Abstract
Generation and destruction of antigenic peptides by ER resident aminopeptidases ERAP1 and ERAP2 have been shown in the last few years to be important for the correct functioning and regulation of the adaptive immune response. These two highly homologous aminopeptidases appear to have evolved complex mechanisms well suited for their biological role in antigen presentation. Furthermore, polymorphic variability in these enzymes appears to affect their function and predispose individuals to disease. This review discusses our current understanding of the molecular mechanisms behind ERAP1/2 function as suggested by several recently determined crystallographic structures of these enzymes.
Biology of ERAP1/2
The human adaptive immune system identifies diseased and aberrant cells by monitoring the cell-surface presentation of the peptide products of proteolytic digestion of intracellular and endocytosed proteins (Rock and Goldberg, 1999), which are indicative of the overall cellular protein content. Peptides are presented by MHC-I and MHC-II proteins for recognition by receptors on T cells. In general, peptides bound by MHC-I proteins require proteolytic processing before binding. The proteolytic pathway that leads to the generation of most antigenic epitopes starts at the proteasome and ends with a series of trimming events in the endoplasmic reticulum by the ER-resident aminopeptidases ERAP1 and/or ERAP2. Peptides longer than 8-10 residues (the optimum length for binding to most MHC-I allelic variants) can be N-terminally processed in the ER by ERAP1 and/or ERAP2 to generate the correct-length mature antigenic peptide, however this processing can also destroy epitopes by trimming to lengths too small to bind onto MHC class I molecules. The importance of ERAP1 and ERAP2 for the generation of several key antigenic epitopes that relate to human disease has been thoroughly demonstrated both in vitro and in vivo [reviewed in (Evnouchidou et al., 2009; Haroon and Inman, 2010) and (Weimershaus et al., 2012)]. Furthermore, the capacity of these two enzymes to destroy epitopes has also been shown to be a fundamental part of their biological role (York et al., 2002). This dual “generate or destroy” role gives these two enzymes the capability to control antigen generation by selecting which epitopes will be efficiently produced and loaded onto MHC class I molecules. As a result ERAP1/2 activity can influence the antigenic peptide repertoire and the resulting immunodominance hierarchy (Blanchard et al., 2008; Hammer et al., 2007; York et al., 2006). These regulatory properties of ERAP1 and ERAP2 have elevated interest in the function of these molecules amongst immunologists but remain poorly understood at both functional and mechanistic levels.
ERAP1 and ERAP2 are both zinc metalloaminopeptidases that belong to the M1 protease family (Rawlings et al., 2012) characterized by GAMEN and HExxHx18E motifs. They are highly homologous (~50% sequence identity) and along with the homologous IRAP (~50 % sequence identity) form a gene cluster on chromosome 5 in humans (in mouse, ERAP1 is on chromosome 13, IRAP is on chromosome 17, and ERAP2 is absent). All three recently have been classified to the oxytocinase subfamily of M1 aminopeptidases (Tsujimoto and Hattori, 2005). ERAP1 was the first to be associated with a role in antigen processing and as a result this enzyme has been more thoroughly characterized. ERAP1 has molecular and enzymatic properties that are well suited to its specific biological role. Many antigenic peptide precursors that enter the ER are too long to bind to MHC-I proteins, having 1-6 or possibly more amino acids that require trimming (Cascio et al., 2001). In general, aminopeptidases show reduced activity for substrates of that length (10-15 amino acids) and display higher activity for shorter substrates, a property that would promote epitope destruction unless a specific protection mechanism was present. Thus, an aminopeptidase with a role in epitope generation would need an unusual length preference. Furthermore, antigenic peptide precursors vary tremendously in terms of peptide sequence and as a result any aminopeptidase activity (or activities) suitable for trimming them would have to be able to deal with a very large number of different peptide sequences. Biochemical analysis showed that indeed ERAP1 was able to trim larger peptides faster than smaller ones, and that residues throughout the whole length of the enzyme could affect processing rates (Chang et al., 2005; Evnouchidou et al., 2008). These properties suggest that ERAP1 has evolved to fill in this specific biological role and imply that the enzyme could impose a bias on the proteolytic fate of possible epitopes depending on their sequences (Georgiadou et al., 2010). In fact, MHC-I proteins from an ERAP1-deficient mouse carry a spectrum of peptides substantially different than in ERAP1-sufficient animals (Blanchard et al., 2010; Hammer et al., 2007). Less is known about the preferences of ERAP2, in part because of its absence in mouse and consequent lack of a gene-deficient model organism. Initial reports suggest that ERAP2 may not share the length preferences of ERAP1 (Chang et al., 2005). However, ERAP2 has been shown to be important for the trimming of precursor sequences that ERAP1 trims inefficiently, possibly by forming ERAP1/2 functional heterodimers (Saveanu et al., 2005). The third aminopeptidase involved in antigen processing, IRAP, has been shown to generate antigenic epitopes on a discreet pathway of cross-presentation independently of ERAP1 and ERAP2, and to share at least the basic functionality of ERAP1 for efficiently trimming larger precursor sequences (Georgiadou et al., 2010; Saveanu et al., 2009).
Structures of ERAP1 and ERAP2
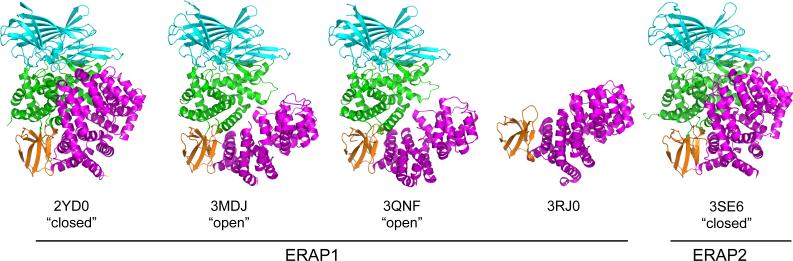
During the last two years several three-dimensional structures of ERAP1 and ERAP2 have been solved, a development that has greatly contributed to our understanding of both the mechanism of peptide trimming and substrate selection preferences for these two enzymes (Ascher et al., 2012; Birtley et al., 2012; Evnouchidou et al., 2012; Gandhi et al., 2011; Kochan et al., 2011; Nguyen et al., 2011). Both enzymes show an overall similar domain topology, having four structural domains folding over a concave structure (Figure 1). Domain II (green in Figure 1) is the catalytic domain that contains the zinc atom and catalytic motifs. Domain I (cyan) caps off the active site. Domain III (orange) is a small sandwich domain between domains II and IV that acts like a hinge facilitating postulated conformational changes between domains II and IV (see below). Finally, domain IV (magenta) has a concave structure comprised by several helical armadillo-type repeats.
Figure 1.
Cartoon representations of known ERAP1/2 crystal structures. Domain I is shown in cyan, domain II in green, domain III in orange and IV in magenta. Note the repositioning of domain IV relative to domains I and II in the “open” versus the “closed” conformations.
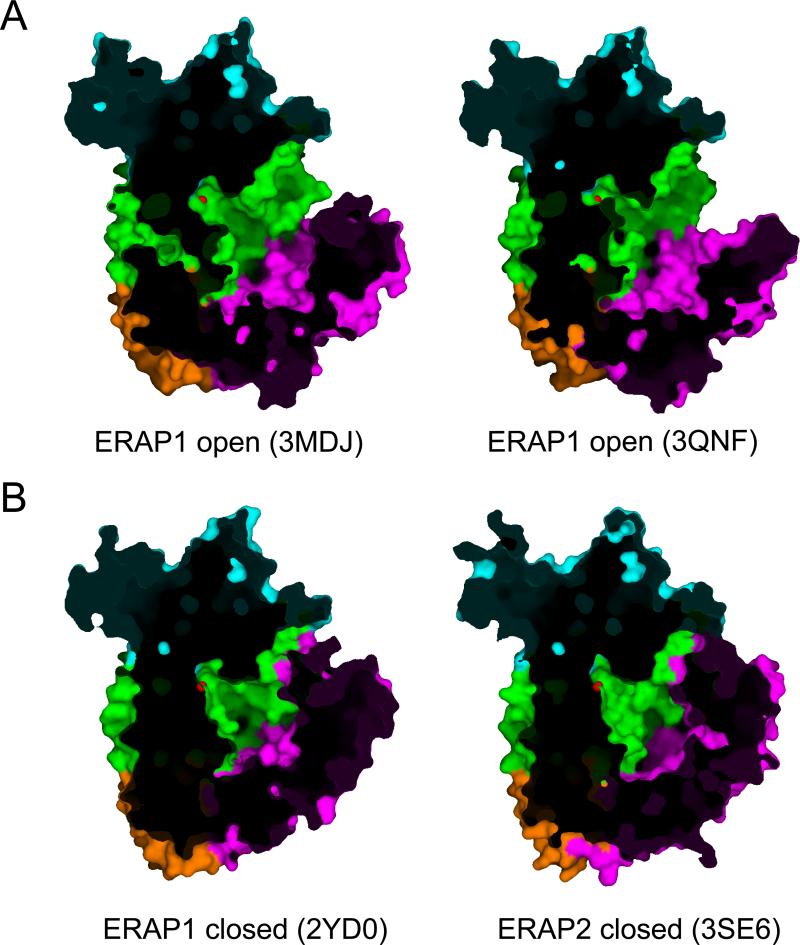
ERAP1 has been shown to adopt at least two distinct conformational states inside the crystal that differ in the orientation of domain IV relative to domains II and I. In one conformation (PDB codes 3MDJ and 3QNF), henceforth termed “open”, domain IV is oriented away from domain II leading to the formation of a large, shallow and solvent exposed cavity starting from the base of the domain I and II interface, extending through the base of the interface of domains III and IV, and ending in the concave section of domain IV (Kochan et al., 2011; Nguyen et al., 2011) (Figure 2A). In another conformation (PDB code 2YDO), henceforth termed “closed”, the edge of domain IV is juxtaposed onto the edge of domain I, also making interactions with domain II (Figure 2B). This topology completely excludes the enzyme's catalytic site from the solvent, but also creates a very large internal cavity, lined by residues of domain II and IV. ERAP2 follows a similar domain topology but has only been crystalized in a configuration similar to the “closed” ERAP1 structure (PDB codes 3SE6 and 4E36) (Birtley et al., 2012; Evnouchidou et al., 2012).
Figure 2.
Surface representation comparing the “open” to the “closed” ERAP1 and ERAP2 structures. Domains are colored as in figure 1. Cutaway views are shown to allow visualization of the internal cavity of the enzyme. The Zn(II) atom is shown in red. Notice how the internal cavity is open towards the solvent on the top-left edge of each figure.
The identification of such a large cavity adjacent to the enzyme's catalytic center has led researchers to hypothesize that it constitutes a binding site for elongated peptides (Birtley et al., 2012; Gandhi et al., 2011; Kochan et al., 2011; Nguyen et al., 2011). Indeed, the size of the cavity is sufficient to accommodate even the largest of ERAP1's substrates (16 amino acid long). A series of shallow pockets could provide atomic interactions that drive substrate binding and at the same time display sufficiently broad specificity to allow processing of the large pool of sequences of peptides that ERAP1 is likely to encounter inside the ER. The overall electrostatic potential of this cavity is negative, something that is consistent with the experimental observation that ERAP1 often prefers peptide substrates with positively charged amino acids at several positions in the sequence (Evnouchidou et al., 2008). Comparison between the structural features of this internal cavity for ERAP1 and ERAP2 has suggested that sufficient differences exist between the two enzymes to support different selectivity for substrates (Birtley et al., 2012). However, the absence of crystal structures for complexes of ERAP1/2 with substrates or inhibitors longer than the dipeptide analog bestatin, makes it difficult to unequivocally identify specificity pockets and compare substrate preferences.
Recently, the crystal structure of the isolated C-terminal domain of ERAP1 has been solved, showing a concave configuration highly similar to the one found in the full-length ERAP1 structures (Gandhi et al., 2011). In this structure, a crystallographic dimer is formed by interactions between the C-terminal polyhistidine tag of one monomer and the interior surface of the other. The nature of the interactions as well as the presence of histidine residues in several naturally occurring epitopes prompted the researchers to propose that this interaction mimics the natural binding of a peptide substrate by the enzyme (Gandhi et al., 2011). Indeed, several of the residues implicated have been previously proposed to constitute the C-terminus binding pocket based on the analysis of a full-length ERAP1 crystal structure (Kochan et al., 2011). The recently solved crystal structure of ERAP2 however revealed that the pocket suggested to be responsible for C-terminus binding in ERAP1 is not conserved in ERAP2, although it should be noted that ERAP2 probably has different length and C-terminal sequence preferences from ERAP1 (Birtley et al., 2012; Chang et al., 2005)
Conformational changes
The observation of two distinct ERAP1 crystal structures (“open” and “closed”) suggested that the enzyme could undergo a significant conformational change during its catalytic cycle. This idea was further supported by the observation that in the closed structure, the catalytic site and S1 specificity pocket were more organized but lack direct access to the solvent that would be necessary to allow both substrate binding and product release (Kochan et al., 2011; Nguyen et al., 2011). Furthermore, a key active site residue universally conserved at this position in M1 family aminopeptidases, Tyr438, is differently oriented between the two ERAP1 conformations. Mechanistic studies of other M1 family members have established the key functional role for a tyrosine at this position (Addlagatta et al., 2006) and in ERAP1 mutagenesis to phenylalanine abolished catalytic activity, suggesting that the functional role was conserved in ERAP1 (Nguyen et al., 2011). In the open form, the tyrosine hydroxyl group is oriented away from the active site Zn(II) atom, moving closer by 5Å in the closed structure presumably to a position appropriate to stabilize the tetrahedral transition state intermediate formed during catalysis (Nguyen et al., 2011). These observations have led us to hypothesize that the open ERAP1 conformation is responsible for initial substrate binding and product release whereas the closed conformation is the one that is catalytically active and responsible for scissile bond cleavage. Only one conformation has been reported for ERAP2, which closely resembles the closed conformation of ERAP1 (Birtley et al., 2012). However, since in this conformation the catalytic site is completely isolated from access to the external solvent, an alternate open conformation should exist for ERAP2 also to allow substrate binding and product release.
Length dependence
A hallmark of ERAP1 function is its ability to trim efficiently large peptides while sparing smaller ones (Chang et al., 2005). This property, termed “the molecular ruler” is highly relevant to its biological function due to the stringent length requirements for MHC class I binding (Engelhard, 1994; Madden, 1995). Although the precise mechanism for length selection is not yet clear, structural analysis has yielded insights on how ERAP1 could achieve this function. As noted above, an elongated channel extending away from the catalytic site with potential side-chain binding subsites could accommodate a long peptide substrate. A C-terminal binding subsite has been proposed that could bind the C-terminus of the bound peptide only when the peptide is of sufficient length to concurrently occupy the catalytic and C-terminal binding site (Gandhi et al., 2011; Kochan et al., 2011). Biochemical evidence has revealed a complex allosteric activation mechanism for ERAP1 that leads to self-activation only for peptides for above a certain length (Nguyen et al., 2011). Interestingly, small peptide products also can participate in this allosteric mechanism suggesting that ERAP1 products might perhaps regulate its activity if present in sufficiently high local concentration. The allosteric activation of ERAP1 has been proposed to arise from the occupation of a regulatory site that is neighboring the catalytic site (Nguyen et al., 2011). This could provide a mechanism for length-dependent cleavage, as only long peptides could concurrently occupy both active and regulatory sites. It is possible that the proposed C-terminal binding site is indeed the regulatory site, although this has not been demonstrated to date.
ERAP1 exhibits substrate inhibition, another enzymatic property that may be related to its biological function. Some substrates can self-inhibit ERAP1 activity when present at higher concentrations, leading to distinct, bell-shaped, Michaelis-Menten kinetics (Evnouchidou et al., 2011). This phenomenon has been previously described in metabolic enzymes and is responsible for the regulation of their activity, leading to buffering of product generation when very high concentrations of substrate are present. ERAP1 however has the additional property of processing a multitude of different substrates (i.e. peptide sequences) indicating that the over-representation of a particular substrate (epitope precursor) may affect (reduce) the processing rate of other peptides. This property, in conjunction with the ability of ERAP1 to be activated by small peptides, suggest that ERAP1 trimming activity could be regulated by the antigenic peptide content of the ER leading to subtle regulation of antigen presentation. The importance of such regulatory events in antigen presentation and immunodominance is not currently understood and requires further investigation.
The enzymatic properties of ERAP2 have been studied much less intensively and ERAP2 appears to share only some of the molecular properties of ERAP1. However ERAP2 can be very important for the generation of specific epitopes (Saveanu et al., 2005) and can be just as efficient in trimming large peptides as ERAP1 (Evnouchidou et al., 2012. Preliminary analysis indicate that ERAP2 does not share the length preferences of ERAP1 {Chang, 2005 #244). Furthermore, it is not known whether ERAP2 can be regulated by substrates or products in a manner similar to ERAP1.
Synergy between ERAP1 trimming and MHCI binding
For ERAP1 products to be successfully presented on the cell surface, they have to be tightly bound by an appropriate MHC class I allele. The close interplay between aminopeptidase specificity and MHC class I binding was initially suggested even before ERAP1 was identified (Falk et al., 1990). Two models have been supported by recent literature to underlie this interplay. In one model, ERAP1 trims the N-terminus of peptides as they are at least partially bound onto MHC class I molecules suggesting that the MHC binding selectivity is the primary driver for ERAP1 trimming (Kanaseki et al., 2006). In another model, the antigenic peptide precursor is trimmed after exclusively interacting with ERAP1 and the released product then proceeds to bind onto MHC (Chang et al., 2005). In the latter model, ERAP1 molecular properties that drive its selectivity can be crucial in determining the availability of certain epitopes. Both models share the notion that a MHC-bound mature antigenic epitope is protected by ERAP1 action. In a recent study, Infantes et al. demonstrated that H2-Ld binding can indeed protect peptides from ERAP1 trimming and that this protection applies even to N-terminally extended peptides, suggesting that trimming of an epitope precursor while bound onto MHC was not necessary at least for the MHC-epitope combination tested (Infantes et al., 2010). However, similar data in a murine system was interpreted as being consistent with a model in which N-terminal trimming of a MHC-bound peptide could occur (Blanchard et al., 2010).
Inspection of the now known structures of ERAP1 can provide insight towards these two models. The observation of an “open” conformation may at first suggest a framework for peptide trimming while bound onto MHC, but the topology of the active site necessitates that the peptide binds in a configuration that orients the C-terminus of the peptide towards the interior of the substrate cavity. This configuration essentially precludes that the peptide main body is bound onto MHCI during catalysis since it would generate a very large number of steric clashes between MHCI and ERAP1. Docking experiments actually suggested that the closest distance possible between the N-terminus of a peptide bound onto MHCI and the Zn(II) catalytic atom of ERAP1 is 20Å (>6 amino acids) making “on MHCI” trimming structurally unlikely unless only 2-3 residues were bound to the MHC(Nguyen et al., 2011). It is conceivable however that additional “open” conformations of ERAP1 exist, in which domain IV is rotated even further away from domains I and II, allowing for closer approach of MHC-bound peptide to the enzyme's active site, or the structural differences between the human and murine homologs allow easier trimming of MHC-bound peptides in the latter case. .
A possible ERAP1/2 heterodimer
ERAP1 and ERAP2 have been proposed to function in concert, supplementing each other's specificity and in that manner facilitate the trimming of the large pool of sequences that can act as their substrates (Saveanu et al., 2005). This concept was further supported by the demonstration that the two enzymes can be co-purified from cells (Saveanu et al., 2005). The topology and molecular properties of such an ERAP1/2 assembly have however remained elusive, possibly because of a weak association constant between the two molecules that hinders biophysical analysis. However, ERAP1 crystallizes as homotrimers and ERAP2 as homodimers, both in configurations that do not occlude the active site, providing topographic models for potential oligomeric form. In particular, in a homodimer observed in the ERAP2 crystal the two ERAP2 monomers interact through domain I and primarily through residues that are conserved amongst ERAP1 and ERAP2 suggesting a possible topology for ERAP1/2 interaction (Birtley et al., 2012). However, to date, no direct experimental evidence exists for this association.
IRAP, the third member of the oxytocinase subfamily of M1 aminopeptidases
Insulin-regulated aminopeptidase (IRAP, also known as placental leucine aminopeptidase, PLAP) is the third member of the oxytocinase subfamily of M1 aminopeptidases (Tsujimoto and Hattori, 2005). Unlike ERAP1 and ERAP2, which are soluble proteins, IRAP is a type II membrane protein, with ~100 amino acids facing the cytosol, a single transmembrane span, and a large extracytoplasmic aminopeptidase domain homologous to ERAP1 and ERAP2. Although already assigned with several biological functions such as the degradation of peptide hormones (Hattori et al., 1999; Keller, 2003; Wallis et al., 2007), IRAP has been shown recently to be directly involved in antigen processing in the cross-presentation pathway (Saveanu et al., 2009; Saveanu and van Endert, 2012). This involvement has been subsequently proposed to only apply to inflammatory and not steady-state dendritic cells suggesting that IRAP participates in a distinct cross-priming mechanism in those cells (Segura et al., 2009). IRAP has been shown to be able to generate antigenic epitopes in vitro although with distinct patterns to ERAP1 (Georgiadou et al., 2010). Furthermore, IRAP has a distinct N-terminal specificity, combining the specificities of ERAP1 and ERAP2 (Zervoudi et al., 2011). Although the three-dimensional structure of IRAP is not currently known, homology modeling of the ectodomain and site-directed mutagenesis have suggested that a single amino acid substitution in the enzyme's S1 specificity pocket, namely Glu541, is responsible for its amino-terminal specificity (Zervoudi et al., 2011).
Insights from structures of other M1 family members
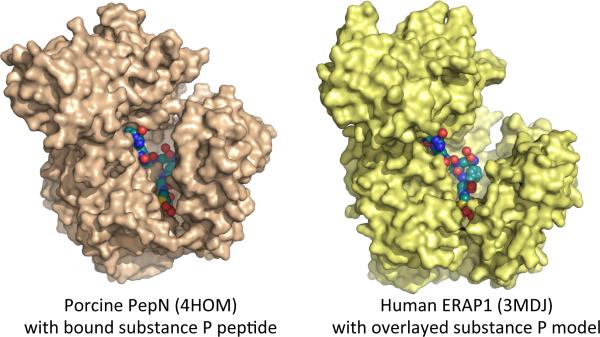
Very recently crystal structures reported for human (Wong et al., 2012) and porcine (Chen et al., 2012) aminopeptidase N have provided additional insights into the structure and enzymology of the M1 family aminopeptidases. Aminopeptidase N, also known as CD13, is a type II membrane protein like IRAP, and for both human and porcine variants the extracytoplasmic domain was isolated and crystallized in complex with a variety of substrates and inhibitors. Although only ~33% identical with ERAP1 or ERAP2, aminopeptidase N adopts a highly similar overall structure. The porcine protein was found in a conformation intermediate between the open and closed conformations observed for ERAP1, whereas the human protein adopted the closed conformation, suggesting that aminopeptidases N may also be able to undergo the large-scale conformational changes hypothesized for ERAP1. Aminopeptidase N has a role in processing angiotensin and other peptide hormones, and can digest peptides at least as long as angiontensin III (8 residues). Both human and porcine variants were crystallized together with long substrates. Human aminopeptidase N apoprotein was crystallized together with angiotensin IV, a hexapeptide. Processing of the substrate was prevented by using the Zn-free apoprotein, which lacks catalytic activity. Porcine aminopeptidase N was crystallized together with substance P, a heptapeptide hormone that acts as aminopeptidase N inhibitor, and also with a heptaalanine peptide substrate. For substance P the proline residue at the second position prevents hydrolysis, but for polyalanine the enzyme was captured in an active form, presumably with crystal lattice contacts preventing the conformational changes needed for hydrolysis (in a second crystal form a shorter hexa-alanine peptide was bound as a substrate inhibitor). Both enzymes were also crystallized with aminopeptide inhibitors amastatin and/or bestatin, and as holoenzymes. The different views of active site geometry provided by these structures generally validate ideas about M1 aminopeptidase enzymatic mechanism derived from earlier studies, although the order of individual protonation and bond breaking steps still remains to be conclusively established. Importantly, the long peptide substrates provide a model for how ERAP1 could accommodate long substrates within its binding site. In fact, docking substance P from the porcine aminopeptidase N onto human ERAP1 results in no substantial clashes, with the peptide oriented to exit the enzyme at the junction of domains II and IV (Figure 3). However, this potential substrate binding mode is different from that predicted for ERAP1 from the domain IV- hexahistidine interaction and additional work will be required to determine whether ERAP1 binds long substrates similarly.
Figure 3.
Surface representations of porcine aminopeptidases N (PDB code 4HOM) and human ERAP1 (PDB code 3MDJ). The structure of aminopeptidases N contains the heptapeptide hormone substance P that was co-crystallized with the enzyme. The same peptide is shown in identical conformation bound onto the known structure of ERAP1.
ERAP1/2 coding SNPs linked with disease
During the last 5 years a plethora of genome-wide association studies have linked several coding single nucleotide polymorphisms near the genes of ERAP1 and ERAP2 with predisposition to human disease. Examples include autoimmune inflammatory diseases such as rheumatopathies, type I diabetes, psoriasis, cancer and susceptibility to viral or parasitic infections (Cagliani et al., 2010; Haroon and Inman, 2010; Mehta et al., 2009; Strange et al., 2010; Sun et al., 2010; Tenzer et al., 2009). The pro-inflammatory function of ERAP1 of promoting shedding of TNFR1 may contribute to the pathogenesis of these diseases (Cui et al., 2002). Enzymatic activity of ERAP1 is however not necessary for this function and no correlation has been found between ERAP1 SNPs and serum cytokine levels in patients suggesting that this may not be the primary link between ERAP1 SNPs and disease (Haroon et al., 2010). Instead, the recently discovered importance of ERAP1/2 in antigen processing has led scientists to hypothesize that the mechanism behind disease association is altered antigen processing by ERAP1/2 alleles. One of the most notable examples has been the association of ERAP1/2 coding SNPs with the inflammatory disease ankylosing spondylitis (AS) (Burton et al., 2007; The Australo-Anglo-American Spondyloarthritis et al., 2011). AS has been proposed to have an autoimmune etiology, and predisposition to this disease is largely dependent on carrying the HLA allele B27. However, most HLA-B27 carriers do not develop the disease, suggesting that other genes in addition to HLA-B27 may be involved. The idea that AS disease pathogenesis results from altered antigen processing of HLA-B27 epitopes by ERAP1 variants recently has been supported by a genetic study demonstrating that the association of ERAP1 SNPs with AS requires the presence of HLA-B27 (Haroon et al., 2012; The Australo-Anglo-American Spondyloarthritis et al., 2011). Although the antigenic epitopes that contribute to AS, pathogenesis have not been unequivocally defined, several recent studies have indicated that disease-associated ERAP1/2 SNPs can alter enzymatic activity, specificity, and even length selection (Evnouchidou et al., 2012; Evnouchidou et al., 2011; Garcia-Medel et al., 2012; Kochan et al., 2011).
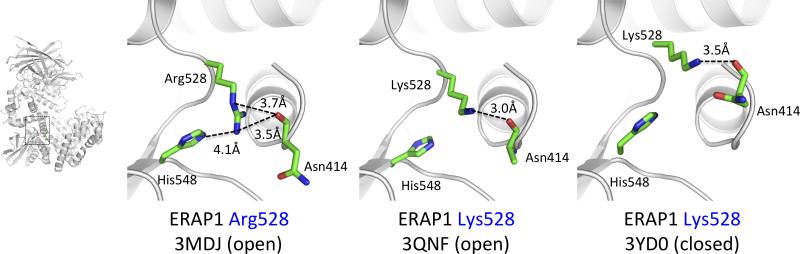
The mechanism by which single nucleotide changes in ERAP1 alter enzymatic properties is not well understood. All disease-associated SNPs in the ERAP1 gene were found on the crystal structure to be located distant from the enzyme's catalytic site (Kochan et al., 2011; Nguyen et al., 2011). The M349V polymorphism is the closest to the catalytic site and could directly participate in interactions with the substrate, although it does not reside inside a clear specificity pocket. Q730E is at the inner surface of the internal cavity of ERAP1, distal from the catalytic site but could still interact with the C-terminal moiety of the peptide substrate. Interestingly K528R, the SNP with the clearest disease-association, is nowhere near the catalytic site of the postulated peptide binding site and instead resides in domain III near the junction with domain IV. Domain III has been proposed as a junction for the conformational change between the ERAP1 open and closed forms. The Q730E and K528R polymorphisms have been shown to affect both the activity and the specificity of ERAP1 as well as its ability to be inhibited by substrates (Evnouchidou et al., 2011). Additionally, the K528R polymorphism has been suggested to affect the length-dependent trimming properties of ERAP1 (Kochan et al., 2011). Although Q730E may alter specificity by directly affecting substrate-enzyme interactions, K528R would have to exert effects on the enzymatic mechanism indirectly, perhaps by affecting the dynamics between the two ERAP1 conformations. Comparison of the configuration of this residue in the open and closed ERAP1 conformation indicates that the nature of the side-chain of residue 528 can affect interactions with residues Asn414 and His548, the former belonging to Helix 4 of domain II and the latter to domain III, both being structural features that reorient during the conformational change (Figure 4). However, the mechanism by which these different interactions are transmitted to the active site and the effects in the catalytic mechanism are unknown.
Figure 4.
Atomic interactions between the side-chain of the polymorphic position 528 in ERAP1. Lys528 makes hydrogen bonding interactions with the backbone of Asn414 that resided in helix 4 of domain II. In contrast Arg528 interacts with His548 on domain III as well.
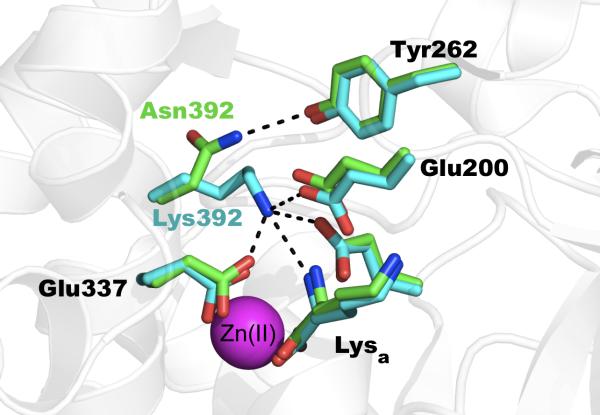
A coding SNP in the gene of ERAP2 has been associated with human disease, namely predisposition to AS, pre-eclampsia, and resistance to HIV infection (Cagliani et al., 2010; Hill et al., 2011; Johnson et al., 2009; Tsui et al., 2010). By comparison with the ERAP1 polymorphisms, and explanation for this SNP appears to be more straightforward. The SNP (rs2549782) codes for the variation N392K, which in the recent ERAP2 crystal structure was shown to reside adjacent to the enzyme's catalytic site. ERAP1 and IRAP have an Asn residue at that location, the related aminopeptidase N E. coli homolog carries a Lys and the mammalian aminopeptidase N homolog carries an Asn, indicating that both permutations are compatible with aminopeptidase functionality (Fournie-Zaluski et al., 2009). The crystal structures of both ERAP2 alleles were recently solved (Birtley et al., 2012; Evnouchidou et al., 2012). Comparison of the two structures indicated that the side-chain of the polymorphic residue assumes distinct conformations in the two variants (Figure 5). In one of the variants the lysine ! -amino group approaches the N-terminus of the bound substrate leading to destabilization and reduced catalytic efficiency. In the other variant the Asn side chain orients away from the substrate N-terminus and hydrogen bonds with the nearby Tyr262. This substitution also leads to rearrangements of the S1 specificity pocket of the enzyme, possibly underlying a distinct change in specificity seen between the two alleles (Evnouchidou et al., 2012).
Figure 5.
Atomic interactions between the side-chain of the polymorphic position 392 in ERAP2. Asn 392 makes hydrogen bonding interactions with Tyr262. Lys392, approaches the N-terminus of the active-site bound lysine amino acid and makes several salt-bridge interactions with catalytically important Glu residues.
Outstanding questions
Despite the recent progress, several outstanding questions remain in understanding the molecular mechanisms of ERAP1 and ERAP2 enzymatic activity. The structural basis for length selection by ERAP1 is not known, as the allosteric regulatory sites have not been localized on the enzyme. The attractive model of alternation between open and closed conformations remains to be verified experimentally for proteins in solution. The binding site(s) for long peptides on ERAP1, and the sites responsible for internal sequence specificity, are not known. The mechanistic basis for disease linkage of ERAP1 and ERAP2 allelic variant still remains to be elucidated. Finally, it is not clear whether or not ERAP1/2 activity is regulated in vivo by substrates and products, as suggested by in vitro enzymatic work, but this has important implications for the roles of these proteins in generating (and destroying) antigenic epitopes.
Highlights.
! ! ERAP1 and ERAP2 process antigenic peptides and can regulate antigen presentation
! ! They do so by complex and poorly understood molecular mechanisms
! ! Several recent crystallographic structures have just been released for these enzymes
! ! These structures provide novel insight onto the mechanism of these enzymes and their role in disease
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Addlagatta A, Gay L, Matthews BW. Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site. Proc Natl Acad Sci U S A. 2006;103:13339–44. doi: 10.1073/pnas.0606167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascher DB, Polekhina G, Parker MW. Crystallization and preliminary X-ray diffraction analysis of human endoplasmic reticulum aminopeptidase 2. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:468–71. doi: 10.1107/S1744309112006963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtley JR, Saridakis E, Stratikos E, Mavridis IM. The Crystal Structure of Human Endoplasmic Reticulum Aminopeptidase 2 Reveals the Atomic Basis for Distinct Roles in Antigen Processing. Biochemistry. 2012;51:286–295. doi: 10.1021/bi201230p. [DOI] [PubMed] [Google Scholar]
- Blanchard N, Gonzalez F, Schaeffer M, Joncker NT, Cheng T, Shastri AJ, Robey EA, Shastri N. Immunodominant, protective response to the parasite Toxoplasma gondii requires antigen processing in the endoplasmic reticulum. Nat Immunol. 2008;9:937–44. doi: 10.1038/ni.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard N, Kanaseki T, Escobar H, Delebecque F, Nagarajan NA, Reyes-Vargas E, Crockett DK, Raulet DH, Delgado JC, Shastri N. Endoplasmic reticulum aminopeptidase associated with antigen processing defines the composition and structure of MHC class I peptide repertoire in normal and virus-infected cells. J Immunol. 2010;184:3033–42. doi: 10.4049/jimmunol.0903712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PR, Clayton DG, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–37. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R, Riva S, Biasin M, Fumagalli M, Pozzoli U, Lo Caputo S, Mazzotta F, Piacentini L, Bresolin N, Clerici M, Sironi M. Genetic diversity at endoplasmic reticulum aminopeptidases is maintained by balancing selection and is associated with natural resistance to HIV-1 infection. Hum Mol Genet. 2010;19:4705–14. doi: 10.1093/hmg/ddq401. [DOI] [PubMed] [Google Scholar]
- Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. Embo J. 2001;20:2357–66. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc Natl Acad Sci U S A. 2005;102:17107–12. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lin YL, Peng G, Li F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc Natl Acad Sci U S A. 2012;109:17966–71. doi: 10.1073/pnas.1210123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Hawari F, Alsaaty S, Lawrence M, Combs CA, Geng W, Rouhani FN, Miskinis D, Levine SJ. Identification of ARTS-1 as a novel TNFR1-binding protein that promotes TNFR1 ectodomain shedding. J Clin Invest. 2002;110:515–26. doi: 10.1172/JCI13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhard VH. Structure of peptides associated with class I and class II MHC molecules. Annu Rev Immunol. 1994;12:181–207. doi: 10.1146/annurev.iy.12.040194.001145. [DOI] [PubMed] [Google Scholar]
- Evnouchidou I, Birtley J, Seregin S, Papakyriakou A, Zervoudi E, Samiotaki M, Panayotou G, Giastas P, Petrakis O, Georgiadis D, Amalfitano A, Saridakis E, Mavridis IM, Stratikos E. A common single nucleotide polymorphism in endoplasmic reticulum aminopeptidase 2 induces a specificity switch that leads to altered antigen processing. J Immunol. 2012;189:2383–92. doi: 10.4049/jimmunol.1200918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evnouchidou I, Kamal RP, Seregin SS, Goto Y, Tsujimoto M, Hattori A, Voulgari PV, Drosos AA, Amalfitano A, York IA, Stratikos E. Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J Immunol. 2011;186:1909–13. doi: 10.4049/jimmunol.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evnouchidou I, Momburg F, Papakyriakou A, Chroni A, Leondiadis L, Chang SC, Goldberg AL, Stratikos E. The Internal Sequence of the Peptide-Substrate Determines Its N-Terminus Trimming by ERAP1. Plos One. 2008;3 doi: 10.1371/journal.pone.0003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evnouchidou I, Papakyriakou A, Stratikos E. A New Role for Zn(II) Aminopeptidases: Antigenic Peptide Generation and Destruction. Curr Pharm Design. 2009;15:3656–3670. doi: 10.2174/138161209789271816. [DOI] [PubMed] [Google Scholar]
- Falk K, Rotzschke O, Rammensee HG. Cellular peptide composition governed by major histocompatibility complex class I molecules. Nature. 1990;348:248–51. doi: 10.1038/348248a0. [DOI] [PubMed] [Google Scholar]
- Fournie-Zaluski MC, Poras H, Roques BP, Nakajima Y, Ito K, Yoshimoto T. Structure of aminopeptidase N from Escherichia coli complexed with the transition-state analogue aminophosphinic inhibitor PL250. Acta Crystallogr D Biol Crystallogr. 2009;65:814–22. doi: 10.1107/S090744490901779X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi A, Lakshminarasimhan D, Sun Y, Guo HC. Structural insights into the molecular ruler mechanism of the endoplasmic reticulum aminopeptidase ERAP1. Sci Rep. 2011;1:186. doi: 10.1038/srep00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Medel N, Sanz-Bravo A, Van Nguyen D, Galocha B, Gomez-Molina P, Martin-Esteban A, Alvarez-Navarro C, Lopez de Castro JA. Functional interaction of the ankylosing spondylitis associated endoplasmic reticulum aminopeptidase 1 polymorphism and HLA-B27 in vivo. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M112.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadou D, Hearn A, Evnouchidou I, Chroni A, Leondiadis L, York IA, Rock KL, Stratikos E. Placental leucine aminopeptidase efficiently generates mature antigenic peptides in vitro but in patterns distinct from endoplasmic reticulum aminopeptidase 1. J Immunol. 2010;185:1584–92. doi: 10.4049/jimmunol.0902502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer GE, Gonzalez F, James E, Nolla H, Shastri N. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat Immunol. 2007;8:101–8. doi: 10.1038/ni1409. [DOI] [PubMed] [Google Scholar]
- Haroon N, Inman RD. Endoplasmic reticulum aminopeptidases: Biology and pathogenic potential. Nat Rev Rheumatol. 2010;6:461–7. doi: 10.1038/nrrheum.2010.85. [DOI] [PubMed] [Google Scholar]
- Haroon N, Tsui FW, Chiu B, Tsui HW, Inman RD. Serum cytokine receptors in ankylosing spondylitis: relationship to inflammatory markers and endoplasmic reticulum aminopeptidase polymorphisms. J Rheumatol. 2010;37:1907–10. doi: 10.3899/jrheum.100019. [DOI] [PubMed] [Google Scholar]
- Haroon N, Tsui FW, Uchanska-Ziegler B, Ziegler A, Inman RD. Endoplasmic reticulum aminopeptidase 1 (ERAP1) exhibits functionally significant interaction with HLA-B27 and relates to subtype specificity in ankylosing spondylitis. Ann Rheum Dis. 2012;71:589–95. doi: 10.1136/annrheumdis-2011-200347. [DOI] [PubMed] [Google Scholar]
- Hattori A, Matsumoto H, Mizutani S, Tsujimoto M. Molecular cloning of adipocyte-derived leucine aminopeptidase highly related to placental leucine aminopeptidase/oxytocinase. J Biochem (Tokyo) 1999;125:931–8. doi: 10.1093/oxfordjournals.jbchem.a022371. [DOI] [PubMed] [Google Scholar]
- Hill LD, Hilliard DD, York TP, Srinivas S, Kusanovic JP, Gomez R, Elovitz MA, Romero R, Strauss JF., 3rd Fetal ERAP2 variation is associated with preeclampsia in African Americans in a case-control study. BMC Med Genet. 2011;12:64. doi: 10.1186/1471-2350-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infantes S, Samino Y, Lorente E, Jimenez M, Garcia R, Del Val M, Lopez D. Cutting Edge: H-2L(d) Class I Molecule Protects an HIV N-Extended Epitope from In Vitro Trimming by Endoplasmic Reticulum Aminopeptidase Associated with Antigen Processing. Journal of Immunology. 2010;184:3351–3355. doi: 10.4049/jimmunol.0901560. [DOI] [PubMed] [Google Scholar]
- Johnson MP, Roten LT, Dyer TD, East CE, Forsmo S, Blangero J, Brennecke SP, Austgulen R, Moses EK. The ERAP2 gene is associated with preeclampsia in Australian and Norwegian populations. Hum Genet. 2009;126:655–66. doi: 10.1007/s00439-009-0714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaseki T, Blanchard N, Hammer GE, Gonzalez F, Shastri N. ERAAP Synergizes with MHC Class I Molecules to Make the Final Cut in the Antigenic Peptide Precursors in the Endoplasmic Reticulum. Immunity. 2006;25:795–806. doi: 10.1016/j.immuni.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SR. The insulin-regulated aminopeptidase: a companion and regulator of GLUT4. Front Biosci. 2003;8:s410–20. doi: 10.2741/1078. [DOI] [PubMed] [Google Scholar]
- Kochan G, Krojer T, Harvey D, Fischer R, Chen L, Vollmar M, von Delft F, Kavanagh KL, Brown MA, Bowness P, Wordsworth P, Kessler BM, Oppermann U. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1101262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden DR. The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- Mehta AM, Jordanova ES, Corver WE, van Wezel T, Uh HW, Kenter GG, Jan Fleuren G. Single nucleotide polymorphisms in antigen processing machinery component ERAP1 significantly associate with clinical outcome in cervical carcinoma. Genes Chromosomes Cancer. 2009;48:410–8. doi: 10.1002/gcc.20648. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Chang SC, Evnouchidou I, York IA, Zikos C, Rock KL, Goldberg AL, Stratikos E, Stern LJ. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol. 2011;18:604–13. doi: 10.1038/nsmb.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–50. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC classI-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, van Endert PM. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–97. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- Saveanu L, Carroll O, Weimershaus M, Guermonprez P, Firat E, Lindo V, Greer F, Davoust J, Kratzer R, Keller SR, Niedermann G, van Endert P. IRAP Identifies an Endosomal Compartment Required for MHC Class I Cross-Presentation. Science. 2009;325:213–7. doi: 10.1126/science.1172845. [DOI] [PubMed] [Google Scholar]
- Saveanu L, van Endert P. The role of insulin-regulated aminopeptidase in MHC class I antigen presentation. Front Immunol. 2012;3:57. doi: 10.3389/fimmu.2012.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E, Albiston AL, Wicks IP, Chai SY, Villadangos JA. Different cross-presentation pathways in steady-state and inflammatory dendritic cells. Proc Natl Acad Sci U S A. 2009;106:20377–81. doi: 10.1073/pnas.0910295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange A, Capon F, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LD, Cheng H, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42:1005–9. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzer S, Wee E, et al. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat Immunol. 2009;10:636–46. doi: 10.1038/ni.1728. [DOI] [PubMed] [Google Scholar]
- The Australo-Anglo-American Spondyloarthritis C. the Wellcome Trust Case Control C. et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui FW, Haroon N, Reveille JD, Rahman P, Chiu B, Tsui HW, Inman RD. Association of an ERAP1 ERAP2 haplotype with familial ankylosing spondylitis. Ann Rheum Dis. 2010;69:733–6. doi: 10.1136/ard.2008.103804. [DOI] [PubMed] [Google Scholar]
- Tsujimoto M, Hattori A. The oxytocinase subfamily of M1 aminopeptidases. Biochim Biophys Acta. 2005;1751:9–18. doi: 10.1016/j.bbapap.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Wallis MG, Lankford MF, Keller SR. Vasopressin is a physiological substrate for the insulin-regulated aminopeptidase IRAP. Am J Physiol Endocrinol Metab. 2007;293:E1092–102. doi: 10.1152/ajpendo.00440.2007. [DOI] [PubMed] [Google Scholar]
- Weimershaus M, Evnouchidou I, Saveanu L, van Endert P. Peptidases trimming MHC class I ligands. Curr Opin Immunol. 2012 doi: 10.1016/j.coi.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Wong AH, Zhou D, Rini JM. The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing. J Biol Chem. 2012;287:36804–13. doi: 10.1074/jbc.M112.398842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci U S A. 2006;103:9202–7. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, Rock KL. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat Immunol. 2002;3:1177–84. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- Zervoudi E, Papakyriakou A, Georgiadou D, Evnouchidou I, Gajda A, Poreba M, Salvesen GS, Drag M, Hattori A, Swevers L, Vourloumis D, Stratikos E. Probing the S1 specificity pocket of the aminopeptidases that generate antigenic peptides. Biochem J. 2011;435:411–20. doi: 10.1042/BJ20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]