Abstract
Dehydroepiandrosterone (DHEA) is a neurosteroid with potential effects on neurogenesis and neuronal survival in humans. However, most studies on DHEA have been performed in rodents, and there is little direct evidence for biological effects on the human nervous system. Furthermore, the mechanism of its action is unknown. Here, we show that DHEA significantly increased the growth rates of human neural stem cells derived from the fetal cortex and grown with both epidermal growth factor (EGF) and leukemia inhibitory factor (LIF). However, it had no effect on cultures grown in either factor alone, suggesting a specific action on the EGF/LIF-responsive cell. Precursors of DHEA such as pregnenolone or six of its major metabolites, had no significant effect on proliferation rates. DHEA did not alter the small number (<3%) of newly formed neuroblasts or the large number (>95%) of nestin-positive precursors. However, the number of glial fibrillary acidic protein-positive cells, its mRNA, and protein were significantly increased by DHEA. We found both N-methyl-d-aspartate and sigma 1 antagonists, but not GABA antagonists, could completely eliminate the effects of DHEA on stem cell proliferation. Finally we asked whether the EGF/LIF/DHEA-responsive stem cells had an increased potential for neurogenesis and found a 29% increase in neuronal production when compared to cultures grown in EGF/LIF alone. Together these data suggest that DHEA is involved in the maintenance and division of human neural stem cells. Given the wide availability of this neurosteroid, this finding has important implications for future use.
Dehydroepiandrosterone (DHEA) and its sulfate are the most abundant steroids in the blood of young adult humans. Levels of DHEA peak at ≈20 years of age and then decline to reach values of 20-30% at 70-80 years of age (1). Significant interest has arisen from the hypothesis that declining DHEA concentrations in adults may serve as an indicator of a number of conditions including the loss of insulin sensitivity, obesity, diabetes, cardiovascular diseases, stress, and aging (2). In clinical studies, DHEA levels were found to decline in mental illnesses such as major depressive disorder or in systematic diseases that respond to DHEA supplementation (3, 4).
While DHEA represents the most abundant steroid product of the adrenal cortex, it has also been identified as a “neurosteroid.” Neurosteroids are synthesized de novo in the central nervous system independent, at least in part, of peripheral organ activity (5, 6). Neurosteroids possess the ability to affect neurons through activation of γ-aminobutyric acid (GABA)A, N-methyl-d-aspartate (NMDA), and sigma receptors (5, 6), although the exact role of each with regard to DHEA is not well established. DHEA has been shown to be neuroprotective after oxidative stress in rat hippocampal neuronal cultures (7) and hippocampal damage induced by NMDA (8). DHEA has also been shown to be a potential signaling molecule in neuronal differentiation during development (9) and has recently been shown to increase neurogenesis in the adult rodent hippocampus (10). In contrast to the human brain, levels of DHEA in the adult rat brain are very low (6, 11). Thus, there is some question as to the relevance of the effect of DHEA on rodents and how this relates to the human situation.
Recent studies have shown that neural precursor cells can be derived from the developing brain allowing the direct examination of proliferation, differentiation, and migration of these cells in the culture dish (12, 13). These cells can be maintained in culture as spherical aggregates termed neurospheres (14-16) that consist of both multipotent stem cells and more restricted progenitor cells responsive to the mitogens epidermal growth factor (EGF) and fibroblast growth factor 2 (FGF-2) (17). Neurospheres are regionally specified based on the brain region from which they were isolated (18, 19) and also differ between species (20). However, recently, we have grown neurospheres from the human fetal cortex for extended periods of time in EGF and LIF and shown that they represent a fairly homogenous population of cells immunoreactive for the stem cell marker nestin (21, 22). After a full genomic analysis of these cells, we have shown them to be stable in culture over time and between lines with regard to global gene expression and termed them long-term human neural stem cells derived from the cortex (ltNSCCTX) (22).
Given the species differences in DHEA and stem cell biology, ltNSCCTX provide an ideal and innovative model to understand developmental and molecular mechanisms of neurosteroid actions in the human central nervous system. We show here that DHEA, but not its derivatives, increased the cell proliferation of ltNSCCTX in vitro. We also established which receptor signaling mediated this DHEA action, and finally showed that ltNSCCTX grown in the presence of DHEA underwent increased neurogenesis. These results have important implications for brain repair and are relevant to understanding the actions of DHEA on the human brain.
Materials and Methods
Neuronal Progenitor Cell Culture. Human embryonic tissue was obtained from the Birth Defects Laboratory at the University of Washington, Seattle. Neurospheres were generated from three cortex samples, M006 (13 weeks postconception male), M007 (18 weeks postconception male), and M009 (16 weeks postconception male). The method of collection conformed to the guidelines recommended by National Institutes of Health for the collection of such tissues and set out by the University of Washington and the University of Wisconsin, Madison. Institutional Review Board approval was obtained for all of these studies.
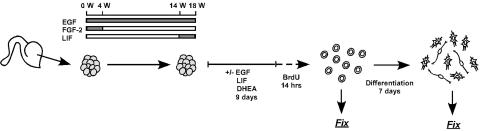
The schematic illustration of the experimental schedule and treatments is summarized in Fig. 1. Cortical progenitors were isolated from embryonic brain and induced to proliferate as free-floating neurospheres (16). Freshly isolated tissue was mechanically chopped into small cubes using a McLwain tissue chopper (Mickle Laboratory Engineering, Surrey, England) and seeded into T75 flasks at an approximate density of 200,000 cells (eight cubes) per ml of basal medium [DMEM/Ham's F-12 (7:3) containing penicillin/streptomycin/amphotericin B (PSA, 1%)] supplemented with B27 (2%; Life Technologies), EGF (20 ng/ml; Sigma), and FGF-2 (20 ng/ml; R & D Systems) with heparin (5 μg/ml). All cultures were maintained in a humidified incubator (37°C) and half the growth medium was replenished every 4-5 days. For passaging of neurospheres, a chopping method was used that does not require trypsin or mechanical dissociation, and cell/cell contact was continuously maintained (16). Neurospheres were passaged every 14 days by chopping them into 200-μm cubes that were seeded into fresh growth medium at a density equivalent to ≈100,000 cells per ml. At 2 weeks after the first passage, cells were switched to basal medium containing N2 supplement (1%; Life Technologies) and 20 ng/ml EGF. After 10 weeks of growth, 10 ng/ml LIF (Chemicon) was added to the cultures, which were then grown for a further 4 weeks before analysis.
Fig. 1.
Schematic describing culture schedules and treatments for ltNSCctx. Neurospheres were generated from three fetal cortex samples, initially grown for 4 weeks in EGF and FGF-2. The cultures were then switched to medium supplemented with EGF alone. After >10 weeks of growth, the cultures were supplemented with EGF and LIF and maintained for a further 4 weeks. Thirty to 40 neurospheres were cultured in coated T25 flasks with basal medium in the presence or absence of EGF, LIF, and DHEA. After 9 days, neurospheres were pulsed with 0.2 μM BrdUrd for 14 h and then dissociated into a single cell suspension with enzyme. Dissociated cells were plated onto poly(L)-lysine/laminin-coated coverslips for 1 h. To check the number of cells immunostained with BrdUrd or other cell markers, cells were fixed 1 h after plating. For differentiation studies, the plated cells were differentiated for 7 days under serum-free conditions and then visualized with anti-TuJ1, GFAP, and Hoechst-labeled nuclei.
Cumulative BrdUrd Pulse Labeling. To identify newly dividing cells in neurospheres, BrdUrd pulse-labeling was performed as described (22). Briefly, neurospheres were collected, medium was removed, and cells were washed three times with DMEM. Thirty to 40 neurospheres were transferred to poly(2-hydroxyethyl methacrylate)-coated T25 flasks containing basal medium. After 9 days of culture with feeding every 3 days, neurospheres were pulsed with 0.2 μM BrdUrd (Sigma) for 14 h, and dispersed into a single cell suspension with Accutase (Innovative Cell Technologies, San Diego) for 10 min at 37°C. Dissociated cells (25,000 cells per 50 μl) were plated onto poly(L)-lysine/laminin-coated glass coverslips in 24-well plates and incubated for 60 min at 37°C. The plating medium consisted of basal medium with B27 (2%; Life Technologies). For differentiation studies, dissociated cells were continuously maintained in the plating medium and differentiated for 7 days. These serum-free conditions allowed for differentiation of dissociated cells (23).
DHEA was purchased from Steraloids (Newport, RI). DHEA derivatives listed in Table 1 were synthesized and characterized as described (24, 25). Stock solutions (10 mM) were prepared in 100% ethanol and added to give a final medium concentration of 1 μM. Ciliary neurotrophic factor (CNTF) or IL-6 (R & D Systems) were used at a final concentration of 10 ng/ml. The NMDA receptor antagonist (+)-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine hydrogen malate (MK801) and GABAA receptor antagonist bicuculline (10 μM each in medium) were purchased from R & D Systems. Sigma 1 receptor antagonists BD1063 or haloperidol (Tocris Cookson, Ellisville, MO) were used at a final concentration of 3 μM.
Table 1. Effects of DHEA and its derivatives on BrdUrd incorporation in human neuropheres.
| Steroid | Common name | % BrdUrd+ cells |
|---|---|---|
| No steroid | 13.8 ± 1.7 | |
| 3β-hydroxyandrost-5-en-17-one | DHEA | 23.6 ± 1.5* |
| 4-pregnene-3,20-dione | Pregnenolone | 17.1 ± 0.8 |
| Androst 5-ene-3β, 17β-diol | Androstenediol | 17.3 ± 0.6 |
| 3β, 7α-dihydroxyandrost-5-en-17-one | 7α-hydroxy DHEA | 15.1 ± 1.7 |
| 3β, 7β-dihydroxyandrost-5-en-17-one | 7β-hydroxy DHEA | 14.0 ± 2.1 |
| 3β-hydroxyandrost-5-ene-7, 17-dione | 7-oxo-DHEA | 12.9 ± 1.7 |
| Androst-5-ene-3β, 7β, 17β-triol | 19.1 ± 2.8 | |
| Androst-5-ene-3β, 16α, 17β-triol | 15.6 ± 2.1 |
P < 0.05 versus all others.
Immunocytochemistry. Cultures were fixed in ice cold methanol for 20 min or 4% paraformaldehyde (PFA in PBS) at room temperature and washed in PBS. The cells were then incubated in 2 M HCl for 20 min at 37°C. The acid was neutralized with 0.1 M sodium borate and washed several times in PBS. Fixed cells were blocked in 5% goat serum with 0.3% Triton X-100 and incubated with anti-BrdUrd antibodies (rat monoclonal, 1:500, Accurate Chemical, Westbury, NY) for 30 min. After incubation with the primary antibodies, either fluorescein or Cy3 (goat anti-rat IgG, 1:1,000, The Jackson Laboratory) was used to visualize the signal. Hoechst 33258 (0.5 μg/ml in PBS, Sigma) was added for 5 min after completion of the secondary antibody incubation as a nuclear stain. For labeling of cell markers, fixed cells were incubated for 30 min with primary antibodies to nestin (monoclonal 1:500, Chemicon), β-tubulin-III (TuJ1; monoclonal IgG2b, 1:500, Sigma), or glial fibrillary acidic protein (GFAP) (rabbit polyclonal, 1:1,000, DAKO) to label neural progenitor cells, neurons, or astrocyte/stem cells, respectively. For double-labeling of BrdUrd in conjunction with either TuJ1 or GFAP, all steps of cell marker staining procedure were completed before commencement of staining for BrdUrd.
Cell Counts. Cell counts were performed by using a Nikon fluorescence microscope (40× objective) and metamorph imaging software (Universal Imaging, Downingtown, PA). Quantification of cells was based on counting the number of Hoechst-stained nuclei and the specific immunostained cells in at least four independent fields (total area >25 mm2) from a minimum of three coverslips. All cell-counting data were expressed as means ± SEM across three separate cell lines derived from the cortex. One-way functional ANOVA with Newman-Keuls post hoc test was used for analysis. Differences were considered significant when P was <0.05.
RT-PCR. Total RNA was isolated from neurospheres by using TRIzol reagent (Life Technologies). For RT-PCR, we used a GeneAmp RNA PCR kit (Roche Molecular Systems). All procedures for total RNA isolation and RT-PCR were performed according to the procedure suggested by the manufacturers. The following primer pairs were used: human GFAP (26), NMDA receptor subunits NR1 and NR2A-D (27), and sigma 1 receptor (28). The primers of cyclophylin were designed on the basis of the sequences of human cDNA (GenBank accession no. Y00052; forward, 5′-GTTTGCAGACAAGGTCCCAAA-3′; reverse, 5′-TGATCTTCTTGCTGGTCTTG-3′), and the predicted size of PCR product is 397 bp. The amplification profile was 94°C for 0.5 min, 58°C for 1 min, and 72°C for 1 min. For PCR of cyclophylin and the others, samples were incubated for 25 and 32 cycles, respectively. PCR products were electrophoresed and analyzed with densitometry by using scion image software (Scion, Frederick, MD).
Western Blotting. Preparation of cell lysates and Western blotting were performed on neurospheres, using monoclonal anti-GFAP (Transduction Laboratories, Lexington, KY) or NR1 (Chemicon), as described (22). Blots were quantitated by scion image.
Results and Discussion
DHEA Significantly Increases Proliferation of EGF/LIF Responsive Cells Within ItNSCctx Cultures. The three lines generated for this study grew as neurospheres as described in detail previously (16, 22). As predicted the cultures became responsive to LIF after 10 weeks and from this point were maintained in EGF/LIF. To confirm the differentiation potential of these lines, individual neurospheres were dissociated and plated to laminin/poly(L)-lysine coated coverslips for 7 days under differentiating conditions. After fixation and immunostaining for the neuronal marker TuJ1 and astrocyte/stem cell marker GFAP, these cultures were able to produce large numbers of well differentiated neurons and astrocyte/stem cells with a more fibroblast-like morphology (data not shown).
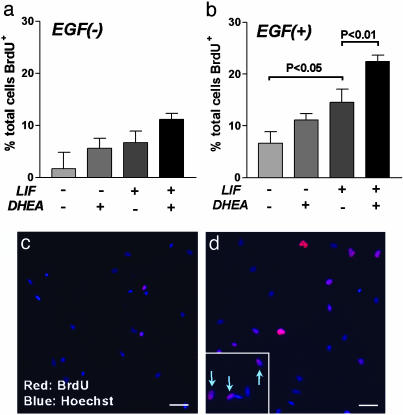
We first asked what role DHEA may play in the growth of these ltNSCctx cultures. To establish whether DHEA could enhance growth rates in the absence of EGF or LIF, we designed a withdrawal experiment (Fig. 1). EGF, LIF or both factors were withdrawn from the medium in the presence or absence of DHEA for 9 days. In the complete absence of EGF, the numbers of BrdUrd+ cells tended to be increased in the presence of either LIF or DHEA, although this did not reach significance (Fig. 2a). This increase may have been due to trace amounts of EGF remaining associated with membranes. In the presence of EGF, LIF significantly increased BrdUrd incorporation (Fig. 2b) as we have shown previously (22), whereas DHEA again tended to increase BrdUrd incorporation but did not reach significance (Fig. 2b). However, in the presence of EGF/LIF, DHEA significantly increased the number of BrdUrd+ cells from ≈14% to 24% (P < 0.05; Fig. 2 b-d). These data show DHEA selectively increases the division of the EGF/LIF-responsive cell within the neurospheres.
Fig. 2.
DHEA increases the number of BrdUrd+ cells in ltNSCctx in the presence of EGF and LIF. When EGF was withdrawn for 9 days, BrdUrd+ cells were not altered by the addition of LIF and DHEA (a). DHEA significantly increased the number of BrdUrd+ cells in the presence of EGF and LIF, although no significant effect was observed with DHEA in the absence of LIF (b). Photomicrographs represent BrdUrd immunoreactivity in cells grown in EGF (c) or EGF/LIF/DHEA (d) for 9 days. (Inset) Arrows designate BrdUrd-labeled cells at higher magnification. (Scale bar, 20 μm.)
Other Cytokines Cannot Substitute for LIF in Making EGF Cells Responsive to DHEA. LIF is a member of a family of IL-6 cytokines that also includes ciliary neurotrophic factor (CNTF), oncostatin, and cardiotrophin 1 (29). These factors signal through a receptor complex made up of unique components specific for the particular cytokines and a common receptor subunit, gp130, but do not have the same effect as LIF on the proliferation of EGF-responsive neural stem cells (22). The addition of CNTF or IL-6 could not mimic the effect of LIF with regard to enabling the EGF-responsive cells to respond to DHEA (see Fig. 6, which is published as supporting information on the PNAS web site).
Other Active Components of the DHEA Pathway Do Not Affect the Growth Rate of ltNSCctx. DHEA is readily converted metabolically to derivatives that are more active in some assays (24, 30, 31). In rat liver homogenate, the sequence is DHEA → 7α-OH-DHEA → 7-oxo-DHEA → 7β-OH-DHEA → androst-5-en-3β,7β,17β-triol. In the human brain, DHEA is synthesized from pregnenolone and metabolized mainly to 7α-OH-DHEA and androst-5-ene-3β,17β-diol (androstenediol) (6, 32). To determine whether DHEA per se or one of its metabolites stimulated cell growth and incorporation of BrdUrd, neurospheres were cultured in the continual presence of individual steroids (1 μM; listed in Table. 1) with basal medium containing EGF/LIF for 9 days as described previously. The cultures were then pulse-labeled with BrdUrd, dissociated, plated for 1 h, and stained with BrdUrd antibody. Neither pregnenolone nor any of the six metabolites affected the number of BrdUrd+ cells (Table 1). These experiments suggest that the effects reported here are due to the DHEA introduced into the culture medium, not to the metabolism of DHEA, because no derivative tested in this experiment was more effective.
The DHEA Responsive Cell Expresses High Levels of GFAP. Although under our culture conditions neurospheres contain >95% nestin-positive precursor cells we consider stem cells, there remain a small number of neuroblasts that spontaneously arise within these cultures (22). To determine whether the ratio of neuroblasts to stem cells was affected by DHEA treatment (i.e., which type of cells were proliferating in response to DHEA), acutely dissociated neurospheres were immunostained for the neuronal marker TuJ1 and the precursor cell marker nestin (see Fig. 7, which is published as supporting information on the PNAS web site). The number of TuJ1+ cells seen at this time (<3% of the population) and nestin+ cells (>95% of the population) were not affected by LIF or DHEA, suggesting that DHEA did not induce the division of a cell that spontaneously generated neuroblasts within the neurospheres. Rather, it was likely that the DHEA-responsive cell remained as a stem cell under these conditions.
Our previous results also indicated that the expression of GFAP was positively regulated by LIF in ltNSCctx (22). We hypothesized that DHEA treatment with LIF might further enhance GFAP expression in ltNSCctx. As expected, LIF increased the number of GFAP-positive cells over EGF alone (Fig. 3a; P < 0.05). DHEA alone or with EGF had no effect on the number of GFAP-positive cells, but in combination with LIF it further increased the total number of GFAP-positive cells from 80% to 90% (Fig. 3a; significantly different from EGF/LIF at P < 0.05). To further determine whether newly dividing cells expressed GFAP, we performed double labeling with GFAP and BrdUrd antibody (Fig. 3 b-d). In the presence of LIF, DHEA significantly increased the number of GFAP+/BrdUrd+ double-positive cells by >30% (P < 0.05; Fig. 3b). Furthermore, we found that >95% of BrdUrd+ cells were also immunostained by GFAP antibody, suggesting that in the presence of EGF/LIF/DHEA the majority of cells dividing within the neurospheres were GFAP-positive.
Fig. 3.
DHEA dynamically regulates GFAP expression. (a) The numbers of GFAP+ cells treated with LIF (P < 0.05) and LIF/DHEA (P < 0.01) were significantly increased compared to that in the absence of both LIF and DHEA. (b) The percentages of double-staining with GFAP and BrdUrd antibody after 9-day culture with LIF and DHEA. Photomicrographs of cells cultured in the absence (c) and presence (d) of LIF/DHEA for 9 days show a difference in the number and intensity of GFAP+/BrdUrd+ cells. (Scale bar, 15 μm.) (e) Semiquantitative RT-PCR revealed that LIF/DHEA treatment increased GFAP mRNA compared to LIF alone. P, a positive control using total RNA isolated from human embryonic brain; N, a negative control, reverse transcriptase withdrawn during RT step. (f) Western blot analysis confirmed the effect of DHEA on GFAP protein expression.
We next determined absolute RNA and protein levels of GFAP in response to LIF and DHEA by using RT-PCR and Western blot. ltNSCctx were grown in basal medium with EGF in the presence or absence of LIF and DHEA for 9 days, followed by total RNA and protein isolation. Semiquantitative analysis of RT-PCR for GFAP revealed that LIF/DHEA treatment increased GFAP mRNA (Fig. 3e). Densitometric analysis showed that the level of GFAP expression was 1.3 times higher in LIF/DHEA/EGF compared to LIF/EGF alone (Fig. 3e). Western blot analysis confirmed the effect of DHEA on GFAP protein expression (Fig. 3f). Using the densitometric analysis of each band, we found that DHEA could increase GFAP protein by ≈40% when compared to LIF/EGF alone (Fig. 3f).
We have previously shown that LIF also has powerful effects on GFAP expression in ltNSCctx (22), presumably through the gp130 receptor system and the Janus kinase-signal transducer and activators of transcription (JAK-STAT) signaling pathway (33, 34). In the current study we confirm these findings and show that although DHEA had no direct effect on EGF-treated cultures, it increased both GFAP expression and the number of GFAP/BrdUrd-positive cells when compared to EGF/LIF. Interestingly, neural stem cells capable of forming large numbers of neurons in adult mammals have been identified as astrocyte-resembling cells that are GFAP- and nestin-positive (35, 36). Given that the DHEA cultures showed increased GFAP production and neurogenesis, our results lend further support to the idea that an “astrocyte-like” cell may act as a stem cell in the human central nervous system (36-39).
DHEA Acts Through both NMDA and Sigma Receptors in ltNSCctx. We next asked which receptors mediated the effect of DHEA in ltNSCctx. It has been reported that neurosteroids are able to modulate NMDA receptor functions through non-genomic action (9, 40). NR1 is common to functional NMDA receptors (41) and was clearly detected in LIF and LIF/DHEA treatments, whereas it was barely detectable in the absence of LIF (Fig. 4a). In contrast, NR2B mRNA was detected in all cultures and not differentially expressed in the presence or absence of LIF/DHEA, whereas no NR2A, NR2C, or NR2D mRNA was detected in any of the neurosphere cultures (Fig. 4a). When using densitometric analysis of each band, the levels of NR1 mRNA were found to be >10-fold greater in the presence of LIF/DHEA when compared to EGF alone (data not shown). Furthermore, Western blot analysis using anti-NR1 antibody revealed that NR1 proteins were strongly induced by LIF and LIF/DHEA treatments (Fig. 4b). Thus, the functional subunits of NMDA receptor on cells within ltNSCctx could be induced by LIF and LIF/DHEA.
Fig. 4.
NMDA and sigma 1 receptor signaling are involved in the effect of DHEA. (a) Gene expression of NMDA receptor subunits. Positive (P) and negative (N) controls were prepared similarly to the protocol mentioned in Fig. 3. (b) Western blot analysis confirmed the effect of LIF on NR1 protein expression in ltNSCctx. (c) Specific NMDA receptor antagonist MK-801 (10 μM) significantly blocks the effect of DHEA. (d) Sigma 1 receptors are also involved in the effect of DHEA. Neurospheres were cultured for 9 days in basal medium with EGF, LIF, and DHEA, plus 3 μM BD1063 or haloperidol. (e) The blockade of GABAA receptors has no effect in ltNSCctx. *, P < 0.05 versus LIF without DHEA. NS, not significant.
We next determined whether inhibition of NMDA receptors using the antagonist MK801 could block the effects of DHEA on proliferation. The addition of 10 μM MK801 in the medium with EGF/LIF for 9 days did not change the basal levels of BrdUrd+ cell number (Fig. 4c). However, the increased proliferation induced by DHEA/LIF based on BrdUrd+ cell number could be completely blocked by MK801 (Fig. 4c). These effects of MK801 suggested that glutamate receptor activation with either NMDA or glutamate may also be able to enhance proliferation of cells within these cultures. To test this, we added glutamate agonists to the culture medium. Glutamate (0.1 μM) and NMDA were both found to significantly increase the number of BrdUrd+ cells, suggesting a direct effect on stem cell proliferation (see Fig. 8, which is published as supporting information on the PNAS web site).
Sigma receptors (a type of opiate receptor) may be able to modify NMDA receptor signaling for cellular effects of neurosteroids (42, 43). To test this possibility, we first checked sigma 1 receptor expression in ltNSCctx by using RT-PCR. Sigma 1 receptor mRNA was detected in all neurosphere cultures and was not seen to be affected by either LIF or DHEA (data not shown). To establish whether these receptors were important for the effects of DHEA, neurospheres were cultured for 9 days in basal medium with EGF/LIF or EGF/LIF/DHEA with or without 3 μM BD1063 or haloperidol, which specifically antagonize sigma 1 receptor functions (42). The blockade of sigma receptor again completely abolished the effects of DHEA on proliferation (Fig. 4d). These experiments suggested that NMDA receptor signaling after sigma 1 receptor-activation might involve DHEA actions on these cells.
Previous reports indicate that GABAA receptors are another possible candidate other than the NMDA receptor to mediate various functions of DHEA in the central nervous system (44, 45). We next tested the role of GABAA receptors in the cell proliferation induced by LIF/DHEA. Neurospheres were cultured for 9 days in basal medium with EGF/LIF or EGF/LIF/DHEA with or without the GABAA receptor antagonist bicuculline. Bicuculline (10 μM) did not reduce the LIF/DHEA-mediated proliferation (Fig. 4e). Moreover, the treatment of 10 or 0.1 μM GABA for 9 days had no effect on the number of BrdUrd+ cells in ltNSCctx (data not shown). From these results, we conclude that GABA receptor signaling is not involved in the effects of DHEA reported here in vitro, although we cannot exclude its importance in other species or in vivo.
ltNSCctx Grown in DHEA Undergo a Greater Level of Neurogenesis After Differentiation. We hypothesized that LIF/DHEA treatment may alter the potential of ltNSCctx to generate neurons. To test this possibility, neurospheres in the continual presence or absence of LIF or LIF/DHEA were cultured in basal medium containing EGF for 9 days. After dissociation and plating onto laminin-coated coverslips, the cultures were allowed to differentiate over 7 days. During this time new neurons are born through the proliferation of neuronal progenitors (T. Ostenfeld and C.N.S., unpublished observations). The differentiated cells were immunostained for TuJ1 (Fig. 5 a, d, and e) and GFAP (Fig. 5b). When the cultures were maintained in the absence of LIF/DHEA for 9 days, 10% of total cells were TuJ1+ after differentiation for 7 days (Fig. 5a). DHEA alone during the growth phase had no effect, whereas LIF treatment tended to increase the number of TuJ1+ cells (P = 0.07). In contrast, combined LIF/DHEA treatment significantly increased the number of TuJ1+ cells after differentiation (P < 0.05 over EGF alone; Fig. 5a). In contrast to TuJ1+ cells, the number of GFAP+ cells was not significantly different between the groups (Fig. 5b). This finding suggests that the increases seen in GFAP levels at 1 h postplating (Fig. 3) in response to DHEA disappear when the cells exit proliferation and are allowed to differentiate. To determine whether the increased number of new neurons were derived from cells induced by DHEA just before plating, neurospheres were pulse-labeled with BrdUrd for 14 h under the various conditions before plating (Fig. 5 c and f). At 7 days of differentiation, the numbers of TuJ1+/BrdUrd+-positive cells were not changed by adding DHEA or LIF in the medium before plating (Fig. 5c). However, the number of double-positive cells was clearly increased after EGF/LIF/DHEA treatment compared to EGF/LIF alone (P < 0.05; Fig. 5c). These results indicate that DHEA positively regulates the number of neurons produced by these cultures.
Fig. 5.
ltNSCCTX grown in the presence of DHEA generated more maturing neurons. Neurospheres were cultured in the presence or absence of EGF, LIF, and DHEA for 9 days, pulsed with BrdUrd, plated, and immunostained with anti-TuJ1, anti-GFAP, and Hoechst-labeled nuclei. (a) TuJ1+ cells were significantly increased by simultaneous treatment of LIF and DHEA, although the numbers of TuJ1+ cells were not altered by either DHEA or LIF alone. Photomicrographs represent TuJ1 immunoreactivity (red) in differentiated cells grown in the absence (d) or presence (e) of LIF/DHEA for 9 days. (b) There were no significant differences in the percentage of GFAP-labeled cells. (c) The numbers of TuJ1+/BrdUrd+ double-positive cells were not affected by either DHEA or LIF alone. However, TuJ1+/BrdUrd+ double-positive cells were increased by LIF/DHEA. (f) After 7 days of differentiation, many TuJ1 (green) and BrdUrd (red) double-positive cells (designated by arrows) were found in cells grown in the presence of LIF/DHEA for 9 days. *, P < 0.01 versus LIF or DHEA alone. (Scale bar, 40 μm.)
Conclusion
In this study, we show that DHEA can increase proliferation of human neural stem cells and positively regulate the number of neurons produced by these cultures. It is known that DHEA amounts fall progressively during aging, and reduced levels of DHEA have been reported in both adolescents and adults with major depressive disorders. Interestingly, the adult human brain also has neural stem cells within specific regions that contribute to divide and make new neurons (46). Furthermore, DHEA has recently been shown to be neuroprotective (8) and increase neurogenesis (10) in the adult rodent hippocampus. Although it is impossible to test the effects of DHEA on the adult human brain in vivo, our results show an effect on human neural stem cells, and this may be relevant to some of the beneficial effects of DHEA in clinical studies (3, 4). More importantly, these direct effects of DHEA (which is currently available as a health supplement in the United States) on human cells warn against its long-term use until the consequences of its actions have been further investigated.
Supplementary Material
Acknowledgments
This work was supported by funds from the University of Wisconsin Foundation and by Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad (to M.S.).
Abbreviations: DHEA, dehydroepiandrosterone; EGF, epidermal growth factor; GABA, γ-aminobutyric acid; GFAP, glial fibrillary acidic protein; LIF, leukemia inhibitory factor; ltNSCCTX, long-term cortical neural stem cells; MK801, (+)-5-methyl-10,11-dihydro-5H-dibenzo[a,d]cyclohepten-5,10-imine hydrogen malate; NMDA, N-methyl-d-aspartate; TuJ1, β-tubulin III.
References
- 1.Orentreich, N., Brind, J. L., Vogelman, J. H., Andres, R. & Baldwin, H. (1992) J. Clin. Endocrinol. Metab. 75, 1002-1004. [DOI] [PubMed] [Google Scholar]
- 2.Celec, P. & Starka, L. (2003) Physiol. Res. 52, 397-407. [PubMed] [Google Scholar]
- 3.Bloch, M., Schmidt, P. J., Danaceau, M. A., Adams, L. F. & Rubinow, D. R. (1999) Biol. Psychiatry 45, 1533-1541. [DOI] [PubMed] [Google Scholar]
- 4.Roshan, S., Nader, S. & Orlander, P. (1999) Eur. J. Clin. Invest. 29, 210-213. [DOI] [PubMed] [Google Scholar]
- 5.Baulieu, E. E. (1997) Recent Prog. Horm. Res. 52, 1-32. [PubMed] [Google Scholar]
- 6.Baulieu, E. E. (1998) Psychoneuroendocrinology 23, 963-987. [DOI] [PubMed] [Google Scholar]
- 7.Bastianetto, S., Ramassamy, C., Poirier, J. & Quirion, R. (1999) Mol. Brain Res. 66, 35-41. [DOI] [PubMed] [Google Scholar]
- 8.Kimonides, V. G., Khatibi, N. H., Svendsen, C. N., Sofroniew, M. V. & Herbert, J. (1998) Proc. Natl. Acad. Sci. USA 95, 1852-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Compagnone, N. A. & Mellon, S. H. (1998) Proc. Natl. Acad. Sci. USA 95, 4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karishma, K. K. & Herbert, J. (2002) Eur. J. Neurosci. 16, 445-453. [DOI] [PubMed] [Google Scholar]
- 11.Corpechot, C., Robel, P., Axelson, M., Sjovall, J. & Baulieu, E. E. (1981) Proc. Natl. Acad. Sci. USA 78, 4704-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKay, R. (1997) Science 276, 66-71. [DOI] [PubMed] [Google Scholar]
- 13.Gage, F. H. (2000) Science 287, 1433-1438. [DOI] [PubMed] [Google Scholar]
- 14.Chalmers-Redman, R. M., Priestley, T., Kemp, J. A. & Fine, A. (1997) Neuroscience 76, 1121-1128. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds, B. A., Tetzlaff, W. & Weiss, S. (1992) J. Neurosci. 12, 4565-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svendsen, C. N., ter Borg, M. G., Armstrong, R. J., Rosser, A. E., Chandran, S., Ostenfeld, T. & Caldwell, M. A. (1998) J. Neurosci. Methods 85, 141-152. [DOI] [PubMed] [Google Scholar]
- 17.Ciccolini, F. & Svendsen, C. N. (1998) J. Neurosci. 18, 7869-7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hitoshi, S., Tropepe, V., Ekker, M. & van der Kooy, D. (2002) Development (Cambridge, U.K.) 129, 233-244. [DOI] [PubMed] [Google Scholar]
- 19.Ostenfeld, T., Joly, E., Tai, Y. T., Peters, A., Caldwell, M., Jauniaux, E. & Svendsen, C. N. (2002) Dev. Brain Res. 134, 43-55. [DOI] [PubMed] [Google Scholar]
- 20.Svendsen, C. N., Skepper, J., Rosser, A. E., ter Borg, M. G., Tyres, P. & Ryken, T. (1997) Dev. Brain Res. 99, 253-258. [DOI] [PubMed] [Google Scholar]
- 21.Lendahl, U., Zimmerman, L. B. & McKay, R. D. (1990) Cell 60, 585-595. [DOI] [PubMed] [Google Scholar]
- 22.Wright, L. S., Li, J., Caldwell, M. A., Wallace, K., Johnson, J. A. & Svendsen, C. N. (2003) J. Neurochem. 86, 179-195. [DOI] [PubMed] [Google Scholar]
- 23.Caldwell, M. A., He, X. L., Wilkie, N., Pollack, S., Marshall, G., Wafford, K. A. & Svendsen, C. N. (2001) Nat. Biotechnol. 19, 475-479. [DOI] [PubMed] [Google Scholar]
- 24.Lardy, H., Kneer, N., Wei, Y., Partridge, B. & Marwah, P. (1998) Steroids 63, 158-165. [DOI] [PubMed] [Google Scholar]
- 25.Marwah, P., Marwah, A., Kneer, N. & Lardy, H. (2001) Steroids 66, 581-595. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez-Ramos, J. R., Song, S., Kamath, S. G., Zigova, T., Willing, A., Cardozo-Pelaez, F., Stedeford, T., Chopp, M. & Sanberg, P. R. (2001) Exp. Neurol. 171, 109-115. [DOI] [PubMed] [Google Scholar]
- 27.Yoshioka, A., Ikegaki, N., Williams, M. & Pleasure, D. (1996) J. Neurosci. Res. 46, 164-178. [DOI] [PubMed] [Google Scholar]
- 28.Shamsul, O. M., Moore, P., El Sherbeny, A., Roon, P., Agarwal, N., Sarthy, V. P., Casellas, P., Ganapathy, V. & Smith, S. B. (2001) Mol. Brain Res. 95, 86-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auernhammer, C. J. & Melmed, S. (2000) Endocr. Rev. 21, 313-345. [DOI] [PubMed] [Google Scholar]
- 30.Lardy, H., Marwah, A. & Marwah, P. (2002) Lipids 37, 1187-1191. [DOI] [PubMed] [Google Scholar]
- 31.Shi, J., Schulze, S. & Lardy, H. A. (2000) Steroids 65, 124-129. [DOI] [PubMed] [Google Scholar]
- 32.Weill-Engerer, S., David, J. P., Sazdovitch, V., Liere, P., Schumacher, M., Delacourte, A., Baulieu, E. E. & Akwa, Y. (2003) Brain Res. 969, 117-125. [DOI] [PubMed] [Google Scholar]
- 33.Bonni, A., Sun, Y., Nadal-Vicens, M., Bhatt, A., Frank, D. A., Rozovsky, I., Stahl, N., Yancopoulos, G. D. & Greenberg, M. E. (1997) Science 278, 477-483. [DOI] [PubMed] [Google Scholar]
- 34.Nakashima, K., Wiese, S., Yanagisawa, M., Arakawa, H., Kimura, N., Hisatsune, T., Yoshida, K., Kishimoto, T., Sendtner, M. & Taga, T. (1999) J. Neurosci. 19, 5429-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Verdugo, J., Doetsch, F., Wichterle, H., Lim, D. A. & Alvarez-Buylla, A. (1998) J. Neurobiol. 36, 234-248. [DOI] [PubMed] [Google Scholar]
- 36.Doetsch, F., Caille, I., Lim, D. A., Garcia-Verdugo, J. M. & Alvarez-Buylla, A. (1999) Cell 97, 703-716. [DOI] [PubMed] [Google Scholar]
- 37.Laywell, E. D., Rakic, P., Kukekov, V. G., Holland, E. C. & Steindler, D. A. (2000) Proc. Natl. Acad. Sci. USA 97, 13883-13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skogh, C., Eriksson, C., Kokaia, M., Meijer, X. C., Wahlberg, L. U., Wictorin, K. & Campbell, K. (2001) Mol. Cell. Neurosci. 17, 811-820. [DOI] [PubMed] [Google Scholar]
- 39.Imura, T., Kornblum, H. I. & Sofroniew, M. V. (2003) J. Neurosci. 23, 2824-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, F. S., Gibbs, T. T. & Farb, D. H. (1991) Mol. Pharmacol. 40, 333-336. [PubMed] [Google Scholar]
- 41.Monyer, H., Burnashev, N., Laurie, D. J., Sakmann, B. & Seeburg, P. H. (1994) Neuron 12, 529-540. [DOI] [PubMed] [Google Scholar]
- 42.Monnet, F. P., Mahe, V., Robel, P. & Baulieu, E. E. (1995) Proc. Natl. Acad. Sci. USA 92, 3774-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurice, T., Phan, V. L., Urani, A., Kamei, H., Noda, Y. & Nabeshima, T. (1999) Jpn. J. Pharmacol. 81, 125-155. [DOI] [PubMed] [Google Scholar]
- 44.Majewska, M. D., Demirgoren, S., Spivak, C. E. & London, E. D. (1990) Brain Res. 526, 143-146. [DOI] [PubMed] [Google Scholar]
- 45.Demirgoren, S., Majewska, M. D., Spivak, C. E. & London, E. D. (1991) Neuroscience 45, 127-135. [DOI] [PubMed] [Google Scholar]
- 46.Eriksson, P. S., Perfilieva, E., Bjork-Eriksson, T., Alborn, A. M., Nordborg, C., Peterson, D. A. & Gage, F. H. (1998) Nat. Med. 4, 1313-1317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.