Abstract
Tissues from 98 human hepatocellular carcinomas (HCCs) obtained from hepatic resections were subjected to somatic copy number variation (CNV) analysis. Most of these HCCs were discovered in livers resected for orthotopic transplantation, although in a few cases, the tumors themselves were the reason for the hepatectomies. Genomic analysis revealed deletions and amplifications in several genes, and clustering analysis based on CNV revealed five clusters. The LSP1 gene had the most cases with CNV (46 deletions and 5 amplifications). High frequencies of CNV were also seen in PTPRD (21/98), GNB1L (18/98), KIAA1217 (18/98), RP1-1777G6.2 (17/98), ETS1 (11/98), RSU1 (10/98), TBC1D22A (10/98), BAHCC1 (9/98), MAML2 (9/98), RAB1B (9/98), and YIF1A (9/98). The existing literature regarding hepatocytes or other cell types has connected many of these genes to regulation of cytoskeletal architecture, signaling cascades related to growth regulation, and transcription factors directly interacting with nuclear signaling complexes. Correlations with existing literature indicate that genomic lesions associated with HCC at the level of resolution of CNV occur on many genes associated directly or indirectly with signaling pathways operating in liver regeneration and hepatocyte growth regulation.
Hepatocellular carcinoma (HCC) is the most common type of liver cancer and is associated with high mortality and morbidity. Several chronic conditions are associated with an increased incidence of HCC, including alcoholism, chronic infections with hepatitis B virus (HBV) or hepatitis C virus (HCV), hemochromatosis, and metabolic diseases. Common to all these conditions is that their chronic forms may cause liver cirrhosis and HCC can often arise on this background. Because most of the previously mentioned conditions do not involve genotoxic agents and none of the associated viruses carry transforming oncogenes, it is reasonable to assume that, for most HCCs, cirrhosis per se and associated irregularities induced in the hepatocyte microenvironment are somehow responsible for generating genomic or epigenetic alterations leading to carcinogenesis. Cirrhosis is associated with a higher rate of hepatocyte proliferation. HCC in rodents appears, in most conditions, associated with long-term hyperproliferation of hepatocytes.1 Several genomic abnormalities have been described in HCC, such as mutations in p53 and β-catenin, deletions in areas of chromosome 1, and activation of specific oncogenes by HBV through insertional mutagenesis.2 Genes expressed by HBV or HCV may cause a long-term promotion effect, also facilitating the evolution of HCC.3
Studies4–6 of gene expression in HCC have demonstrated subgroups of three clusters with differing gene expression profiles and prognosis. Despite several studies related to gene expression profiling of hepatocyte-derived tumors (adenomas or HCCs), there has been little analysis of genome-wide alterations underpinning the altered patterns of gene expression.7 We performed this study to assess genetic alterations (deletions or amplifications) detectable by analysis of copy number variation (CNV) and to examine the relationships, if any, to the known pathways controlling hepatocyte growth during liver regeneration and in hepatocyte cultures.8–14 We have used the Affymetrix (Santa Clara, CA) 6.0 single-nucleotide polymorphism (SNP) oligonucleotide arrays for these studies. The method provides useful information on the types of deletions and amplifications larger than approximately 1000 bp and is much more detailed than studies of large areas of loss of heterozygosity. In addition, this approach provides detailed information about the site of the genomic alterations within specific genes, allowing correlations between the expected changes in protein structure and the change in function.
Materials and Methods
Case Material
Formalin-fixed, paraffin-embedded (FFPE) tissue specimens were obtained from the Surgical Pathology Archives of the University of Pittsburgh Medical Center, Pittsburgh, PA, in accordance with Institutional Review Board regulations (PRO-08050186). Anonymized samples spanned a 26-year period, from 1981 through 2006. This time interval partially antedated establishment of the Milan criteria15 and spanned a period during which HBV infection was a more frequent indication for transplantation at our center.
Clinical information was obtained via Honest Broker from the EDIT Transplant Database and Surgical Pathology records. This included patient survival, tumor size, tumor recurrence after transplantation, underlying disease, and α-fetoprotein levels. The 98 adult patients included 22 females and 76 males. Samples were derived from native livers at transplantation in all but six patients who underwent surgical resection. Matched control tissue was obtained from noncirrhotic tissue, usually from hilar tissue, and did not include lymph nodes. Underlying diseases included HBV infection (n = 22), HCV infection (n = 21), ethanol use (n = 13), hemochromatosis (n = 3), and primary sclerosing cholangitis (n = 2). Six patients had multiple underlying disorders (3 HBV and HCV, 2 HCV and ethanol use, and 1 HBV, HCV, and ethanol use), and one patient each had sarcoid, granulomatous disease not further specified, type II glycogen storage disease, autoimmune hepatitis, and nonalcoholic steatohepatitis. In 26 patients, there was either no known underlying disorder or insufficient data to establish an underlying disease. Tumors were solitary in 30 patients and multiple in 66 patients. No determination was possible in the remaining two patients. All tumors were HCCs, with five representing the fibrolamellar variant. Histological grading was performed by one of the authors (M.A.N.), and tumors were evaluated for the presence of steatosis, inflammation, giant cells, fibrosis, bile production, cytoplasmic inclusions, and Mallory hyaline.
Tissue Processing, DNA Extraction, Amplicon Generation, and Labeling
FFPE tissues were microdissected to achieve tumor purity >85%. The dissected tissues were incubated in xylene solution overnight to remove residual paraffin. DNA was then extracted using the Qiagen tissue kit (Qiagen, Valencia, CA). Amplicons were generated by performing linear PCR using random 12mers on 2-μg DNA templates using the following program: 94°C for 1 minute and then 40 cycles of 94°C for 30 seconds, 45°C for 1 minute, and 72°C for 2 minutes. The PCR products were then purified and digested with DNase1 for fragmentation. The fragmented DNA was then labeled with biotinylated cytosine.
Hybridization, Washing, and Scanning of the SNP 6.0 Chip
Fragmented DNA (250 μg) was hybridized with a pre-equilibrated Affymetrix chip at 50°C for 16 hours. After the hybridization cocktails were removed, the chips were then washed in a fluidic station with low-stringency buffer (six times SSPE, 0.01% Tween 20, and 0.005% antifoam) for 10 cycles (two mixes per cycle) and stringent buffer (100 mmol/L MES, 0.1 mol/L NaCl, and 0.01% Tween 20) for 4 cycles (15 mixes per cycle) and then stained with streptavidin-phycoerythrin. This was followed by incubation with biotinylated mouse anti-avidin antibody and restaining with streptavidin-phycoerythrin. The chips were scanned in a G7 scanner (Affymetrix Inc., Santa Clara, CA) to detect hybridization signals.
SYBR Green Real-Time Quantitation PCR
The LightCycler FastStart DNA Master SYBR-Green I kit (Roche Applied Science, Indianapolis, IN) was used for real-time PCR amplification. The reaction was performed in a LightCycler machine (Roche Applied Science, Indianapolis, IN). A quantitation standard curve of normal male DNA from 50,000 to 500,000 copies of genome was generated using known amounts of template copies. Genomic DNA (100 ng) was used for all of the experimental and control samples. HotStarTaq DNA polymerase (Qiagen, Valencia, CA) was activated with a 10-minute pre-incubation step at 95°C. Amplification for LSP1 (leukocyte-specific protein 1; 5′-CAATGCTCCTCTATCCAGCC-3′ and 5′-ATGGAGGGGCACTGATTGGA-3′) and PTPRD (protein tyrosine phosphatase, receptor type, D; 5′-CGTAGAGATGCCCAGTATCTGC-3′ and 5′-CTCTGATGAGGCTGCCATTGTG-3′) was performed with 45 cycles of the following program: 94°C for 10 seconds, 62°C for 5 seconds, and 72°C for 10 seconds. LightCycler data software version 3.5 (Roche Applied Science) was used to quantify and fit the data with a standard curve. A separate β-actin DNA quantification was also performed in parallel with either LSP1 or PTPRD analysis.
Determination of CNV
To analyze the data for CNV, CEL files of 196 samples (98 tumor and matched healthy pairs) were imported into Partek GenomeSuite 6.5 (Partek Inc., St. Louis, MO). Determining copy number from the summarized hybridization intensities of each tumor sample was accomplished by normalizing each sample to the matched healthy sample reference. The genomic segmentation algorithm detects a segmentation according to the following criteria: i) neighboring regions have statistically significantly different average intensities, ii) break points (region boundaries) are chosen to give optimal statistical significance, and iii) detected regions must contain a user-specified minimum number of SNPs. To detect regions of deletion or amplification in the genome, a threshold of a minimum 10 markers (P < 0.001) in a region of >500 bp is set. The detected amplification or deletion regions were then mapped to genome regions where known genes reside. The changes in copy number are based on an expected normal number of two. The detail data per case are shown on Supplemental Table S1 (available at http://ajp.amjpathol.org). Based on the previously mentioned criteria, the lowest number above which Partek GenomeSuite 6.5 was judging for amplification was 2.51 (highest seen, 12.29). The highest number below which a deletion was judged was 1.53 (lowest seen, 0.47). The data reflect the presence of both tumor and healthy cells in the tumor sample, although microdissection was used to restrict the presence of nontumor cells to the minimum possible.
Hierarchical Clustering of Cases
Cases were clustered according to 1324 CNV conditions in the exons or introns of 473 known genes from Partek preprocessing (see Supplemental Table S1 at http://ajp.amjpathol.org), ignoring areas that do not contain expressed genes. We filtered out genes with CNV changes in <10% (ie, 9) samples. The filtering reduced the data from 93 samples and 473 genes to 78 samples and 15 genes. The distance measure between two cases was defined as the reciprocal of a CNV similarity score, which was calculated as the total number of concordant gene aberrations in two patients. For example, if two patients have three concordant gene aberrations, their distance is defined as one third. When two patients have no concordant gene aberration, the distance is defined as a large constant (a constant of five was used herein); varying this constant does not change the clustering result. Hierarchical clustering was then performed using average linkage by the hclust function in R software (http://www.r-project.org).
The testing dependence of two variable Fisher's exact tests was used to test the dependence of two variables in a 2 × 2 or larger contingency table. For example, it could be used to test the dependence of patients in clusters and their deletion in PTPRD or the dependence of LSP1 alteration and tumor size. For comparing continuous observations in multiple groups (eg, tumor size of patients in the five clusters), an analysis of variance model and an F-test were used. Both tests were performed using R software.
Moving-Window Procedure for CNV Histogram
For genomic visualization of CNV regions cumulated across cases, we used moving windows of 100 kb and counted the number of cases that contain any CNV region overlapped with the window. The genome-wide CNV map was then plotted by R software with chromosomal locations on the x axis and number of CNV-change patients on the y axis; amplifications and deletions were separated by upward blue and downward red colors.
Results
Complete Data from the Study
The data set derived from the CNV determination procedure and subjected to analysis is shown in Supplemental Table S1 (available at http://ajp.amjpathol.org). The table provides a complete reference to CNV changes for each specific gene and each case. There are instructions for the use of the data set and explanations for the nomenclature used to identify individual columns provided in the table.
Size of the DNA Fragments Used for Analysis
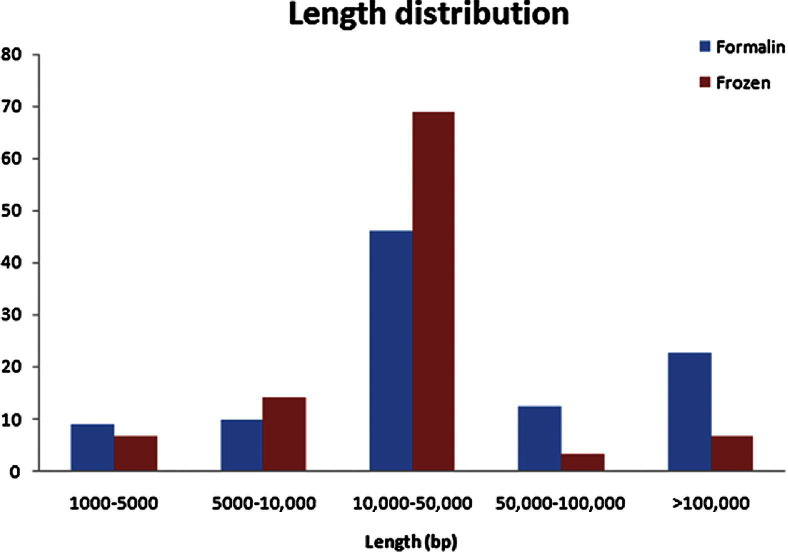
The DNA used for the studies was extracted from FFPE tissue available in our archives. To investigate the effect of the fixation and the prolonged embedding in paraffin, a comparison was made between the length of the DNA used in our study and DNA extracted from a few freshly frozen nonfixed HCC samples (Figure 1). There is close proximity in the frequencies of DNA fragments associated with the specific lengths, suggesting no demonstrable interference in our studies from the effects of paraffin embedding and fixation.
Figure 1.
Comparison of DNA fragmentation between fresh-frozen and FFPE tissues. Copy number distribution of frozen sample and its matched FFPE sample.
Genes Identified as Having a Higher CNV Change Frequency
CNV was seen in some genes in more than two cases (Table 1), demonstrating the total CNV as the sum of the observed deletions and amplifications. The sum of deletions and amplifications is used as the basis for the ranking, with each of these two types of change also shown in separate columns. In addition, the type of CNV observed is provided for each gene and each case. In terms of total CNV (in 98 cases), amplifications/deletions for the two top-ranking genes were as follows: LSP1 (51: 5/46) and PTPRD (21: 4/17). These were followed by GNB1L (18: 0/18), ETS1 (11: 0/11), RSU1 (Ras suppressor 1; 10: 3/7), TBC1D22A (10: 2/8), BAHCC1 (9: 0/9), CNIH2 (9: 5/4), KLC2 (9: 5/4), MAML2 (mastermind-like 2; 9: 2/7), PACS1 (9: 5/4), RAB1B (9: 5/4), and YIF1A (9: 5/4). (The KIAA1217 and RP1-177G6.2 genes will not be discussed, because they have no known function and are listed as hypothetical loci in the UniGene database.) Several other genes directly associated with hepatocyte growth biological characteristics also had CNV. These include PTCH1 (Patched-1; 3: 0/3), Ctnnb1 (2: 0/2), KFL6 (2: 0/2), and EGFR (2: 0/2). The relationship of some of these genes to the known intracellular signaling cascades associated with hepatocyte growth will be described in Discussion.
Table 1.
Number of Cases with CNV per Gene
| Chromosome location of gene | Gene symbol | CNV cases per gene | Amplifications | Deletions |
|---|---|---|---|---|
| 11 | LSP1 | 51 | 5 | 46 |
| 9 | PTPRD | 21 | 4 | 17 |
| 22 | GNB1L | 18 | 0 | 18 |
| 10 | KIAA1217 | 18 | 5 | 13 |
| X | RP1-177G6.2 | 17 | 4 | 13 |
| 11 | ETS1 | 11 | 0 | 11 |
| 10 | RSU1 | 10 | 3 | 7 |
| 22 | TBC1D22A | 10 | 2 | 8 |
| 17 | BAHCC1 | 9 | 0 | 9 |
| 11 | CNIH2 | 9 | 5 | 4 |
| 11 | KLC2 | 9 | 5 | 4 |
| 11 | MAML2 | 9 | 2 | 7 |
| 11 | PACS1 | 9 | 5 | 4 |
| 11 | RAB1B | 9 | 5 | 4 |
| 11 | YIF1A | 9 | 5 | 4 |
| 7 | ETV1 | 8 | 2 | 6 |
| 6 | FUCA2 | 8 | 2 | 6 |
| 1 | NBPF10 | 8 | 4 | 4 |
| 16 | PDXDC1 | 8 | 0 | 8 |
| 8 | RNF139 | 8 | 0 | 8 |
| 18 | BCL2 | 7 | 0 | 7 |
| 1 | CLCC1 | 7 | 0 | 7 |
| 1 | PDE4DIP | 7 | 0 | 7 |
| 12 | PTPN11 | 7 | 3 | 4 |
| 15 | ZNF592 | 7 | 0 | 7 |
| 1 | ABL2 | 6 | 0 | 6 |
| 1 | CAMTA1 | 6 | 0 | 6 |
| 3 | ERC2 | 6 | 0 | 6 |
| 9 | JAK2 | 6 | 3 | 3 |
| 7 | LHFPL3 | 6 | 0 | 6 |
| 1 | NBPF1 | 6 | 2 | 4 |
| 8 | RUNX1T1 | 6 | 2 | 4 |
| 10 | VIM | 6 | 0 | 6 |
| 5 | APC | 5 | 2 | 3 |
| 2 | ASB18 | 5 | 0 | 5 |
| 19 | BBC3 | 5 | 0 | 5 |
| 19 | CCDC9 | 5 | 0 | 5 |
| X | CT45-2 | 5 | 3 | 2 |
| X | CT45-4 | 5 | 3 | 2 |
| 19 | NOTCH3 | 5 | 0 | 5 |
| 2 | TACR1 | 5 | 2 | 3 |
| 8 | TATDN1 | 5 | 0 | 5 |
| 9 | ABL1 | 4 | 4 | 0 |
| 12 | BICD1 | 4 | 0 | 4 |
| 8 | DEFB4 | 4 | 2 | 2 |
| 5 | EPB41L4A | 4 | 2 | 2 |
| 1 | FCGR1C | 4 | 4 | 0 |
| 6 | GSTA4 | 4 | 0 | 4 |
| 1 | HFM1 | 4 | 0 | 4 |
| 11 | JAM3 | 4 | 0 | 4 |
| 8 | KCNB2 | 4 | 4 | 0 |
| 17 | KCNH6 | 4 | 0 | 4 |
| 8 | KCNQ3 | 4 | 4 | 0 |
| 15 | LOC283755 | 4 | 0 | 4 |
| 3 | LSAMP | 4 | 0 | 4 |
| 8 | MFHAS1 | 4 | 0 | 4 |
| 16 | MGC34761 | 4 | 0 | 4 |
| 11 | MPZL3 | 4 | 0 | 4 |
| 20 | PTPN1 | 4 | 0 | 4 |
| 10 | RAB11FIP2 | 4 | 2 | 2 |
| 1 | SELENBP1 | 4 | 3 | 1 |
| 17 | WDR68 | 4 | 0 | 4 |
| 11 | ALDH3B2 | 3 | 3 | 0 |
| 1 | ATG4C | 3 | 0 | 3 |
| 11 | CABP2 | 3 | 3 | 0 |
| 12 | CCDC91 | 3 | 0 | 3 |
| 11 | CCND1 | 3 | 3 | 0 |
| 1 | CD48 | 3 | 0 | 3 |
| 1 | CLK2 | 3 | 0 | 3 |
| 4 | CLOCK | 3 | 0 | 3 |
| 19 | DPF1 | 3 | 0 | 3 |
| 7 | DPY19L1 | 3 | 0 | 3 |
| 17 | ETV4 | 3 | 0 | 3 |
| 17 | FAM18B2 | 3 | 0 | 3 |
| 18 | FECH | 3 | 0 | 3 |
| 11 | FGF19 | 3 | 3 | 0 |
| 9 | GABBR2 | 3 | 3 | 0 |
| 1 | HCN3 | 3 | 0 | 3 |
| 7 | HOXA10 | 3 | 0 | 3 |
| 7 | HOXA11 | 3 | 0 | 3 |
| 7 | HOXA9 | 3 | 0 | 3 |
| 1 | KHDRBS1 | 3 | 0 | 3 |
| 16 | KIAA0182 | 3 | 0 | 3 |
| 8 | KIAA0196 | 3 | 3 | 0 |
| 12 | LOH12CR1 | 3 | 0 | 3 |
| 19 | MUC16 | 3 | 0 | 3 |
| 19 | NFIX | 3 | 0 | 3 |
| 8 | NSMCE2 | 3 | 3 | 0 |
| 11 | ORAOV1 | 3 | 3 | 0 |
| 3 | PARL | 3 | 0 | 3 |
| 3 | PIK3CA | 3 | 0 | 3 |
| 2 | PIP5K3 | 3 | 0 | 3 |
| 9 | PTCH1 | 3 | 0 | 3 |
| 7 | PTPRN2 | 3 | 0 | 3 |
| 12 | PWP1 | 3 | 0 | 3 |
| 14 | RAD51L1 | 3 | 0 | 3 |
| 13 | RB1 | 3 | 0 | 3 |
| 3 | SH3BP5 | 3 | 0 | 3 |
| 1 | SORT1 | 3 | 0 | 3 |
| X | SPACA5 | 3 | 0 | 3 |
| X | SPACA5B | 3 | 0 | 3 |
| 8 | STK3 | 3 | 3 | 0 |
| Y | UTY | 3 | 0 | 3 |
Chromosomal Localization of CNV Alterations
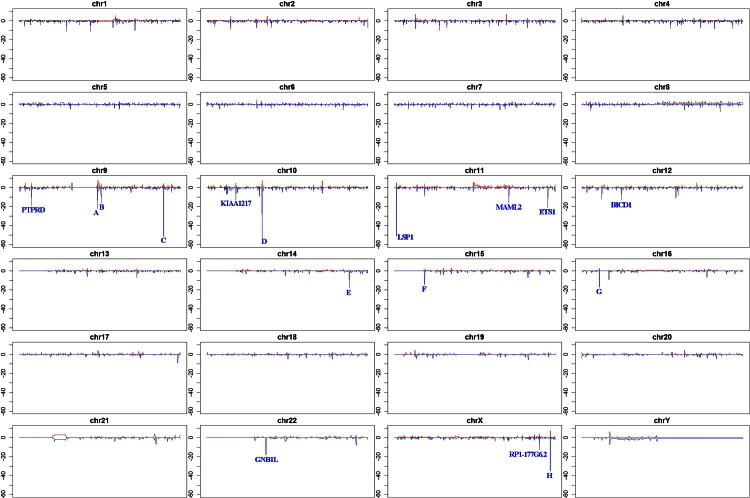
The deletions and amplifications for each chromosome are illustrated (Figure 2). The size of the individual bars shows, on the y axis, the number of cases associated with the particular CNV. The sites labeled as A (23 cases, chromosome 9), B (18 cases, chromosome 9), C (52 cases, chromosome 9), D (54 cases, chromosome 10), E (18 cases, chromosome 14), F (16 cases, chromosome 15), G (17 cases, chromosome 16), and H (39 cases, chromosome X) are deletions in areas of the chromosome containing no known genes. The significance of the high frequency of deletions in these areas of the genome with no known gene associations is not clear. The top-ranked eight genes associated with CNV are also marked in their chromosomal locations. Three cases are associated with extensive amplifications in the Y chromosome. Only the male cases were used to derive the data for the Y chromosome.
Figure 2.
Localization of deletions and amplifications for each chromosome (chr) by moving-window analysis. For genomic visualization of CNV regions cumulated across sample population, we used moving windows of 100 kb and counted the number of cases that contained any CNV region overlapping with the window. The genome-wide CNV map is plotted with chromosomal locations on the x axis [arranged from the beginning of the chromosome (p region) to the end of the chromosome (q region)] and number of cases with CNV on the y axis. The number of cases (of a total of 98) with amplifications and deletions on the specific site is illustrated by red (upward) and blue (downward), respectively. The eight genomic locations (A–H) that contain no known genes and that have a significant number of deletions are noted.
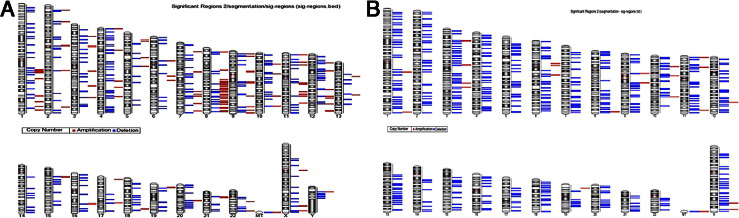
Ideograms demonstrating all of the CNV seen in the study, regardless of their frequency, on their chromosomal locations from male and female cases are presented (Figure 3, A and B, respectively). Y-chromosome data were obtained using only male patients. There were 76 CNV cases from male patients and 22 CNV cases from female patients. Contiguous areas with deletions are seen in some male cases in chromosomes 4, 10, 12, 20, and Y, with contiguous areas of amplification seen in chromosomes 1, 8, and 11 (Figure 3A). Overall, there are no cases with contiguous sites of amplification in female cases, with cases having contiguous areas of deletion seen in chromosomes 5, 9, 11, 13, 19, and 22 (Figure 3B). The precise location and frequency (number of cases) for each deletion and amplification per chromosome are shown in Supplemental Table S1 (available at http://ajp.amjpathol.org).
Figure 3.
Ideogram of chromosome deletions or amplifications. Each bar next to the specific chromosome ideogram demonstrates that at least one locus was detected to have a genome copy number alteration. The frequency of the events is not demonstrated but is shown in Figure 2. Red bars indicate amplifications; blue bars, deletions. The chromosome number is indicated at the bottom of each ideogram. A: Cases from male patients. B: Cases from female patients. The data for male patients were calculated while excluding the female patients, to avoid erroneous detection of a deficient amount of Y-chromosome genomic material because the Y chromosome is absent in females.
Cluster Analysis and Correlation with Tumor Attributes
Clustering analysis of 78 cases was based on CNV present in exons and introns of known expressed genes, ignoring CNV in areas that do not contain expressed genes. Five clear clusters of patients were identified (Figure 4): clusters A to C mainly contain LSP1 deletions, and clusters D1 to D2 contain deletions in five genes on chromosome 11. More specifically, cluster A is characterized by only an LSP1 deletion. Cluster B contains cases with deletions in LSP1 plus GNB1L, PTPRD, and RP1-177G6.2. Cluster C contains a deletion on LSP1 plus KIAA1217. Clusters D1 and D2 have an amplification or a deletion in five genes (YIF1A, RAB1B, PACS1, CNH2, and KLC2) on chromosome 11.
Figure 4.
Clustering of the HCC cases based on CNV (amplifications and deletions). Cases were clustered according to 1324 CNV conditions in the exons or introns of 473 known genes from Partek preprocessing, ignoring areas that do not contain expressed genes. Genes that have CNV changes in <10% (or 9) samples were filtered out. The filtering reduces the data from 93 samples and 473 genes to 78 samples and 15 genes. For details, see Materials and Methods.
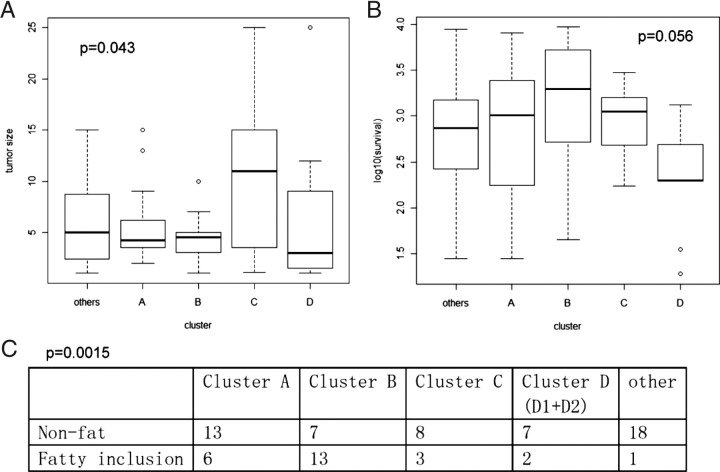
We correlated the cluster assignment with tumor attributes using Fisher's exact test (for discrete attributes) or the F-test from an analysis of variance model (for continuous attributes). A total of 13 attributes were tested (Figure 5). Tested parameters were as follow: sex (P = 0.09), number of tumors (P = 0.7), tumor size (cluster C had a larger tumor size, P = 0.043; Figure 5A), survival (clusters D1 + D2 had shorter survival, P = 0.056; Figure 5B), recurrence (P = 0.4), presence of Mallory bodies (P = 0.53), fat (cluster B had more fatty inclusions, P = 0.0015; Figure 5C), number of fibrolamellar HCCs (P = 0.09), cytoinclusions (P = 0.7), inflammation (P = 0.45), giant cells (P = 0.28), fibrosis (P = 0.23), and bile plugs (P = 0.89).
Figure 5.
Box plots and a contingency table demonstrate tumor attributes correlated with cluster assignment. A: Tumor size. B: Log-transformed survival (not statistically significant). C: Fatty cell inclusion.
Correlations were also seen with some of the attributes and specific genes as follows: tumor size, MAML2 (P = 0.02); inflammation, ETS1 (P = 0.04); presence of fat in tumor cells, GNB1L (P = 0.02) and MAML2 (P = 0.00018); fibrosis, PTPRD (P = 0.02); and bile plugs and cholestasis, RSU1 (P = 0.025).
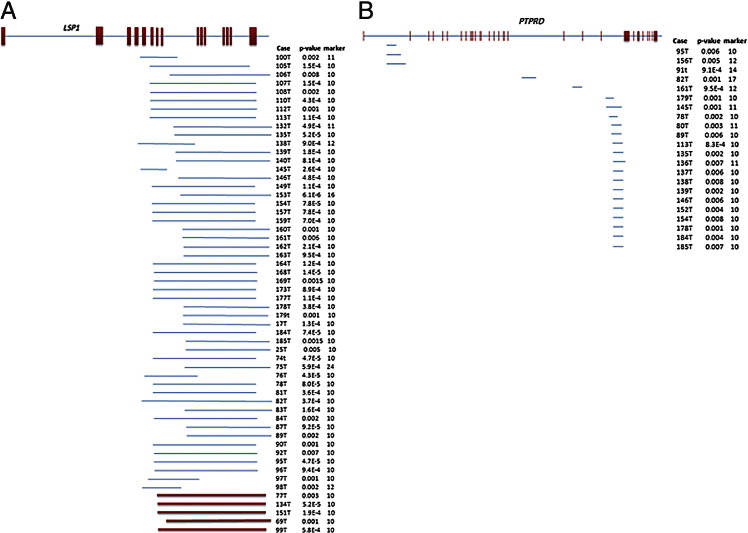
Deletion Mapping for LSP1 and PTPRD
The mapping of the observed deletions in LSP1 and PTPRD (Figure 6, A and B, respectively) demonstrated that deletions of LSP1 occur in the C-terminal region, associated with binding to F-actin filaments.16 The amplifications observed also occur in the same region (as described in Discussion). The deletions seen in PTPRD all seem to occur in a specific intron site of the gene, toward the C terminus (as described in Discussion). To validate the genomic alteration of LSP1 and PTPRD, quantitative PCR was performed on 21 separate cases of liver cancer with matched healthy tissues. As shown in Table 2, 10 of 21 cases showed deletion of LSP1, whereas 2 cases showed amplification. On the other hand, only 5 of 19 cases showed PTPRD deletion, and 1 case had amplification. Of five cases containing PTPRD deletions, four also contain an LSP1 deletion. The data obtained by quantitative PCR show CNV change ratios in agreement with the results obtained by the SNP array.
Figure 6.
Genomic mapping of the deletions (blue) and amplifications (red) in LSP1 and PTPRD. Exons are represented by vertical bar; introns, horizontal line. The spans of deletions of LSP1 (A) or PTPRD (B) are indicated by a blue line; amplifications in LSP1 (A) are shown in red. The minimal number of markers used for detection and P values are indicated. All LSP1 CNVs are located in the basic C-terminal portion of the resulting protein. Most PTPRD deletions occur in a site between exons 24 and 25.
Table 2.
Genome Copy Numbers of LSP1 and PTPRD in a Separate Set of HCC Cases, Assessed by Quantitative PCR
| Case no. | LSP1 gene copies matched T/N⁎ | PTPRD gene copies matched T/N⁎ |
|---|---|---|
| 105 | 0.57/2 | 1.96/2 |
| 107 | 0.81/2 | 2.11/2 |
| 108 | 1.08/2 | NA |
| 109 | 2.32/2 | 4.21/2 |
| 111 | 2.07/2 | NA |
| 121 | 2.25/2 | 1.84/2 |
| 134 | 4.59/2 | 2.01/2 |
| 138 | 0.85/2 | 0.63/2 |
| 139 | 1.09/2 | 1.04/2 |
| 149 | 1.18/2 | 2.18/2 |
| 151 | 3.55/2 | 2.08/2 |
| 118 | 2.06/2 | 2.16/2 |
| 119 | 2.12/2 | 2.18/2 |
| 144 | 2.05/2 | 2.02/2 |
| 145 | 0.76/2 | 0.46/2 |
| 153 | 0.79/2 | 1.94/2 |
| 163 | 1.02/2 | 2.15/2 |
| 184 | 0.89/2 | 0.41/2 |
| 154 | 0.79/2 | 0.10/2 |
| 157 | 1.92/2 | 1.89/2 |
| 165 | 1.95/2 | 2.21/2 |
The 21 cases used for assessment of the gene copy numbers of LSP1 and PTPRD were collected from an existing tissue microarray totally separate from the ones used in this study.
N, normal tissue; NA, not available; T, tissue from tumor.
For each case, T was compared with matched N from the same case.
Genomic Analysis of Regions A to H not Associated with Known Genes
The data shown in Figure 2 list eight genomic locations (A to H) that apparently contain no known genes and that have a significant number of deletions. The data associated with this analysis are given in detail in Supplemental Figure S1 (available at http://ajp.amjpathol.org). In summary, areas upstream and downstream of the deletions were analyzed for the presence of specific genes, listed per region. However, further analysis identified the presence of five motifs present in common in all of these regions (see Supplemental Figure S2 at http://ajp.amjpathol.org). In regions A, B, E, F, and G, these motifs are interspersed. In regions C, D, and H, these motifs are tightly clustered, suggesting the possibility of a promoter enhancer region with binding sites for transcription factors (see Supplemental Figure S3 at http://ajp.amjpathol.org). Of the eight sites, C, D, and H are associated with the most cases (51, 53, and 35, respectively). These areas will be further investigated for the possibility of controlling regions of genes important in HCC growth dysregulation. Genes associated in proximity to these three regions include the following: for C, the DENN/MADD domain containing 1A, LIM homeobox 2; for D, synaptotagmin XV, G-protein–regulated inducer of neurite outgrowth 2; and for H, high-mobility group box 3 (see Supplemental Table S2 at http://ajp.amjpathol.org). Gene ontology analysis for potential associations of the motifs with genes of different functions was also performed, and the results indicate a range of possibilities for involvement of these motifs with expression of specific types with different functions (see Supplemental Table S3 at http://ajp.amjpathol.org). The chromatin status using the ENCODE database shows a weak transcription signal for these areas, as expected (see Supplemental Table S4 at http://ajp.amjpathol.org).
Discussion
The complete list of affected genes and their distribution in the 98 cases is shown in Table 1. Overall, 103 genes had more than three CNVs, and the residual 358 genes had only two CNVs. Given that the total number of genes involved (N = 461) is a small fraction of the genome (estimated at approximately 33,000 genes), we considered even the presence of the same gene on two occasions as worthy of mention. In contrast, the involvement of the same gene in four or more cases, given the odds from the total gene number, warrants explicit consideration as to the biological meaning of the finding in the context of what is known related to hepatocyte growth regulation.
Liver regeneration8,9,13 and growth of hepatocytes in culture17 have been the sources of most information related to hepatocyte growth regulation. The combination of these studies has primarily implicated two growth factor/receptor systems, hepatocyte growth factor (HGF) receptor (MET) and epidermal growth factor receptor and all its ligands, primarily epidermal growth factor, transforming growth factor-α, and amphiregulin, as the primary mitogens for hepatocytes.17 Weaker mitogenic effects have been demonstrated in culture for some ligands of the fibroblast growth factor receptor family members.18 Many cytokines, however, play important roles in liver regeneration (less so in hepatocyte cultures) in priming early stages of the regenerative response. These include norepinephrine,19–21 TNF,22–24 IL-6,25 bile acids,26 serotonin,27 leptin,28 and Notch1.29,30 The combined effect of growth factors and cytokines drives hepatocytes into the cell cycle, resulting in a multiplicity of events within the first hours after partial hepatectomy and causing activation of STAT3, NF-κB, Notch-dependent genes, β-catenin, and cyclin D1.8,9
With the exception of β-catenin (two deletions), Notch-3 (five deletions), FGF19 (three amplifications), none of the previously mentioned genes is directly affected by CNV events, even though they play an important role in the regulation of hepatocyte growth. On the other hand, most of the genes involving many cases identified in this study (Table 1) can be meaningfully and speculatively correlated with components of internal signaling cascades activated by the previously mentioned signaling molecules. The potential correlations discussed later are based on existing published literature about the functions of these genes in hepatocytes or other cell types, not on direct experimentation. The results overall indicate that the primary genetic copy number alterations associated with the development of liver cancer do not target the extracellular signals or the initiators of the intracellular signaling pathways known to be driving hepatocyte growth; rather, they affect the molecules in downstream signaling pathways. These will be described.
LSP1 Gene
This gene, located on chromosome 11 and not so far involved in hepatocyte growth biological features, had the most CNVs (46 deletions and 5 amplifications). LSP1 has been primarily studied in hematopoietic tissues. The final protein is composed of 339 amino acids, with an acidic N terminal and a basic C terminal. The latter contains amino acid sequences highly homologous to actin-binding domains of two F-actin–binding proteins, caldesmon and the villin headpieces (CI, CII, VI, and VII).16 The C-terminal site binds to F-actin sites and is involved in mediation of mobility-associated responses in activated leukocytes.31 Not much is known about binding partners of the N-terminal part. LSP1 is a substrate of p38 mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 and protein kinase C.32 Studies in hematopoietic cells have shown that LSP1 is a cytoskeletal targeting protein for the extracellular signal–regulated kinase/mitogen activated protein kinase pathway. The findings suggest a model in which mitogen activated protein kinase kinase 1 and extracellular signal–regulated kinase 2 are organized in a cytoskeletal signaling complex, together with kinase suppressor of Ras (KSR), protein kinase C, and LSP1.33 Mice genetically deficient in LSP1 exhibit accelerated healing of skin wounds and increased re-epithelialization rates, collagen synthesis, and angiogenesis.34 The results suggest that LSP1 plays an inhibitory role in cell motility, and deletions associated with loss of function may result in enhanced motility and migration. LSP1 loss of function may mediate effects through KSR, a protein directly binding to LSP1.33 KSR was recently described as a major regulator of cell growth, because it acts as a scaffold for mitogen-activated protein kinase, RAF, and mitogen activated protein kinase.35,36 Of interest, all deletions of LSP1 affect the C terminal, containing the F–actin binding region (Figure 3A). LSP1 amplifications, however, also affect the same site. If amplifications were to result in excess production of a truncated protein containing only the C terminal, this may have a decoy function, blocking the binding sites on the F–actin filaments and preventing binding of the full–length molecule. In other studies, certain polymorphisms of LSP1 are associated with enhanced probability of breast cancer development in women who carry germ–line mutations of the BRCA1 gene.37 We have shown that integrin-linked kinase (ILK), another protein associated with the cytoskeleton and controlling function of another Ras suppressor gene (RSU1, see later), through its partner PINCH, also has growth-suppressing effects on healthy hepatocytes.38 LSP1 and ILK may constitute a dual redundant system for control of activity of Ras proteins, thus acting as brakes for hepatocyte growth. Loss of function of either one should have enhancing effects on hepatocyte growth. This has been demonstrated for healthy liver for ILK.38–42
PTPRD Gene
This was the second most affected gene in our study, with 4 amplifications and 17 deletions. Located on chromosome 14, PTPRD is a known tumor suppressor gene, with deletions and inactivating mutations in several human cancers, including lung cancer and glioblastomas.43 Deletions of PTPRD have been shown in rat hepatomas and in the human hepatoblastoma HepG2 cell line.44 Another member of the family, PTPRT, is the most frequently mutated protein tyrosine phosphatase receptor in human cancer.45 Both PTPRD and PTPRT negatively regulate STAT3.45 The activated form of STAT3 is phosphorylated in tyrosine sites by janus kinase kinases. After tyrosine phosphorylation, STAT3 moves to the nucleus, where it functions as a transcription factor and is involved in enhanced transcription of many cell cycle–related genes. This has been demonstrated for hepatocytes during liver regeneration.46,47 PTPRD (and PTRPT) dephosphorylate tyrosine residues responsible for STAT3 activation and, thus, suppress cell growth. In addition to the 21 deletions seen in PTPRD, deletions were also found in the following: PTPN11, four cases (PTPN11 is associated with Noonan syndrome48); PTPRN2, three cases; PTPRT, two cases; PTPRCAP, two cases; and PTPRK, two cases. These findings demonstrate that deletions in members of the PTPR family and, especially, PTPRD contribute to the development of the malignant phenotype in hepatocytes. Of interest, STAT3 is a major component of the signaling machinery involved in the entry of hepatocytes into the S phase during liver regeneration.49 The activation of STAT3 is seen within 1 to 3 hours after partial hepatectomy. IL-6 is a major regulator of the timing of activation of STAT3, and its receptor (gp130) activates STAT3 via activation of JAK kinases.25 There is a delay in liver regeneration in mice deficient in IL-6.25 Growth factor receptors (HGF and epidermal growth factor)50 and the α-1 adrenergic receptor51 also activate STAT3 in a time-delayed fashion in hepatocytes, and they are likely to take over activation of STAT3 in IL-6–deficient mice. All 21 deletions observed in our study occur at an intron site between exons 24 and 25 (Figure 5B).
Ets1 Gene
V-ets erythroblastosis E26 oncogene homolog 1 (Ets1) is a member of the ETS transcription factor family. These proteins regulate numerous genes and are involved in stem cell development, cell senescence and death, and tumorigenesis.52 The conserved ETS domain is a winged helix-turn-helix DNA-binding motif that recognizes the core consensus DNA sequence GGAA/T. There were 11 deletions in the 98 cases (no amplifications) detected in our study. Ets1 is involved in several malignancies as a regulator of key progression-linked determinants, such as osteopontin.53 Most reports relate to overexpression of Ets1 in tumor cells. It is not clear whether the deletions we detected would have a gain or loss-of-function effect. However, Ets1 is a key mediator of the effects of HGF and its receptor, MET. Many HGF effects on induction of metalloproteinases, osteopontin, and angiogenesis are mediated by Ets1.54,55 HGF induces expression of Ets1 via an Ras-dependent pathway.56 Given the key role of HGF as a major hepatocyte mitogen, the findings related to Ets1 need to be placed in perspective with the known studies of HGF related to hepatocyte growth.9
MAML2 Gene
There were seven deletions and two amplifications seen in this gene. MAML2 and associated family members 1 and 3 participate in the formation of the Notch-associated RBP-J/CBF complex, which mediates effects of the intracellular domain of Notch after it migrates to the nucleus.57 It is not clear what effect the observed deletions in MAML2 would have for Notch function. Notch1 is a key regulator of mitogenic signals at the earliest stages of liver regeneration. The intracellular domain of Notch migrates to hepatocyte nuclei within 15 minutes after partial hepatectomy and causes expression of the dependent genes Hes1 and Hes3.30 Inhibition of Notch expression by silencing RNA in all liver cells is associated with a blunted regenerative response.30 On the other hand, germ-line elimination of Notch1 is associated with a diffuse increase in hepatocyte proliferation,29 even though the regenerative response in these animals is also blunted. The findings link MAML2 and, thus, Notch, a regulator of liver regeneration, as a causative contributor to hepatoma malignant behavior. There is extensive literature connecting MAML2 with mucoepidermoid carcinomas, leukemias, lymphomas, and hidradenomas.58
RSU1 Gene
There were seven deletions and three amplifications seen in this gene. RSU1 is a protein expressed in most cells. It has a leucine-rich repeat domain. It causes suppression of Ras-related functions. RSU1 has suppressive effects on the growth of cancer cells.59 It binds to the LIM5 domain of the protein PINCH.60 The latter is part of the complex formed with ILK and Parvin (IPP complex). We have shown, in recent studies,38 that targeted genetic loss of ILK in hepatocytes is associated with enhanced basal levels of hepatocyte proliferation, increased liver size, and enhanced deposition of extracellular matrix. Livers with this defect have an enhanced regenerative response and end up with a significantly increased liver/body weight ratio at the end of regeneration after partial hepatectomy.61 Elimination of ILK results in dissociation of the IPP complex and, potentially, loss of action of RSU1, which is normally bound to the IPP complex via PINCH. We have speculated that loss of function of RSU1, caused by the dissociation of the IPP complex, may be the reason for the enhanced proliferation of hepatocytes after targeted elimination of ILK.39,40 The deletions in RSU1 found in this study connect this work with the findings of abnormally enhanced hepatocyte growth related to targeted ILK removal from hepatocytes. Of interest, the much more frequent deletions in LSP1 (previously described) may also involve the similar loss of KSR (previously described). In fact, six of the seven deletions in RSU1 were seen in cases that also had deletions in LSP1.
PTCH1 Gene
This gene expresses the receptor of Hedgehog, Patched-1, which inhibits Smoothened, a protein associated with initiation of the signaling cascade of the Hedgehog pathway. Hedgehog proteins bind to Patched-1 and prevent it from inhibiting Smoothened. There were three deletions associated with PTCH1. Deletions of Patched-1 are likely to relieve Smoothened from inhibition and, thus, cause continuous activation of the Hedgehog pathway. There is already emerging literature for a role of Hedgehog in regulation of liver regeneration62 and of the growth of hepatic progenitor cells.63 A recent study64 has also documented dysregulation of the Hedgehog pathway in human liver cancer. There is extensive literature65 on the role of this pathway in the development of the basal cell–nevus syndrome and associated neoplasias.
Other Gene Mutations and Genomic Changes
Several other genes listed in Table 1 are also associated with regulation of cell growth in epithelial cells, including hepatocytes. These genes include ABL1, ABL2, Bcl2, APC, JAK2, CCND1, RB1, and BBC3 (alias PUMA), among others.9,10 Other genes, such as GNB1L (18 deletions), TBC1D22A(two amplifications and eight deletions), BAHCC1 (nine deletions), CNIH2 (five amplifications and four deletions), and KLC2 (five amplifications and four deletions), have not been associated with either HCC or cancer biological characteristics in the literature.66 These genes need to be further investigated for their precise role in HCC. Although we see deletions in several of these genes, it is not clear from our data whether these deletions result in a loss or gain of function.
There is extensive literature on chromosomal aberrations in HCC. Previous studies67,68 have focused on deletions on chromosome 1 (both p and q arms) and amplifications in the q region. Our results confirm these studies. Allelic gains have also been described for chromosome 8q, which are also confirmed from our results showing extensive amplifications (in males) of multiple regions in the q arm of chromosome 8.69 Chromosomes 9, 11, and 12 also showed areas of gene amplification (CNV gains). There were several cases with contiguous deletions on the Y chromosome (Figures 2 and 3). The functions of the affected genes are not well understood. Given the high preponderance of HCC in males versus females, the functions of these genes in hepatocyte growth biological features should be investigated.
The clustering of HCC based on CNV of expressed genes is shown in Figure 4. LSP1 plays a dominant role in defining the clustering of the cases. Clusters A through C are mainly associated with LSP1 deletions. Cluster B is distinguished from A and C by the deletions seen in PTPRD. Cluster C is associated with deletions in gene KIAA1217 (a hypothetical locus of unknown function). Cluster D is composed of two parts. Genes YIF1A, RAB1B, PACS1,CNIH2, and KLC2 bear amplifications in cluster 4A and deletions in cluster 4B. CNVs undoubtedly are acting in concert with other genomic changes of smaller size (eg, mutations, such as in β-catenin63 or p5370) to contribute to HCC behavior. Recent studies71 with rodent livers have also demonstrated that hepatocytes become spontaneously aneuploid at a high frequency with increasing age of the animals. It is not clear whether this occurs in human hepatocytes.
Because the time from resection of the tissue samples used in this study spans a wide spectrum, we addressed the possibility of artifacts that should be considered for FFPE material. DNA fragmentation is often seen in FFPE tissue samples. In Figure 1, the distribution of the size of DNA fragments seen in a fresh-frozen HCC versus that of an FFPE sample is almost indistinguishable. In Supplemental Table S1 (available at http://ajp.amjpathol.org), the range of the size of deletions is from 32,854,362 to 535 bp. The method applied for detection of CNV is performed by comparison of the tumor samples with the DNA of the corresponding healthy tissues. Thus, it is unlikely that larger deletions, if present in our sample size, would have been missed due to DNA fragmentation, because fragmentation does not occur on a deleted area (physically paradoxical) but rather on existing DNA fragments. If larger deletions existed in our samples, the method applied would have reported them as such.
Concluding Remarks
The absence of a strong correlation of CNV alterations with many specific tumor parameters, such as multiplicity, histological variables, and survival, in this series is not totally unexpected for several reasons. First, the presence of a few cases with no evidence of CNV suggests that full tumor development may occur in the complete absence of such changes. Our retrospective series is limited in number, is restricted to a single transplantation center, and spans several decades during which clinical practice evolved considerably. Therefore, it would be unlikely that correlation with any single CNV subset would be of sufficient strength to emerge as a dominant predictor of tumor behavior. Rather, the results indicate the need for integrating such studies with other modalities of tumor cell genomic analysis in many patients to construct a comprehensive roadmap of the multiplicity of pathways leading to HCC pathogenesis. On the other hand, studies6,72 of the patterns of gene expression of HCC have shown clustering into two to three overall patterns. The combined studies suggest that alignments of multiple genomic alterations may activate one of the two to three specific gene expression patterns and that neoplastic behavior of the affected hepatocytes ensues once such threshold change has been achieved. Also, we did not observe any correlation between the genomic alterations observed and the associated etiology that led to the cirrhosis pattern that caused liver resection, including alcoholic liver disease and viral causes. A similar lack of correlations has been noted with such potentially associated pathogenetic factors and the associated gene expression pattern of HCC in previous studies.
Footnotes
Supported by the Rangos Fund for Enhancement of Pathology Research (RSG-08-137-01-CNE to Y.P.Y.) and grants from the NIH (5R01CA103958-05 and 5R01CA035373-27 to G.K.M. and R01CA098249 to J.-H.L.).
M.A.N. and G.T. contributed equally to this work.
Supplemental material for this article can be found on http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.12.021.
Supplementary data
Determination of the eight frequent CNV regions. A: From the eight regions highlighted (A–H), the regions that repeatedly show CNV deletion patterns among different cases are shown. B: The frequent CNV regions are determined as between the blue dashed lines. The exact start and end chromosomal positions and the number of cases associated with each CNV region are shown in the table.
Identifying sequence motifs in the eight frequent CNV regions. We performed motif finding in the eight frequent CNV regions using the MEME web tool. The identified motifs provide information of potential transcription binding sites that explains frequent deletion in patients with cancer. The top five motifs, ranked by the q values and the motifs, are shown.
Location and distribution of the five motifs in the eight frequent CNV regions. Regions C and H clearly have many motif 1 appearances assembled closely together with a few other motifs. Region D has many motif 1, 4, and 5 appearances, also assembled closely. Regions C, D, and H have the most cases containing a deletion in the regions (51, 53, and 35 cases, respectively).
References
- 1.Pitot H.C. Adventures in hepatocarcinogenesis. Ann Rev Pathol. 2007;2:1–29. doi: 10.1146/annurev.pathol.2.010506.092027. [DOI] [PubMed] [Google Scholar]
- 2.Thomas M.B., Jaffe D., Choti M.M., Belghiti J., Curley S., Fong Y., Gores G., Kerlan R., Merle P., O'Neil B., Poon R., Schwartz L., Tepper J., Yao F., Haller D., Mooney M., Venook A. Hepatocellular carcinoma: consensus recommendations of the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol. 2010;28:3994–4005. doi: 10.1200/JCO.2010.28.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feitelson M.A., Reis H.M., Liu J., Lian Z., Pan J. Hepatitis B virus X antigen (HBxAg) and cell cycle control in chronic infection and hepatocarcinogenesis. Front Biosci. 2005;10:1558–1572. doi: 10.2741/1640. [DOI] [PubMed] [Google Scholar]
- 4.Lee J.S., Thorgeirsson S.S. Genetic profiling of human hepatocellular carcinoma. Sem Liver Dis. 2005;25:125–132. doi: 10.1055/s-2005-871192. [DOI] [PubMed] [Google Scholar]
- 5.Lee J.S., Chu I.S., Heo J., Calvisi D.F., Sun Z., Roskams T., Durnez A., Demetris A.J., Thorgeirsson S.S. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 6.Luo J.H., Ren B., Keryanov S., Tseng G.C., Rao U.N., Monga S.P., Strom S., Demetris A.J., Nalesnik M., Yu Y.P., Ranganathan S., Michalopoulos G.K. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44:1012–1024. doi: 10.1002/hep.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo H.G., Park E.S., Lee J.S., Lee Y.H., Ishikawa T., Kim Y.J., Thorgeirsson S.S. Identification of potential driver genes in human liver carcinoma by genomewide screening. Cancer Res. 2009;69:4059–4066. doi: 10.1158/0008-5472.CAN-09-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michalopoulos G.K., DeFrances M.C. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 9.Michalopoulos G.K. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalopoulos G.K. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalopoulos G.K. Liver regeneration: alternative epithelial pathways. Int J Biochem Cell Biol. 2011;43:173–179. doi: 10.1016/j.biocel.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fausto N. Liver regeneration. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 13.Fausto N., Campbell J.S., Riehle K.J. Liver regeneration. Hepatology. 2006;43:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 14.Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology. 2004;39:1477–1487. doi: 10.1002/hep.20214. [DOI] [PubMed] [Google Scholar]
- 15.Mazzaferro V., Regalia E., Doci R., Andreola S., Pulvirenti A., Bozzetti F., Montalto F., Ammatuna M., Morabito A., Gennari L. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334:693–699. doi: 10.1056/NEJM199603143341104. [DOI] [PubMed] [Google Scholar]
- 16.Jongstra-Bilen J., Janmey P.A., Hartwig J.H., Galea S., Jongstra J. The lymphocyte-specific protein LSP1 binds to F-actin and to the cytoskeleton through its COOH-terminal basic domain. J Cell Biol. 1992;118:1443–1453. doi: 10.1083/jcb.118.6.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Block G.D., Locker J., Bowen W.C., Petersen B.E., Katyal S., Strom S.C., Riley T., Howard T.A., Michalopoulos G.K. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J Cell Biol. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houck K.A., Zarnegar R., Muga S.J., Michalopoulos G.K. Acidic fibroblast growth factor (HBGF-1) stimulates DNA synthesis in primary rat hepatocyte cultures. J Cell Physiol. 1990;143:129–132. doi: 10.1002/jcp.1041430117. [DOI] [PubMed] [Google Scholar]
- 19.Cruise J.L., Houck K.A., Michalopoulos G.K. Induction of DNA synthesis in cultured rat hepatocytes through stimulation of alpha 1 adrenoreceptor by norepinephrine. Science. 1985;227:749–751. doi: 10.1126/science.2982212. [DOI] [PubMed] [Google Scholar]
- 20.Cruise J.L., Knechtle S.J., Bollinger R.R., Kuhn C., Michalopoulos G. Alpha 1-adrenergic effects and liver regeneration. Hepatology. 1987;7:1189–1194. doi: 10.1002/hep.1840070604. [DOI] [PubMed] [Google Scholar]
- 21.Houck K.A., Cruise J.L., Michalopoulos G. Norepinephrine modulates the growth-inhibitory effect of transforming growth factor-beta in primary rat hepatocyte cultures. J Cell Physiol. 1988;135:551–555. doi: 10.1002/jcp.1041350327. [DOI] [PubMed] [Google Scholar]
- 22.Webber E.M., Bruix J., Pierce R.H., Fausto N. Tumor necrosis factor primes hepatocytes for DNA replication in the rat. Hepatology. 1998;28:1226–1234. doi: 10.1002/hep.510280509. [DOI] [PubMed] [Google Scholar]
- 23.Yamada Y., Fausto N. Deficient liver regeneration after carbon tetrachloride injury in mice lacking type 1 but not type 2 tumor necrosis factor receptor. Am J Pathol. 1998;152:1577–1589. [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada Y., Webber E.M., Kirillova I., Peschon J.J., Fausto N. Analysis of liver regeneration in mice lacking type 1 or type 2 tumor necrosis factor receptor: requirement for type 1 but not type 2 receptor. Hepatology. 1998;28:959–970. doi: 10.1002/hep.510280410. [DOI] [PubMed] [Google Scholar]
- 25.Cressman D.E., Greenbaum L.E., DeAngelis R.A., Ciliberto G., Furth E.E., Poli V., Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 26.Huang W., Ma K., Zhang J., Qatanani M., Cuvillier J., Liu J., Dong B., Huang X., Moore D.D. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 27.Lesurtel M., Graf R., Aleil B., Walther B., Tian Y., Jochum W., Gaget C., Bader M., Clavien P. Platelet-derived serotonin mediates liver regeneration. Science. 2006;312:104–107. doi: 10.1126/science.1123842. [DOI] [PubMed] [Google Scholar]
- 28.Yamauchi H., Uetsuka K., Okada T., Nakayama H., Doi K. Impaired liver regeneration after partial hepatectomy in db/db mice. Exp Toxicol Pathol. 2003;54:281–286. doi: 10.1078/0940-2993-00265. [DOI] [PubMed] [Google Scholar]
- 29.Croquelois A., Blindenbacher A., Terracciano L., Wang X., Langer I., Radtke F., Heim M.H. Inducible inactivation of Notch1 causes nodular regenerative hyperplasia in mice. Hepatology. 2005;41:487–496. doi: 10.1002/hep.20571. [DOI] [PubMed] [Google Scholar]
- 30.Kohler C., Bell A.W., Bowen W.C., Monga S.P., Fleig W., Michalopoulos G.K. Expression of Notch-1 and its ligand Jagged-1 in rat liver during liver regeneration. Hepatology. 2004;39:1056–1065. doi: 10.1002/hep.20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jongstra-Bilen J., Jongstra J. Leukocyte-specific protein 1 (LSP1): a regulator of leukocyte emigration in inflammation. Immunol Res. 2006;35:65–74. doi: 10.1385/IR:35:1:65. [DOI] [PubMed] [Google Scholar]
- 32.Huang C.K., Zhan L., Ai Y., Jongstra J. LSP1 is the major substrate for mitogen-activated protein kinase-activated protein kinase 2 in human neutrophils. J Biol Chem. 1997;272:17–19. doi: 10.1074/jbc.272.1.17. [DOI] [PubMed] [Google Scholar]
- 33.Harrison R.E., Sikorski B.A., Jongstra J. Leukocyte-specific protein 1 targets the ERK/MAP kinase scaffold protein KSR and MEK1 and ERK2 to the actin cytoskeleton. J Cell Sci. 2004;117:2151–2157. doi: 10.1242/jcs.00955. [DOI] [PubMed] [Google Scholar]
- 34.Wang J., Jiao H., Stewart T.L., Lyons M.V., Shankowsky H.A., Scott P.G., Tredget E.E. Accelerated wound healing in leukocyte-specific, protein 1-deficient mouse is associated with increased infiltration of leukocytes and fibrocytes. J Leukoc Biol. 2007;82:1554–1563. doi: 10.1189/0507306. [DOI] [PubMed] [Google Scholar]
- 35.McKay M.M., Ritt D.A., Morrison D.K. Signaling dynamics of the KSR1 scaffold complex. Proc Natl Acad Sci U S A. 2009;106:11022–11027. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKay M.M., Ritt D.A., Morrison D.K. RAF inhibitor-induced KSR1/B-RAF binding and its effects on ERK cascade signaling. Curr Biol. 2011;21:563–568. doi: 10.1016/j.cub.2011.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antoniou A.C., Sinilnikova O.M., McGuffog L., Healey S., Nevanlinna H., Heikkinen T. CIMBA: Common variants in LSP1, 2q35 and 8q24 and breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum Mol Genet. 2009;18:4442–4456. doi: 10.1093/hmg/ddp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gkretsi V., Apte U., Mars W.M., Bowen W.C., Luo J.H., Yang Y., Yu Y.P., Orr A., St-Arnaud R., Dedhar S., Kaestner K.H., Wu C., Michalopoulos G.K. Liver-specific ablation of integrin-linked kinase in mice results in abnormal histology, enhanced cell proliferation, and hepatomegaly. Hepatology. 2008;48:1932–1941. doi: 10.1002/hep.22537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apte U., Gkretsi V., Bowen W.C., Mars W.M., Luo J.H., Donthamsetty S., Orr A., Monga S.P., Wu C., Michalopoulos G.K. Enhanced liver regeneration following changes induced by hepatocyte-specific genetic ablation of integrin-linked kinase. Hepatology. 2009;50:844–851. doi: 10.1002/hep.23059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donthamsetty S., Bowen W., Mars W., Bhave V., Luo J.H., Wu C., Hurd J., Orr A., Bell A., Michalopoulos G. Liver-specific ablation of integrin-linked kinase in mice results in enhanced and prolonged cell proliferation and hepatomegaly after phenobarbital administration. Toxicol Sci. 2010;113:358–366. doi: 10.1093/toxsci/kfp281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gkretsi V., Bowen W.C., Yang Y., Wu C., Michalopoulos G.K. Integrin-linked kinase is involved in matrix-induced hepatocyte differentiation. Biochem Biophys Res Commun. 2007;353:638–643. doi: 10.1016/j.bbrc.2006.12.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gkretsi V., Mars W.M., Bowen W.C., Barua L., Yang Y., Guo L., St-Arnaud R., Dedhar S., Wu C., Michalopoulos G.K. Loss of integrin linked kinase from mouse hepatocytes in vitro and in vivo results in apoptosis and hepatitis. Hepatology. 2007;45:1025–1034. doi: 10.1002/hep.21540. [DOI] [PubMed] [Google Scholar]
- 43.Veeriah S., Brennan C., Meng S., Singh B., Fagin J.A., Solit D.B., Paty P.B., Rohle D., Vivanco I., Chmielecki J., Pao W., Ladanyi M., Gerald W.L., Liau L., Cloughesy T.C., Mischel P.S., Sander C., Taylor B., Schultz N., Major J., Heguy A., Fang F., Mellinghoff I.K., Chan T.A. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci U S A. 2009;106:9435–9440. doi: 10.1073/pnas.0900571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urushibara N., Karasaki H., Nakamura K., Mizuno Y., Ogawa K., Kikuchi K. The selective reduction in PTPdelta expression in hepatomas. Int J Oncol. 1998;12:603–607. [PubMed] [Google Scholar]
- 45.Zhang X., Guo A., Yu J., Possemato A., Chen Y., Zheng W., Polakiewicz R.D., Kinzler K.W., Vogelstein B., Velculescu V.E., Wang Z.J. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc Natl Acad Sci U S A. 2007;104:4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cressman D.E., Diamond R.H., Taub R. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology. 1995;21:1443–1449. [PubMed] [Google Scholar]
- 47.Li W., Liang X., Kellendonk C., Poli V., Taub R. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J Biol Chem. 2002;277:28411–28417. doi: 10.1074/jbc.M202807200. [DOI] [PubMed] [Google Scholar]
- 48.Dahlgren J. GH therapy in Noonan syndrome: review of final height data. Horm Res. 2009;72(Suppl 2):46–48. doi: 10.1159/000243779. [DOI] [PubMed] [Google Scholar]
- 49.Li W., Liang X., Kellendonk C., Poli V., Taub R. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J Biol Chem. 2002;277:28411–28417. doi: 10.1074/jbc.M202807200. [DOI] [PubMed] [Google Scholar]
- 50.Runge D.M., Runge D., Foth H., Strom S.C., Michalopoulos G.K. STAT 1alpha/1beta, STAT 3 and STAT 5: expression and association with c-MET and EGF-receptor in long-term cultures of human hepatocytes. Biochem Biophys Res Commun. 1999;265:376–381. doi: 10.1006/bbrc.1999.1681. [DOI] [PubMed] [Google Scholar]
- 51.Han C., Bowen W.C., Michalopoulos G.K., Wu T. Alpha-1 adrenergic receptor transactivates signal transducer and activator of transcription-3 (Stat3) through activation of Src and epidermal growth factor receptor (EGFR) in hepatocytes. J Cell Physiol. 2008;216:486–497. doi: 10.1002/jcp.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wai P.Y., Mi Z., Gao C., Guo H., Marroquin C., Kuo P.C. Ets-1 and runx2 regulate transcription of a metastatic gene, osteopontin, in murine colorectal cancer cells. J Biol Chem. 2006;281:18973–18982. doi: 10.1074/jbc.M511962200. [DOI] [PubMed] [Google Scholar]
- 54.Jinnin M., Ihn H., Mimura Y., Asano Y., Yamane K., Tamaki K. Matrix metalloproteinase-1 up-regulation by hepatocyte growth factor in human dermal fibroblasts via ERK signaling pathway involves Ets1 and Fli1. Nucleic Acids Res. 2005;33:3540–3549. doi: 10.1093/nar/gki648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomita N., Morishita R., Taniyama Y., Koike H., Aoki M., Shimizu H., Matsumoto K., Nakamura T., Kaneda Y., Ogihara T. Angiogenic property of hepatocyte growth factor is dependent on upregulation of essential transcription factor for angiogenesis, ets-1. Circulation. 2003;107:1411–1417. doi: 10.1161/01.cir.0000055331.41937.aa. [DOI] [PubMed] [Google Scholar]
- 56.Paumelle R., Tulasne D., Kherrouche Z., Plaza S., Leroy C., Reveneau S., Vandenbunder B., Fafeur V. Hepatocyte growth factor/scatter factor activates the ETS1 transcription factor by a RAS-RAF-MEK-ERK signaling pathway. Oncogene. 2002;21:2309–2319. doi: 10.1038/sj.onc.1205297. [DOI] [PubMed] [Google Scholar]
- 57.Enlund F., Behboudi A., Andren Y., Oberg C., Lendahl U., Mark J., Stenman G. Altered Notch signaling resulting from expression of a WAMTP1-MAML2 gene fusion in mucoepidermoid carcinomas and benign Warthin's tumors. Exp Cell Res. 2004;292:21–28. doi: 10.1016/j.yexcr.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 58.Behboudi A., Enlund F., Winnes M., Andrén Y., Nordkvist A., Leivo I., Flaberg E., Szekely L., Mäkitie A., Grenman R., Mark J., Stenman G. Molecular classification of mucoepidermoid carcinomas: prognostic significance of the MECT1-MAML2 fusion oncogene. Genes Chromosomes Cancer. 2006;45:470–481. doi: 10.1002/gcc.20306. [DOI] [PubMed] [Google Scholar]
- 59.Vasaturo F., Dougherty G.W., Cutler M.L. Ectopic expression of Rsu-1 results in elevation of p21CIP and inhibits anchorage-independent growth of MCF7 breast cancer cells. Breast Cancer Res Treat. 2000;61:69–78. doi: 10.1023/a:1006462323260. [DOI] [PubMed] [Google Scholar]
- 60.Dougherty G.W., Jose C., Gimona M., Cutler M.L. The Rsu-1-PINCH1-ILK complex is regulated by Ras activation in tumor cells. Eur J Cell Biol. 2008;87:721–734. doi: 10.1016/j.ejcb.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Apte U., Gkretsi V., Bowen W.C., Mars W.M., Luo J.H., Donthamsetty S., Orr A., Monga S.P., Wu C., Michalopoulos G.K. Enhanced liver regeneration following changes induced by hepatocyte-specific genetic ablation of integrin-linked kinase. Hepatology. 2009;50:844–851. doi: 10.1002/hep.23059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ochoa B., Syn W.K., Delgado I., Karaca G.F., Jung Y., Wang J., Zubiaga A.M., Fresnedo O., Omenetti A., Zdanowicz M., Choi S.S., Diehl A.M. Hedgehog signaling is critical for normal liver regeneration after partial hepatectomy in mice. Hepatology. 2010;51:1712–1723. doi: 10.1002/hep.23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Omenetti A., Diehl A.M. The adventures of sonic hedgehog in development and repair, II: sonic hedgehog and liver development, inflammation, and cancer. Am J Physiol Gastrointest Liver Physiol. 2008;294:G595–G598. doi: 10.1152/ajpgi.00543.2007. [DOI] [PubMed] [Google Scholar]
- 64.Sicklick J.K., Li Y.X., Jayaraman A., Kannangai R., Qi Y., Vivekanandan P., Ludlow J.W., Owzar K., Chen W., Torbenson M.S., Diehl A.M. Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis. 2006;27:748–757. doi: 10.1093/carcin/bgi292. [DOI] [PubMed] [Google Scholar]
- 65.Saran A. Basal cell carcinoma and the carcinogenic role of aberrant Hedgehog signaling. Future Oncol. 2010;6:1003–1014. doi: 10.2217/fon.10.49. [DOI] [PubMed] [Google Scholar]
- 66.Gong L., Liu M., Jen J., Yeh E.T. GNB1L, a gene deleted in the critical region for DiGeorge syndrome on 22q11, encodes a G-protein beta-subunit-like polypeptide. Biochim Biophys Acta. 2000;1494:185–188. doi: 10.1016/s0167-4781(00)00189-5. [DOI] [PubMed] [Google Scholar]
- 67.Kuroki T., Fujiwara Y., Tsuchiya E., Nakamori S., Imaoka S., Kanematsu T., Nakamura Y. Accumulation of genetic changes during development and progression of hepatocellular carcinoma: loss of heterozygosity of chromosome arm 1p occurs at an early stage of hepatocarcinogenesis. Genes Chromosomes Cancer. 1995;13:163–167. doi: 10.1002/gcc.2870130305. [DOI] [PubMed] [Google Scholar]
- 68.Bilger A., Bennett L.M., Carabeo R.A., Chiaverotti T.A., Dvorak C., Liss K.M., Schadewald S.A., Pitot H.C., Drinkwater N.R. A potent modifier of liver cancer risk on distal mouse chromosome 1: linkage analysis and characterization of congenic lines. Genetics. 2004;167:859–866. doi: 10.1534/genetics.103.024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patil M.A., Gutgemann I., Zhang J., Ho C., Cheung S.T., Ginzinger D., Li R., Dykema K.J., So S., Fan S.T., Kakar S., Furge K.A., Buttner R., Chen X. Array-based comparative genomic hybridization reveals recurrent chromosomal aberrations and Jab1 as a potential target for 8q gain in hepatocellular carcinoma. Carcinogenesis. 2005;26:2050–2057. doi: 10.1093/carcin/bgi178. [DOI] [PubMed] [Google Scholar]
- 70.Hsia C.C., Nakashima Y., Thorgeirsson S.S., Harris C.C., Minemura M., Momosaki S., Wang N.J., Tabor E. Correlation of immunohistochemical staining and mutations of p53 in human hepatocellular carcinoma. Oncol Rep. 2000;7:353–356. [PubMed] [Google Scholar]
- 71.Duncan A.W., Taylor M.H., Hickey R.D., Hanlon Newell A.E., Lenzi M.L., Olson S.B., Finegold M.J., Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee J.S., Thorgeirsson S.S. Functional and genomic implications of global gene expression profiles in cell lines from human hepatocellular cancer. Hepatology. 2002;35:1134–1143. doi: 10.1053/jhep.2002.33165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Determination of the eight frequent CNV regions. A: From the eight regions highlighted (A–H), the regions that repeatedly show CNV deletion patterns among different cases are shown. B: The frequent CNV regions are determined as between the blue dashed lines. The exact start and end chromosomal positions and the number of cases associated with each CNV region are shown in the table.
Identifying sequence motifs in the eight frequent CNV regions. We performed motif finding in the eight frequent CNV regions using the MEME web tool. The identified motifs provide information of potential transcription binding sites that explains frequent deletion in patients with cancer. The top five motifs, ranked by the q values and the motifs, are shown.
Location and distribution of the five motifs in the eight frequent CNV regions. Regions C and H clearly have many motif 1 appearances assembled closely together with a few other motifs. Region D has many motif 1, 4, and 5 appearances, also assembled closely. Regions C, D, and H have the most cases containing a deletion in the regions (51, 53, and 35 cases, respectively).