Abstract
The 1918 influenza A H1N1 virus caused the worst pandemic of influenza ever recorded. To better understand the pathogenesis and immunity to the 1918 pandemic virus, we generated recombinant influenza viruses possessing two to five genes of the 1918 influenza virus. Recombinant influenza viruses possessing the hemagglutinin (HA), neuraminidase (NA), matrix (M), nonstructural (NS), and nucleoprotein (NP) genes or any recombinant virus possessing both the HA and NA genes of the 1918 influenza virus were highly lethal for mice. Antigenic analysis by hemagglutination inhibition (HI) tests with ferret and chicken H1N1 antisera demonstrated that the 1918 recombinant viruses antigenically most resembled A/Swine/Iowa/30 (Sw/Iowa/30) virus but differed from H1N1 viruses isolated since 1930. HI and virus neutralizing (VN) antibodies to 1918 recombinant and Sw/Iowa/30 viruses in human sera were present among individuals born before or shortly after the 1918 pandemic. Mice that received an intramuscular immunization of the homologous or Sw/Iowa/30-inactivated vaccine developed HI and VN antibodies to the 1918 recombinant virus and were completely protected against lethal challenge. Mice that received A/PR/8/34, A/Texas/36/91, or A/New Caledonia/20/99 H1N1 vaccines displayed partial protection from lethal challenge. In contrast, control-vaccinated mice were not protected against lethal challenge and displayed high virus titers in respiratory tissues. Partial vaccine protection mediated by baculovirus-expressed recombinant HA vaccines suggest common cross-reactive epitopes on the H1 HA. These data suggest a strategy of vaccination that would be effective against a reemergent 1918 or 1918-like virus.
During 1918 and 1919, the “Spanish” influenza pandemic killed up to forty million people worldwide (1-4). The exceptionally high mortality rate, especially among young adults, was not observed during later influenza pandemics of 1957 and 1968 (5, 6). It was estimated that ≈30% of the world's population was clinically infected during the 1918 pandemic (7). Sequence analysis of the 1918 influenza virus from fixed and frozen lung tissue has provided molecular characterization and phylogenetic analysis of this strain. The complete coding sequences of the 1918 nonstructural (NS), hemagglutinin (HA), neuraminidase (NA), and matrix (M) genes have been determined (8-14); however, the sequences of these genes did not reveal features that could account for its high virulence. The sequence analysis combined with the laboratory method of reverse genetics has allowed for the generation of recombinant viruses containing one or more 1918 influenza virus genes entirely from cloned cDNAs (14-16). This technology was applied to determine whether existing antiinfluenza drugs would be effective against a reemergent 1918 influenza virus. We found that a recombinant virus possessing the 1918 M segment was inhibited effectively both in tissue culture and in vivo by the M2 ion-channel inhibitors amantadine and rimantadine (15). Moreover, a recombinant virus bearing the surface glycoproteins, HA and NA, of the 1918 pandemic influenza virus (1918 HA/NA:WSN) with the remaining genes of influenza A/WSN/33 virus was found to be sensitive in vitro and in vivo to the NA inhibitors zanamivir and oseltamivir. The 1918 HA/NA:WSN virus had a high virulence phenotype on intranasal (i.n.) infection in mice without prior adaptation in that species. In contrast, a control H1N1 recombinant virus with both the HA and NA of the A/New Caledonia/20/99 (New Cal HA/NA:WSN) virus was highly attenuated relative to the 1918 HA/NA:WSN or parental WSN virus (15).
The HA and NA transmembrane glycoproteins are the major viral surface antigens that define an influenza virus strain and are important virulence factors in birds and mice (17-21). These glycoproteins evolve simultaneously creating balanced HA-NA functional interactions important for efficient replication of influenza A viruses (22). Indeed, our previous observations demonstrated that the 1918 HA and NA proteins appear to be compatible with each other as recombinant viruses possessing either the 1918 HA or 1918 NA individually led to attenuation in mice (15). The HA is also the principal target of the host's immune system and protective immunity provided by current influenza vaccines is largely based on the induction of strain-specific IgG neutralizing antibodies directed against the HA. Major antigenic changes through HA and NA gene reassortment have occurred to create new human pandemic viruses that possess the ability to evade existing immunity. Although evidence suggests that the 1957 Asian and 1968 Hong Kong pandemic strains emerged after genetic reassortment between human and animal influenza viruses (20, 23), the origin of the 1918 pandemic virus has not been precisely elucidated. Phylogenetic and sequence analysis placed the 1918 viral HA within the mammalian group of influenza A viruses and having a close genetic relationship with the oldest available swine influenza strain, A/Swine/Iowa/30 (Sw/Iowa/30, H1N1).
The basis for the exceptional virulence of the 1918 pandemic virus has remained elusive because no influenza virus isolates from the period have been available for study. In the present investigation, we generated recombinant influenza viruses possessing from two to five gene segments of the 1918 pandemic influenza virus. We report here that these recombinant viruses replicated efficiently in mouse lungs, without prior adaptation and were highly lethal for BALB/c mice. These results indicate that the mouse model of influenza virus infection might give insights into the pathogenicity of the 1918 virus. We also have antigenically characterized these recombinant viruses and identified vaccine strategies capable of inducing protective immunity against viruses with antigenic determinants derived from the 1918 virus in mice. These strategies could be used as prophylactic measures in the case of reemergence of new 1918-like viruses (24).
Materials and Methods
Generation of 1918 HA, NA, NP, NS, and M cDNAs and Recombinant Viruses. The 1918 HA, NA, NP, M, and NS cDNAs were constructed by PCR, using overlapping deoxyoligonucleotides corresponding to the published sequence of the influenza A/South Carolina/1/18 (H1N1) virus HA (12) ORF, the influenza A/Brevig Mission/1/18 (BM/1/18, H1N1) virus NA ORF (11), the BM/1/18 virus NP ORF (A. H. Reid, R. Lourens, T. A. Janczewski, T. G. Fanning, and J.K.T., unpublished work), the influenza BM/1/18 virus M ORF (8), or the influenza BM/1/18 virus NS ORF (14). The noncoding regions of each segment are identical to that of the corresponding segment of influenza A/WSN/33 (WSN) H1N1 virus. Primer sequences and PCR reaction conditions are available on request. Recombinant viruses were generated by using the reverse genetics system of Fodor et al. (25), following the methods of Basler et al. (14). Generation of viruses possessing 1918 genes in a WSN background was performed under biosafety level 3 Ag containment (26) to ensure the safety of laboratory workers, environment, and the public. All subsequent laboratory and animal work with live virus was also performed under these high containment conditions. The identity of the 1918 influenza virus genes in the recombinant viruses was confirmed by RT-PCR and sequencing.
Infection of Mice. Male BALB/c mice, 6-7 weeks old (Simonsen Laboratories, Gilroy, CA), were anesthetized with ketamine-xylazine (1.98 and 0.198 mg per mouse, respectively), and 50 μl of infectious virus diluted in PBS was inoculated i.n. LD50 titers were determined by inoculating groups of three mice i.n. with serial 10-fold dilutions of virus. LD50 titers were calculated by the method of Reed and Muench (27). Seven additional mice were infected with the highest inoculating dose (106 plaque-forming units) for determination of weight loss and virus titers in lungs. Whole lungs were removed on day 4 postinfection (p.i.) and homogenized in 1 ml of PBS. After which, homogenates were titrated for virus infectivity in 10-day-old embryonated eggs (15). Egg 50% infectious dose (eID50/ml) titers were calculated by the method of Reed and Muench (27).
Human Serum Samples. For the first serology test, nine human sera were obtained from randomly chosen volunteers (age range 36-93 years) in March of 2002 and stored at -70°C before influenza hemagglutination inhibition (HI) and virus neutralization (VN) analysis. In the second serology test, serum samples were obtained from individuals pre- and postvaccination. All subjects were organ transplant patients who received vaccine as a prophylactic measure before the 2001 influenza season. A total of 15 subjects, born between 1936 and 1956, received inactivated New Cal/99 vaccine or placebo control.
Vaccine Preparation. Viruses used as vaccines were concentrated from allantoic fluid and purified by equilibrium density centrifugation through a 30-60% linear sucrose gradient as described (28). For inactivation, purified whole viruses were adjusted to a protein concentration of 1 mg/ml and treated with 0.025% formalin at 4°C for 3 days. The vaccine doses given throughout are expressed as amounts of total protein measured by Bradford assay (Bio-Rad).
Immunization of Mice. Groups of BALB/c mice (n = 13) were injected i.m. with a single dose of 10 μg of formalin-inactivated vaccine as described (29). Vaccines were suspended in sterile PBS, and a volume of 0.1 ml was injected in the left hind leg. Mock control mice received PBS in place of H1N1 vaccine. Subtype control mice received a similar dose of X-31 (which possesses the surface glycoprotein genes of A/Aichi/2/68 [H3N2] and the internal protein genes of A/Puerto Rico/8/34) vaccine virus (30). Baculovirus-expressed recombinant HA (rHA) protein, corresponding to the HA of A/Texas/36/91 (Tx/91), New Cal/99, or A/Panama/2007/99 (H3N2) virus, was acquired from Protein Sciences (Meriden, CT). Three weeks after vaccination, sera from nine individual mice per group were collected for antibody studies.
Antibody Assays. All sera were initially diluted 1:10 in receptor-destroying enzyme from Vibrio cholerae (Denka Seiken, Tokyo). HI assays were performed with 0.5% chicken erythrocytes by standard methods (31). Titers of VN antibody were determined essentially as described (32) and were determined as the reciprocal of the highest dilution of serum that neutralized 100 plaque-forming units of virus in Madin-Darby canine kidney (MDCK) cell cultures. Antigenic analysis of the H1N1 viruses was performed with reference H1N1 virus stocks and corresponding p.i. ferret antisera, generously provided by N. J. Cox (Centers for Disease Control and Prevention, Atlanta). Chicken antisera were generated by infecting animals i.v. with 106 eID50 of virus in a 0.2-ml volume, followed by a s.c. boost 3 weeks later.
Viral Challenge. Three weeks after vaccination, mice were challenged i.n. with 100 LD50 of 1918 HA/NA:WSN or 1918 HA/NA/M/NP/NS:WSN virus in a volume of 50 μl. After infection, nine mice were monitored daily for disease signs for 14 days p.i. To evaluate protection of the nose, lung, and brains from infection, tissue samples of four mice per group were removed on day 5 p.i. and titrated for virus infectivity as described above.
Results
Construction and Characterization of Recombinant Viruses with 1918 Influenza Virus Genes. Genes encoding the 1918 pandemic influenza virus were reconstructed from deoxyoligonucleotides and corresponded to the reported 1918 virus coding sequences (8-14). Those viral segments not derived from the 1918 influenza virus were derived from the mouse-adapted WSN virus. Recombinant influenza viruses were created expressing two to five genes of the 1918 influenza virus. All recombinant influenza viruses used in this study had high infectivity titers in MDCK cells (Table 1) and in 10-day-old embryonated eggs (7.2-8.8 log10 eID50/ml). The 1918 HA/NA:WSN virus (15) was included in these pathotyping studies for comparison against recombinant viruses additionally expressing the M, NS, or NP genes. The mean virus titers in the lungs were determined on day 4 p.i., when titers were maximal. All four 1918 recombinant viruses replicated in the mouse lungs to high titers and caused lethal disease without prior host adaptation (Table 1). The LD50 titers and percent weight loss observed in mice infected with each of the 1918 recombinant viruses were not significantly different from each other or mice infected with the parental WSN virus (Table 1). Mice infected with lethal doses of the 1918 recombinant or WSN virus began to lose weight 2 days after infection and died 5-9 days p.i.
Table 1. Properties of recombinant influenza viruses used in this study.
| Virus* | Titer†, plaque-forming units/ml | % Weight loss‡ | Lung titers§ | LD50¶ |
|---|---|---|---|---|
| Parental WSN virus | 2.2 × 107 | 28.4 | 6.7 ± 0.2 | 2.5 |
| 1918 HA/NA | 2.1 × 107 | 20.9 | 7.3 ± 0.1 | 2.75 |
| 1918 HA/NA/M | 1.4 × 108 | 28.8 | 7.9 ± 0.2 | 2.75 |
| 1918 HA/NA/M/NS | 2.1 × 107 | 23.7 | 7.3 ± 0.2 | 3.25 |
| 1918 HA/NA/M/NS/NP | 1.4 × 108 | 24.5 | 7.4 ± 0.2 | 1.75 |
All viral genomic segments were derived from the WSN virus unless otherwise indicated.
Titer of virus stocks prepared on MDCK cells.
Average percent weight loss on day 4 p.i. (five mice per group).
Average lung titers of four mice on day 4 p.i. expressed as elD50/ml ± SE.
Expressed as the log10 plaque-forming units required to give 1 LD50.
Antigenic Reactivity of Selected Viruses in HI Test with Ferret and Chicken Serum. Antigenic characterization by HI, using p.i. ferret and chicken antisera, was performed with the 1918 HA/NA recombinant virus and reference variants representing early and recent H1N1 viruses. The ferret antisera revealed that the 1918 HA/NA recombinant virus was antigenically most related to Sw/Iowa/30 virus but distinct from all other influenza A (H1N1) viruses examined (Table 2). Similarly, chicken H1N1 antisera confirmed the antigenic cross-reactivity observed among the 1918 and Sw/Iowa/30 strains, with progressive diminution of inhibition with subsequent H1N1 strains (Table 3). There was low reactivity to the 1918 HA/NA viruses with ferret and chicken antisera to WS/33, PR/8/34, and Tx/91 viruses.
Table 2. HI reactions of H1N1 virus variants with ferret antisera.
| HI titer with ferret antisera
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Virus | 1918 HA | Sw/la/30 | WS/33 | PR/8/34 | USSR/77 | Chili/83 | Tx/91 | New Cal/99 |
| 1918 HA | 2,560 | 1,280 | 320 | 40 | <10 | 10 | 80 | 20 |
| Sw/la/30 | 1,280 | 2,560 | 20 | 320 | 80 | 10 | 80 | 20 |
| WS/33 | <10 | <10 | 640 | 40 | <10 | <10 | <10 | 40 |
| PR/8/34 | 20 | <10 | 160 | 2,560 | 10 | <10 | 10 | 10 |
| USSR/77 | <10 | <10 | 10 | <10 | 1,280 | 20 | <10 | <10 |
| Chili/83 | <10 | <10 | 10 | <10 | 40 | 320 | 20 | 10 |
| Tx/91 | <10 | <10 | 20 | <10 | <10 | <10 | 2,560 | 40 |
| New Cal/99 | 10 | <10 | 10 | 20 | <10 | <10 | 40 | 1,280 |
Serum samples were from ferrets infected with indicated H1N1 viruses. HI titers represent reciprocal of highest dilution of sera inhibiting agglutination of 0.5% chicken erythrocytes by 4 hemagglutination units of virus.
Table 3. HI reactions of H1N1 virus variants with p.i. chicken antisera.
| HI titer with chicken antisera
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Virus | 1918 HA | Sw/la/30 | WS/33 | PR/8/34 | USSR/77 | Chili/83 | Tx/91 | New Cal/99 |
| 1918 HA | 2,560 | 1,280 | 160 | 80 | 20 | 10 | 160 | 20 |
| Sw/la/30 | 640 | 2,560 | <10 | 40 | 10 | 10 | 80 | 10 |
| WS/33 | <10 | <10 | 320 | <10 | <10 | <10 | 10 | <10 |
| PR/8/34 | 10 | <10 | 80 | 2,560 | <10 | <10 | 10 | 20 |
| USSR/77 | <10 | <10 | <10 | <10 | 320 | 20 | 10 | <10 |
| Chili/83 | <10 | <10 | <10 | <10 | 40 | 320 | <10 | <10 |
| Tx/91 | 80 | <10 | 80 | <10 | <10 | <10 | 640 | <10 |
| New Cal/99 | 10 | <10 | 10 | 320 | <10 | <10 | 80 | 2,560 |
Serum samples from chickens infected with indicated H1N1 viruses. HI titers represent reciprocal of highest dilution of sera inhibiting agglutination of 0.5% chicken erythrocytes by 4 hemagglutination units of virus.
Serological Reactivity of Human Sera with 1918 HA/NA Recombinant Virus. Sera from nine humans ranging from 36 to 93 years of age were tested for HI activity against Sw/Iowa/30, PR/8/34, New Cal/99, or 1918 HA/NA recombinant virus. A virus neutralization assay was also included, because with some influenza viruses it provides greater sensitivity than the commonly used HI test (33). Individuals born before 1918 have the highest levels of HI and VN antibodies to the 1918 HA/NA recombinant virus but no reactivity to New Cal/99 virus (Table 4). It was assumed that the 1918-specific antibody in these human sera was generated by natural infection. Two (serum C and E) of three individuals born between 1928 and 1933 possessed HI and VN antibodies to 1918 HA but also possessed cross-reactive antibodies to the three other viruses tested. Individuals born after 1962 (G and H) had HI and VN antibody titers of ≤20 antibodies to 1918 HA. Overall, the four individuals (A-C and E) with detectable HI and neutralization antibodies to the 1918 HA/NA virus also possessed reactivity to Sw/Iowa/30 virus, indicating the antigenic relatedness of the two viruses. In a second study, the effect of New Cal/99 influenza vaccination on the induction of cross-reactive antibodies to 1918 HA/NA virus was investigated. Volunteers, born between 1936 and 1956, were administered influenza vaccine following standard vaccination procedures, and serum was collected from 15 subjects before and after a single-dose of inactivated trivalent influenza New Cal/99 vaccine. Individuals with paired serum samples were tested for HI activity to New Cal/99, 1918 HA, and Sw/Iowa/30 viruses. Before vaccination, the majority of individuals had low HI serum antibody titers (≤20) against all three viruses tested. Sera collected three weeks postvaccination showed increases in HI antibody titers to New Cal/99 virus of 4-fold or greater in all vaccinated individuals (Table 5). Although, the HI antibody response to Sw/Iowa/30 and 1918 HA antigens was considerably lower in comparison to New Cal/99 virus antigen, HI titer increases to these viruses were also observed after New Cal/99 vaccination.
Table 4. VN and HI antibody responses to H1N1 viruses detected in human sera.
| Neutralization† and HI‡ antibody titer
|
|||||
|---|---|---|---|---|---|
| Serum sample* | Year of birth | 1918 HA/NA | Sw/la/30 | PR/8/34 | New Cal/99 |
| A | 1910 | 160 (80) | 40 (40) | 20 (10) | <10 (<10) |
| B | 1911 | 320 (160) | 80 (40) | 80 (20) | <10 (<10) |
| C | 1928 | 160 (160) | 160 (80) | 40 (40) | 40 (40) |
| D | 1932 | 10 (10) | <10 (<10) | 40 (20) | <10 (<10) |
| E | 1933 | 160 (80) | 80 (80) | 160 (40) | 80 |
| F | 1944 | <10 (<10) | <10 (<10) | <10 (<10) | 10 |
| G | 1962 | 10 (10) | <10 (<10) | <10 (<10) | 80 |
| H | 1966 | 20 (20) | <10 (<10) | <10 (<10) | 20 |
| I | 1977 | 10 (10) | <10 (<10) | <10 (<10) | 40 |
Serum samples from individuals ranging from 36 to 93 years of age.
Reciprocal dilution endpoint in VN titers.
Reciprocal dilution in HI titration (in parentheses).
Table 5. HI antibody responses to H1N1 viruses before and after A/New Cal/20/99 vaccination.
| HI antibody titer†
|
||||||
|---|---|---|---|---|---|---|
| Serum sample* | New Cal/99 prevaccination | New Cal/99 postvaccination | Sw/la/30 prevaccination | Sw/la/30 postvaccination | 1918 HA prevaccination | 1918 HA postvaccination |
| 4 | <10 | 1,280 | <10 | 20 | 10 | 40 |
| 5 | <10 | 320 | <10 | <10 | <10 | <10 |
| 6 | 10 | 160 | <10 | 20 | 10 | 40 |
| 7 | 160 | 640 | 10 | 40 | 10 | 40 |
| 9 | <10 | 320 | <10 | 40 | 10 | 40 |
| 12 | 20 | 320 | <10 | 20 | <10 | 40 |
| 17 | <10 | 1,280 | <10 | <10 | <10 | 20 |
| 22 | 10 | 1,280 | <10 | 20 | <10 | 40 |
| 23 | <10 | 1,280 | <10 | <10 | <10 | 20 |
| 33 | 40 | 320 | 20 | 40 | 40 | 80 |
| 1 | <10 | <10 | <10 | <10 | <10 | 10 |
| 16 | <10 | <10 | <10 | <10 | <10 | <10 |
| 18 | <10 | <10 | 10 | <10 | 10 | 10 |
| 21 | <10 | 10 | <10 | 10 | 10 | 10 |
| 50 | <10 | 10 | <10 | 10 | 10 | 20 |
Serum samples from individuals born between 1936 and 1956 tested for seroconversion rates to A/New Cal/20/99 virus. Serum samples 1, 16, 18, 21, and 50 were from placebo controls.
Reciprocal dilution in HI titration.
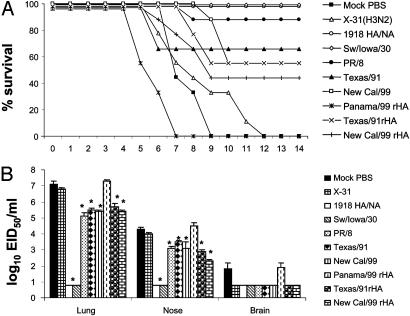
Protective Efficacy of H1N1 Vaccines. The mouse model was used to evaluate a strategy of vaccination against the lethal 1918 recombinant virus. In the first vaccine experiment, we tested the ability of three inactivated H1N1 vaccines to induce protection against the lethal 1918 HA/NA recombinant virus. Vaccinated mice received 10 μg of formalin-inactivated whole H1N1 or control (H3N2) vaccine, and 3 weeks after inoculation mice received a lethal i.n. challenge with 100 LD50 of 1918 HA/NA:WSN recombinant virus. The extent of vaccine efficacy was measured as (i) weight loss and survival over a 14-day postchallenge (p.c.) period and (ii) virus titers in the upper respiratory tract (nose), lower respiratory tract (lung), and brain tissue of individual mice. Immunization with PR/8/34 or homologous 1918 HA/NA:WSN virus vaccine protected the mice against lethal virus challenge, whereas 75% of New Cal/99-vaccinated mice were protected (Fig. 1A). Although PR/8/34- and New Cal/99-vaccinated mice were mostly protected against death, they all had significant weight loss (data not shown) and listlessness the first week of infection; these were taken as signs of morbidity. Lethal H1N1 challenge of H3N2-vaccinated or unvaccinated (mock) control mice resulted in a progressive loss of body weight from day 2 p.c. and death after virus challenge. Furthermore, control mice had high titers of virus in the lung and nose tissue at day 5 p.c. (Fig. 1B). Infectious virus was also present in the brain tissue of 2 of 4 mock-control and X-31-vaccinated mice, but the titers of virus were considerably lower in comparison to respiratory tissues. Protection against infection was incomplete in mice vaccinated with either New Cal/99 or PR/8/34 vaccine, although these mice displayed significant reductions (16- and 500-fold, respectively) in lung virus titers compared to the unvaccinated control mice. In contrast, the 1918 HA/NA:WSN homologous vaccine protected mice from upper and lower respiratory tract infection and vaccinated mice had undetectable virus in brain tissues on day 5 p.c. (Fig. 1B).
Fig. 1.
Protective efficacy of influenza H1N1-inactivated vaccine against lethal challenge with 1918 HA/NA:WSN recombinant influenza virus. Groups of BALB/c mice received a single i.m. inoculation of H1N1 or H3N2 (X-31) vaccine. Control mice received PBS in place of vaccine. Twenty-two days after vaccination, mice were challenged i.n. with 100 LD50 of 1918 HA/NA:WSN recombinant virus. Mice were monitored for survival (A) or killed 5 days later, and virus titers in individual lung, nose, and brain tissue were determined (B). Virus endpoint titers are expressed as mean log10 eID50/ml.
In a second vaccine study, mice received 10 μg of inactivated H1N1 vaccines or purified H1 rHA proteins generated in insect cells by using the recombinant baculovirus system (34). Three weeks after inoculation, mice received a lethal challenge with 100 LD50 of 1918 HA/NA/M/NP/NS:WSN recombinant virus. Mice that received homologous 1918 HA/NA vaccine virus or Sw/Iowa/30 whole virus vaccine were completely protected from death, whereas 50-90% of mice administered PR/8/34, New Cal/99, or Tx/91 whole virus vaccine survived the lethal challenge (Fig. 2A). Despite the degree of resistance to lethal virus challenge observed in PR/8/34-, New Cal/99-, or Tx/91-immunized mice, the lung and nose virus titers at 5 days p.c. were 200- to 50,000-fold higher than virus titers observed in Sw/Iowa/30-vaccinated mice. Infectious virus was undetectable in brain and respiratory tissues of mice administered homologous 1918 HA/NA or Sw/Iowa/30 whole virus vaccine (Fig. 2B). By contrast, 100% of the control mice succumbed to the lethal H1N1 infection and had high titers in the respiratory tissues and low titers in brain tissues at 5 days p.c. Mice were vaccinated with rHA from H1N1 viruses to determine whether the partial cross-protection induced by these vaccines was due to anti-HA immunity. An influenza recombinant protein derived from a different subtype virus, A/Panama/207/99 (H3N2), served as a control. Immunization with rHA protein corresponding to the HA of Tx/91 or New Cal/99 virus resulted in 50-60% survival. As with Tx/91 and New Cal/20/99 whole virus vaccinates, mice corresponding to the rHA-immunized groups had high titers of infectious virus in the respiratory tissues (Fig. 2B).
Fig. 2.
Sw/Iowa/30 vaccine provides protection against lethal challenge with a recombinant influenza virus possessing five 1918 virus genes. Groups of BALB/c mice were vaccinated as described in the legend of Fig. 1. Twenty-two days after vaccination, mice were challenged i.n. with 100 LD50 of 1918 HA/NA/M/NS/NP:WSN recombinant virus. Mice were monitored for survival (A) or killed 5 days later, and virus titers in individual lung, nose, and brain tissue were determined (B). An asterisk indicates that the H1N1-vaccinated group was significantly (P < 0.05) different from the control groups by ANOVA.
The prechallenge antibody responses to the 1918 HA/NA recombinant virus was measured in individual serum samples collected before lethal virus challenge. It was demonstrated previously that administration of this vaccine dose (10 μg) resulted in 100% of mice achieving an HI titer of ≥40 to the homologous virus (29). Each inactivated or rHA vaccine elicited HI titers of 40 or greater to the homologous virus (data not shown). Immunization with 1918 homologous or Sw/Iowa/30 vaccine resulted in the induction of HI and VN antibody responses to the 1918 HA/NA challenge virus. In contrast, the PR/8/34, Tx/91, and New Cal/99 vaccines failed to induce HI and VN antibodies to the 1918 HA/NA:WSN recombinant virus (data not shown). Taken together, these data show that among the H1N1 viruses tested, the Sw/Iowa/30 virus possesses the greatest antigenic similarity to the 1918 influenza virus.
Discussion
Sequence analysis of the 1918 “Spanish” influenza virus genes have not revealed any obvious features that could account for its high virulence thus far. By contrast, the analysis in mice of recombinant WSN viruses containing two to five genes derived from the 1918 virus point to a critical role of the 1918 HA and NA genes in virulence, at least in the mouse model. Our previous observations demonstrated that replacement of the HA and NA genes of WSN virus by those of the 1918 virus did not decrease virulence in mice, an unusual outcome due to the absence of previous mouse adaptation of these genes (15). To understand the contribution of additional 1918 genes in the mouse model of influenza virus pathogenicity, recombinant viruses possessing one to three additional (M, NS, and NP) 1918 genes were generated. Each of the recombinant viruses possessing two to five genes of the 1918 pandemic virus replicated efficiently in mouse respiratory tissues and were highly lethal for this species. Introduction into the 1918 HA/NA:WSN virus of the 1918 M, 1918 M/NS, or 1918 M/NS/NP genes did not significantly increase the virulence of this virus. The ability of the 1918 recombinant viruses to cause lethal disease in mice without prior adaptation was remarkable given that the 1918 genes were derived directly from sequences corresponding to a human virus. Generally, prior adaptation is required before influenza A viruses can replicate efficiently and induce disease in mice (35). Exceptions include some of the H5N1 viruses, which were highly virulent in mice without prior host adaptation, isolated in Hong Kong during 1997 (36-38).
Previously, we have described that the introduction of the NS1 gene of the 1918 virus into a WSN virus background results in attenuation in mice, suggesting that the 1918 NS1 protein is not adapted to the mouse host (14). It is therefore interesting that the additional introduction of the HA, NA, and M genes of 1918 overcomes the loss of pathogenicity in mice associated with the 1918 NS gene alone in a WSN background. Because the 1918NS:WSN virus is less virulent than the 1918 HA/NA/M/NS:WSN virus, these results point to the 1918 HA/NA genes, and perhaps M, as responsible for increased pathogenicity in mice.
An interesting feature of the highly pathogenic 1918 recombinant viruses was the presence of infectious virus in the brain tissues of mice after lethal i.n. virus challenge. The mouse-adapted WSN virus, which is recognized as a neurovirulent strain (39), could also be recovered from mouse brain tissue after i.n. immunization on days 4 and 5 p.i. (data not shown). It has been demonstrated previously that the NA gene of WSN virus plays a critical role in its neurovirulence, most likely by facilitating HA cleavage without the requirement of exogenous trypsin (17). Like the WSN strain, the 1918 HA/NA:WSN recombinant viruses did not require exogenous trypsin to grow in MDCK cells. By contrast, the control H1N1 recombinant virus with both the HA and NA of the New Cal/99 (New Cal HA/NA:WSN) required exogenous trypsin in MDCK cells (data not shown). Interestingly, the 1918 NA protein does not contain the Δ146 mutation associated with this feature in WSN (11). Thus, the genetic basis by which the 1918 constructs share these features with WSN is not known.
The key prevention strategy to reduce influenza pandemic-associated morbidity and mortality will be the implementation of inactivated influenza virus vaccines effective against the pandemic strain. There are no influenza vaccines currently available that could efficiently be used as prophylactic measures if a 1918-like virus reemerges. Therefore, we sought to identify candidate immunogens and evaluate the vaccine efficacy of these immunogens against the highly pathogenic 1918 HA/NA recombinant viruses. Our studies demonstrate that mice which had been inoculated once i.m. with a formalin-fixed homologous (1918 HA/NA:WSN) whole virus vaccine exhibited significant resistance against subsequent challenge with a 1918 HA/NA recombinant virus. Because production of a 1918 recombinant influenza vaccine would be complicated by the higher levels of biosafety containment required, we selected an antigenically related nonpathogenic virus that could be handled under biosafety level 2 conditions. The protection afforded by the biosafety level 2 virus, Sw/Iowa/30 vaccine was similar to that observed in mice that received homologous (1918 HA/NA:WSN) virus vaccine. Sw/Iowa/30-immune mice were protected against mortality and significant weight loss and had undetectable virus in respiratory tissues on day 5 p.i. The high level of protection induced by Sw/Iowa/30 virus vaccine correlated with detectable HI and virus neutralizing antibodies measured in vitro.
Because mouse models are useful as preliminary screens for candidate vaccines and may not be the definitive model for vaccine efficacy in humans, we also carried out HI tests with a panel of p.i. ferret antisera generated against seven influenza A (H1N1) viruses and the 1918 HA/NA:WSN recombinant virus. Ferrets are used to produce p.i. antisera for the determination of antigenic drift among human influenza viruses (40). HI data obtained with ferret antisera illustrate that the HA gene of the 1918 virus was most similar to Sw/Iowa/30 H1N1 virus. The relationship to swine influenza was until now based partly on historical accounts documenting widespread severe influenza-like disease outbreaks in swine during the fall of 1918 (41, 42). Because the HA genes of swine viruses undergo limited variation in antigenic sites (43, 44) compared to human H1N1 viruses, very few genetic changes might be expected of the swine H1N1 influenza virus isolated 12 years later. In fact, we found that survivors of the 1918 influenza pandemic had antibodies that neutralized both the 1918 HA/NA:WSN and Sw/Iowa/30 virus. This finding was consistent with archeoserological data demonstrating that survivors of the 1918 pandemic had antibodies that neutralized classic swine influenza virus (3, 45, 46). The Sw/Iowa/30 and 1918 HAs were found to be easily distinguished from the WS/33, PR/8/34, and other human H1N1 viruses isolated after this time period (Tables 2 and 3). Previous HA protein sequence comparisons between 1918, Sw/Iowa/30, and PR/8/34 viruses support the results of our antigenic analysis (1, 9, 12). There are twenty-two amino acid differences in the HA protein between the 1918 and Sw/Iowa/30 viruses. Only four of these amino acid changes were located in the antigenic sites. In contrast, the antigenic sites of PR/8/34 HA had 15 amino acid changes from the 1918 HA (12). The rate of genetic drift in the HA1 segments of the H1N1 human influenza viruses along with the acquisition of glycosylation sites to mask antigenic sites (47) most likely accounts for the antigenic variation observed between the 1918 HA and other human HIN1 viruses isolated since 1933.
Although PR/8/34, Tx/91, and New Cal/99 viruses differed antigenically from the 1918 HA, vaccines prepared from these H1N1 viruses were able to provide some degree of protection against lethal 1918 recombinant virus challenge in mice. The partial protection afforded by these vaccines cannot be explained by the presence of detectable HI or neutralizing antibodies. However, the virus neutralization activity in vitro may not be an adequate measure of virus neutralization in vivo (48). Thus, other factors in vivo may influence or enhance antibody activity, such as Fc or complement receptor expressing cells types may facilitate the opsonization of virus (49, 50). Although there are multiple cross-reactive viral determinants on influenza A viruses, we hypothesize that the partial protection observed in PR/8/34-, Tx/91-, and New Cal/99-vaccinated mice is largely due to anti-HA immunity. This hypothesis is supported by the partial protection conferred by rHA protein derived from the 1981 and 1999 H1 influenza strains.
Because the genetic structure of the 1918 “Spanish” influenza virus is becoming fully known, questions arise regarding the pathogenesis, antigenicity, and immunity to the pandemic virus. The generation of 1918 recombinant influenza A viruses that are pathogenic in mice provides a reliable model system to test vaccine candidates and identify the viral genes associated with pathogenicity. This study helps to further define the pathogenic nature of this virus, the antigenic characteristics, and vaccine strategies to the 1918 pandemic influenza virus. The identification of effective vaccine strategies should provide additional prophylaxis for laboratory workers and the public if a virus emerged through natural or some other means.
Acknowledgments
This work was partially supported by grants from the National Institutes of Health to P.P., C.F.B., and J.K.T. P.P. is a senior fellow of the Ellison Medical Foundation New Scholar in Global Infectious Diseases. C.F.B. is a New Scholar of the Ellison Medical Foundation Program in Global Infectious Diseases. J.K.T. was supported by National Institutes of Health Grant 5R01 AI0506919-02. This work was also supported by U.S. Department of Agriculture/Agricultural Research Service Current Research Information System Project Number 6612-32000-022-93.
Abbreviations: eID50, egg 50% infectious dose; HA, hemagglutinin; HI, hemagglutination inhibition; i.n., intranasal(ly); M, matrix; MDCK, Madin-Darby canine kidney; NA, neuraminidase; New Cal/99, influenza A/New Caledonia/20/99 (H1N1) virus; NP, nucleoprotein; NS, nonstructural; p.c., postchallenge; p.i., postinfection; Sw/Iowa/30, influenza A/Swine/Iowa/30 (H1N1) virus; rHA, recombinant HA; WSN, influenza A/WSN/33 (H1N1) virus; VN, virus neutralization.
References
- 1.Reid, A. H., Taubenberger, J. K. & Fanning, T. G. (2001) Microbes Infect. 3, 81-87. [DOI] [PubMed] [Google Scholar]
- 2.Kilbourne, E. D. (1975) in The Influenza Viruses (Academic, New York), pp. 483-538.
- 3.Taubenberger, J. K., Reid, A. H., Fanning, T. G., Janczewski, T. A., (2001) Philos. Trans. R Soc. London B 356, 1829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crosby, A. (1989) America's Forgotten Pandemic (Cambridge Univ. Press, Cambridge, U.K.).
- 5.Noble, G. R. (1982) in Basic and Applied Influenza Research, ed. Beere, A. S. (CRC, Boca Raton, FL), pp. 11-50.
- 6.Glezen, N. P. (1996) Epidemiol. Rev. 18, 64-76. [DOI] [PubMed] [Google Scholar]
- 7.Frost, W. (1920) Public Health Rep. 35, 584-597. [Google Scholar]
- 8.Reid, A. H., Fanning, T. G., Janczewski, T. A., McCall, S. & Taubenberger, J. K. (2002) J. Virol. 76, 10717-10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taubenberger, J. K., Reid, A. H. & Fanning, T. G. (2000) Virology 274, 241-245. [DOI] [PubMed] [Google Scholar]
- 10.Taubenberger, J. K., Reid, A. H., Krafft, A. E., Bijwaard, K. E. & Fanning, T. G. (1997) Science 275, 1793-1796. [DOI] [PubMed] [Google Scholar]
- 11.Reid, A. H., Fanning, T. G., Janczewski, T. A. & Taubenberger, J. K. (2000) Proc. Natl. Acad. Sci. USA 97, 6785-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reid, A. H., Fanning, T. G., Hultin, J. V. & Taubenberger, J. K. (1999) Proc. Natl. Acad. Sci. USA 96, 1651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reid, A. H., Taubenberger, J. K. & Fanning, T. G. (2001) Microbes Infect. 3, 81-87. [DOI] [PubMed] [Google Scholar]
- 14.Basler, C., Reid, A., Dybing, J., Janczewski, T., Fanning, T., Zheng, H., Salvatore, M., Perdue, M., Swayne, D., Garcia-Sastre, A., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tumpey, T. M., Garcia-Sastre, A., Mikulasova, A., Taubenberger, J. K., Swayne, D. E., Palese, P. & Basler, C. F. (2002) Proc. Natl. Acad. Sci. USA 99, 13849-13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiss, G. K., Salvatore, M., Tumpey, T. M., Carter, V. S., Wang, X., Basler, C. F., Taubenberger, J. K., Bumgarner, R. E., Palese, P., Katze, M. G. & Garcia-Sastre, A. (2002) Proc. Natl. Acad. Sci. USA 99, 10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goto, H. & Kawaoka, Y. (1998) Proc. Natl. Acad. Sci. USA 95, 10224-10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goto, H., Wells, K., Takada, A. & Kawaoka, Y. (2001) J. Virol. 75, 9297-9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatta, M., Gao, P., Halfmann, P. & Kawaoka, Y. (2001) Science 293, 1840-1842. [DOI] [PubMed] [Google Scholar]
- 20.Wright, P. F. & Webster, R. G. (2001) in Field's Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadelphia), pp. 1533-1579.
- 21.Perdue, M. L. & Suarez, D. L. (2000) Vet. Microbiol. 74, 77-86. [DOI] [PubMed] [Google Scholar]
- 22.Mitnaul, L. J., Matrosovich, M. N., Castrucci, M. R., Tuzikov, A. B., Bovin, N. V., Kobosa, D. & Kawoaka, Y. (2000) J. Virol. 74, 6015-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawaoka, Y., Krauss, S. & Webster, R. G. (1989) J. Virol. 63, 4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lederberg, J. (2001) Proc. Natl. Acad. Sci. USA 98, 2115-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fodor, E., Devenish, L., Engelhardt, O. G., Palese, P., Brownlee, G. G. & Garcia-Sastre, A. (1999) J. Virol. 73, 9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbeito, M. S., Abraham, G., Best, M., Cairns, P., Langevin, P., Sterritt, W. G., Barr, D., Meulepas, W., Sanchez-Vizcaino, J. M. & Saraza, M. (1995) Rev. Sci. Tech. 14, 873-887. [DOI] [PubMed] [Google Scholar]
- 27.Reed, L. J. & Muench, H. (1938) Am. J. Hyg. 27, 493-497. [Google Scholar]
- 28.Cox, N. J. & Kendal, A. P. (1984) J. Infect. Dis. 149, 194-200. [DOI] [PubMed] [Google Scholar]
- 29.Lu, X., Renshaw, M., Tumpey, T. M., Kelly, G. D., Hu-Primmer, J. & Katz, J. M. (2001) J. Virol. 75, 4896-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baez, M., Palese, P. & Kilbourne, E. D. (1980) J. Infect. Dis. 141, 362-365. [DOI] [PubMed] [Google Scholar]
- 31.World Health Organization Collaborating Centers for Reference and Research on Influenza (1982) Concepts and Procedures for Laboratory-Based Influenza Surveillance, eds. Kendal, A. P., Skehel, J. J. & Pereira, M. S. (Centers for Disease Control and Prevention, Atlanta), pp. B17-B35.
- 32.Mozdzanowska, K., Furchner, M., Washko, G., Mozdzanowski, J. & Gerhard, W. (1997) J. Virol. 71, 4347-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowe, T., Abernathy, R. A., Hu-Primmer, J., Thompson, W. W., Lu, X., Lim, W., Fukuda, K., Cox, N. J. & Katz, J. M. (1999) J. Clin. Microbiol. 37, 937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuroda, K., Hauser, C., Rott, R., Klenk, H. D. & Doerfler, W. (1986) EMBO J. 5, 1359-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoyle, L. (1968) in The Influenza Viruses, eds. Gard, S., Hallauer, C. & Meyer, K. F. (Springer, New York), pp. 170-171.
- 36.Lu, X., Tumpey, T. M., Morken, T., Zaki, S. R., Cox, N. J. & Katz, J. M. (1999) J. Virol. 73, 5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dybing, J. K., Schultz-Cherry, S., Swayne, D. E., Suarez, D. L. & Perdue, M. L. (2000) J. Virol. 74, 1443-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao, P., Watanabe, S., Ito, T., Goto, H., Wells, K., McGregor, M., Cooley, A. J. & Kawaoka, Y. (1999) J. Virol. 73, 3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castrucci, M. R. & Kawaoka, Y. (1993) J. Virol. 67, 759-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweet, C., Fenton, R. J. & Price, G. E. (1999) in Handbook of Animal Models of Infection, eds. Zak, O. & Sande, M. A. (Academic, New York), pp. 989-998.
- 41.Beveridge, W. (1977) Influenza: The Last Great Plaque, an Unfinished Story of Discovery (Prodist, New York).
- 42.Koen, J. S. (1919) Am. J. Vet. Med. 14, 468-470. [Google Scholar]
- 43.Sugita, S., Yoshioka, Y., Itamura, S., Kanegae, Y., Oguchi, K., Gojobori, T., Nerome, K. & Oya, A. (1991) J. Mol. Evol. 32, 16-32. [DOI] [PubMed] [Google Scholar]
- 44.Brown, I. H., Ludwig, S., Olsen, C. W., Hannoun, C., Scholtissek, C., Hinshaw, V. S., Harris, P. A., McCauley, J. W., Strong, I. & Alexander, D. J. (1997) J. Gen. Virol. 78, 553-562. [DOI] [PubMed] [Google Scholar]
- 45.Shope, R. E. (1936) J. Exp. Med. 63, 669-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davenport, F. M., Hennessy, A. V. & Francis, T., Jr. (1953) J. Exp. Med. 98, 641-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inkster, M. D., Hinshaw, V. S. & Schulze, I. T. (1993) J. Virol. 67, 7436-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCullough, K. C. (1986) Arch. Virol. 87, 1-36. [DOI] [PubMed] [Google Scholar]
- 49.Schlesinger, J. J. & Chapman, S. (1995) J. Gen. Virol. 76, 217-220. [DOI] [PubMed] [Google Scholar]
- 50.Gerhard, W., Mozdanowska, K., Furchner, M., Washko, G. & Maiese, K. (1997) Immunol. Rev. 159, 95-103. [DOI] [PubMed] [Google Scholar]