiss, D. G., Peterson, D. E., Hoxie, N. J., Kazmierczak, J. J. & Davis, J. P. (1995) Clin. Infect. Dis. 21, 57-62. [DOI] [PubMed] [Google Scholar]
- 2.DuPont, H. L., Chappell, C. L., Sterling, C. R., Okhuysen, P. C., Rose, J. B. & Jakubowski, W. (1995) N. Engl. J. Med. 332, 855-859. [DOI] [PubMed] [Google Scholar]
- 3.Hunter, P. R. & Nichols, G. (2002) Clin. Microbiol. Rev. 15, 145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mead, J. R. (2002) Drug Resist. Update. 5, 47-57. [DOI] [PubMed] [Google Scholar]
- 5.Striepen, B., He, C. Y., Matrajt, M., Soldati, D. & Roos, D. S. (1998) Mol. Biochem. Parasitol. 92, 325-338. [DOI] [PubMed] [Google Scholar]
- 6.Arrowood, M. J., Mead, J. R., Xie, L. & You, X. (1996) FEMS Microbiol. Lett. 136, 245-249. [DOI] [PubMed] [Google Scholar]
- 7.Roos, D. S., Donald, R. G., Morrissette, N. S. & Moulton, A. L. (1994) Methods Cell Biol. 45, 27-63. [DOI] [PubMed] [Google Scholar]
- 8.Gubbels, M. J., Li, C. & Striepen, B. (2003) Antimicrob. Agents Chemother. 47, 309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Striepen, B., White, M. W., Li, C., Guerini, M. N., Malik, S. B., Logsdon, J. M., Jr., Liu, C. & Abrahamsen, M. S. (2002) Proc. Natl. Acad. Sci. USA 99, 6304-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gish, W. (2003) wublast 2.0 (Washington University, St. Louis).
- 11.Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G. & Gibson, T. J. (1998) Trends Biochem. Sci. 23, 403-405. [DOI] [PubMed] [Google Scholar]
- 12.Swofford, D. L. (2001) paup*: Phylogenic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA).
- 13.Retief, J. D. (2000) Methods Mol. Biol. 132, 243-258. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. (2002) Bioinformatics 18, 502-504. [DOI] [PubMed] [Google Scholar]
- 15.Darling, J. A., Sullivan, W. J., Jr., Carter, D., Ullman, B. & Roos, D. S. (1999) Mol. Biochem. Parasitol. 103, 15-23. [DOI] [PubMed] [Google Scholar]
- 16.Berens, R. L., Krug, E. C. & Marr, J. J. (1995) in Biochemistry and Molecular Biology of Parasites, eds. Marr, J. J. & Müller, M. (Academic, London), pp. 89-117.
- 17.Puiu, D., Enomoto, S., Buck, G. A., Abrahamsen, M. & Kissinger, J. C. (2004) Nucleic Acids Res. 32, D344-D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox, B. A. & Bzik, D. J. (2002) Nature 415, 926-929. [DOI] [PubMed] [Google Scholar]
- 19.Donald, R. G. & Roos, D. S. (1995) Proc. Natl. Acad. Sci. USA 92, 5749-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu, G., Marchewka, M. J. & Keithly, J. S. (2000) Microbiology 146, 315-321. [DOI] [PubMed] [Google Scholar]
- 21.Fast, N. M., Kissinger, J. C., Roos, D. S. & Keeling, P. J. (2001) Mol. Biol. Evol. 18, 418-426. [DOI] [PubMed] [Google Scholar]
- 22.Stechmann, A. & Cavalier-Smith, T. (2002) Science 297, 89-91. [DOI] [PubMed] [Google Scholar]
- 23.Pfefferkorn, E. R. & Pfefferkorn, L. C. (1976) J. Parasitol. 62, 993-999. [PubMed] [Google Scholar]
- 24.Woods, K. M. & Upton, S. J. (1998) FEMS Microbiol. Lett. 168, 59-63. [DOI] [PubMed] [Google Scholar]
- 25.Cheng, Y. C., Huang, E. S., Lin, J. C., Mar, E. C., Pagano, J. S., Dutschman, G. E. & Grill, S. P. (1983) Proc. Natl. Acad. Sci. USA 80, 2767-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McHutchison, J. G. & Fried, M. W. (2003) Clin. Liver Dis. 7, 149-161. [DOI] [PubMed] [Google Scholar]
- 27.Allison, A. C. & Eugui, E. M. (2000) Immunopharmacology 47, 85-118. [DOI] [PubMed] [Google Scholar]
- 28.Doyle, P. S., Kanaani, J. & Wang, C. C. (1998) Exp. Parasitol. 89, 9-15. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan, W. J., Jr., Chiang, C. W., Wilson, C. M., Naguib, F. N., el Kouni, M. H., Donald, R. G. & Roos, D. S. (1999) Mol. Biochem. Parasitol. 103, 1-14. [DOI] [PubMed] [Google Scholar]
- 30.Donald, R. G. & Roos, D. S. (1998) Mol. Biochem. Parasitol. 91, 295-305. [DOI] [PubMed] [Google Scholar]
- 31.Donald, R. G. K., Carter, D., Ullman, B. & Roos, D. S. (1996) J. Biol. Chem. 271, 14010-14019. [DOI] [PubMed] [Google Scholar]
- 32.Lee, H. J., Pawlak, K., Nguyen, B. T., Robins, R. K. & Sadee, W. (1985) Cancer Res. 45, 5512-5520. [PubMed] [Google Scholar]
- 33.Vasquez, J. R., Gooze, L., Kim, K., Gut, J., Petersen, C. & Nelson, R. G. (1996) Mol. Biochem. Parasitol. 79, 153-165. [DOI] [PubMed] [Google Scholar]
- 34.Hannaert, V., Saavedra, E., Duffieux, F., Szikora, J. P., Rigden, D. J., Michels, P. A. & Opperdoes, F. R. (2003) Proc. Natl. Acad. Sci. USA 100, 1067-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McFadden, D. C., Seeber, F. & Boothroyd, J. C. (1997) Antimicrob. Agents Chemother. 41, 1849-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Widmer, G., Lin, L., Kapur, V., Feng, X. & Abrahamsen, M. S. (2002) Microbes Infect. 4, 1081-1090. [DOI] [PubMed] [Google Scholar]
- 37.Tzipori, S. & Griffiths, J. K. (1998) Adv. Parasitol 40, 5-36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.