Abstract
Botulinum neurotoxin (BoNT) is a potent biological substance used to treat neuromuscular and pain disorders. Both BoNT type A and BoNT type E display high-affinity uptake into motor neurons and inhibit exocytosis through cleavage of the synaptosome-associated protein of 25 kDa (SNAP25). The therapeutic effects of BoNT/A last from 3 to 12 months, whereas the effects of BoNT/E last less than 4 weeks. Using confocal microscopy and site-specific mutagenesis, we have determined that the protease domain of BoNT/A light chain (BoNT/A-LC) localizes in a punctate manner to the plasma membrane, colocalizing with the cleaved product, SNAP25197. In contrast, the short-duration BoNT/E serotype is cytoplasmic. Mutations in the BoNT/A-LC have revealed sequences at the N terminus necessary for plasma membrane localization, and an active dileucine motif in the C terminus that is likely involved in trafficking and interaction with adaptor proteins. These data support sequence-specific signals as determinants of intracellular localization and as a basis for the different durations of action in these two BoNT serotypes.
Keywords: SNARE, subcellular localization, dileucine motif, duration of action, Clostridium
Botulinum neurotoxins (BoNTs) are the most potent of all biological substances (1). Although BoNTs are well publicized as a potential biological weapon and as the causative agents in clinical botulism, the potency and myorelaxant actions of BoNTs have been exploited clinically in more than 100 indications, including muscle hyperactivity in cerebral palsy and cervical dystonia, migraines, myofacial pain, and focal hyperhidrosis (2-5). These toxins are specific endoproteases that, collectively, target several distinct proteins in nerve terminals. Motor nerve terminals at neuromuscular junctions are particularly sensitive to these neurotoxins, resulting in a transient and reversible muscle relaxation through inhibition of acetylcholine release. The Clostridium neurotoxin family includes seven serotypes of BoNT (A-G), and a single form of toxin produced by Clostridium tetani (TeNT). These toxins consist of a heavy chain (HC, 100 kDa) and light chain (LC, 50 kDa) linked by a disulfide bond (6, 7). The three-dimensional crystal structures of BoNT/A (8) and BoNT/B (9) have been resolved, providing a basis for understanding the structure/function mechanism of BoNT action. The BoNT-LCs are zinc-dependent endoproteases that specifically cleave one of three soluble N-ethylmaleimide-sensitive factor-attachment protein-receptor (SNARE) proteins (10) involved in synaptic vesicle docking and fusion at the nerve terminal (11). The synaptosome-associated protein of 25 kDa (SNAP25) is cleaved at distinct sites near the C terminus by BoNT/A (Q197-R198) and BoNT/E (R180-I181), generating truncated SNAP25197 (12) and SNAP25180 (13), respectively. A ratio of cleaved to intact SNAP25 of as little as 0.1 to 0.35 is sufficient to inhibit exocytosis (14, 15).
BoNTs display long but variable durations of action that are serotype specific. The clinical therapeutic effect of BoNT/A lasts approximately 3 months for neuromuscular disorders and 6-12 months for hyperhidrosis (16). In cultured murine embryonic spinal cord cells treated with BoNT/A, SNAP25197 can be detected for more than 80 days (17). Because SNAP25206 and SNAP25197 display similar half-lives (24 h) (6, 18), a prolonged persistence of BoNT/A-LC within neurons has been suggested to be responsible for the long duration of action (17). Interestingly, the half-lives of most endogenous and exogenous cellular proteins range in length up to a few days (19). No explanation is readily available for the long duration of BoNT/A-LC. Toxin may escape degradation pathways in neuronal cells through subcellular localization into specific cellular structures and/or may be sequestered and protected through interactions with select cellular proteins. In the present study, we demonstrate that BoNT/A-LC localizes to the plasma membrane of neuronal cells through signals present at the N and C termini of the protein. In contrast, BoNT/E-LC, with the shortest duration of action, is cytoplasmic. These data provide insight toward the understanding of the long duration of action of BoNT/A versus BoNT/E at the neuromuscular junction.
Methods
Plasmids. The LCs of BoNT/A (Allergan Hall A), and BoNT/E (Beluga) were cloned in pQBI50 plasmids (Qbiogene, Carlsbad, CA) by PCR amplification of genomic DNA. Constructs expressing BoNT/A-LC (LC/A) and BoNT/E-LC (LC/E) with the GFP at the N terminus were generated: GFP-LC/A and GFP-LC/E. Truncations in the BoNT/A-LC were as follows: GFP-LC/A(ΔN8/ΔC22) with 8 aa deleted from the N terminus and 22 aa from the C terminus, GFP-LC/A(ΔN8) with 8 aa deleted from the N terminus, and GFP-LC/A(ΔC22) with 22 aa deleted from the C terminus. The dileucine mutant, GFP-LC/A(LL→AA) with L427A and L428A, was generated by site-directed mutagenesis (Stratagene) using GFP-LC/A as a template. Wild-type LC/A and the mutants described above were cloned into pET-30b(+) vectors containing polyhistidine affinity tags (Novagen). rLC/A, rLC/A(ΔN8/ΔC22), and rLC/A(ΔC22) were generated by PCR from genomic DNA. rLC/A(LL→AA) was generated by site-directed mutagenesis using rLC/A as a template.
Cell Culture and Transfection. The rat pheochromocytoma cell line PC12 was obtained from ATCC (CRL-1721) and was cultured in collagen IV-coated dishes (BD Biocoat, Bedford, MA) in RPMI medium 1640 (Invitrogen) containing 10% heat-inactivated horse serum, 5% FBS, 10 mM Hepes, 1 mM sodium pyruvate, 0.45% glucose, and 50 units of penicillin/50 μg of streptomycin. Differentiation of PC12 into sympathetic-like neurons was achieved in differentiation medium containing RPMI medium 1640, N2 supplement, sodium pyruvate, glucose, 0.5% BSA, and penicillin/streptomycin supplemented with nerve growth factor (NGF; Harlan, Indianapolis) at 50 ng/ml. Transfection was performed by using Lipofectamine 2000 (Invitrogen). Cells were plated 1 day before transfection at a density of 1.2 × 106 cells per 60-mm dish and were transfected with 10 μg of DNA in 25 μl of Lipofectamine mixed for 20 min in Opti-MEM medium (Invitrogen). The DNA mixture was added to cells in regular culture medium and was incubated overnight. Cells were then differentiated in NGF-containing medium and visualized with an inverted fluorescence microscope (Leica, Bannockburn, IL). Typical transfection efficiencies were 35-50%.
Antibodies, Immunoprecipitation, and Western Blot Analysis. Antibodies to GFP were monoclonal 3E6 (Qbiogene) and rabbit polyclonal (Santa Cruz Biotechnology). Antibody to SNAP25 was SMI-81 (Sternberger, Lutherville, MD). Rabbit polyclonal antibodies that specifically recognize the BoNT/A cleavage product, SNAP25197, and BoNT/E product, SNAP25180, were custom generated for Allergan. These antibodies do not crossreact with the uncleaved substrate SNAP25206.
For immunoprecipitation studies, cells were washed with PBS and lysed in 50 mM Hepes buffer containing 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 1 mM EGTA, and 1% Triton X-100 plus Complete protease inhibitors (Roche, Switzerland). Precleared lysates were incubated with antibody 3E6 at 4°C for 2 hr. Immobilized protein G (Pierce) was then added and incubated for 1 hr at 4°C. Beads were washed in lysis buffer, centrifuged, resuspended in sample loading buffer, and boiled before electrophoresis in Criterion Tris·HCl 4-15% gels (Bio-Rad). Protein expression was analyzed by Western blotting. Cells were washed and lysed in the 1% Triton X-100/50 mM Hepes buffer for 1 hr at 4°C. Sample loading buffer was added to the cleared lysate and the protein was separated in Criterion Tris·HCl 4-15% gels. Proteins were transferred to poly(vinylidene difluoride) (PVDF) membranes, blocked with 1% milk in TBST (25 mM Tris·HCl, pH 7.2/150 mM NaCl/0.1% Tween 20), and incubated with the primary antibody followed by the corresponding secondary antibody. Blots were developed with SuperSignal chemiluminescence substrates (Pierce). When both full-length and cleaved SNAP25 were detected in the same Western blot with SMI-81 antibody, protein was extracted from cells with chloroform/methanol (20). Proteins were separated by SDS/PAGE and blotted as described above.
Immunocytochemistry. Transfected and control PC12 cells were fixed with 4% paraformaldehyde and washed with PBS. Cells were permeablized in 1% Triton X-100 in PBS followed by methanol at -20°C. Blocking was achieved by incubating the cells in 100 mM glycine for 30 min followed by a 30-min incubation in 0.5% BSA/PBS. Staining was performed with the following primary antibodies: polyclonal antibody to GFP (Santa Cruz Biotechnology), monoclonal antibody to SNAP25206 (StressGen Biotechnologies, Victoria, Canada), monoclonal antibody to SNAP25197 (Allergan), and purified polyclonal antibody to SNAP25180 (Allergan). Primary antibodies were incubated at room temperature for 2 hr in a humidified chamber. After washing, Alexa Fluor (480 or 568) -conjugated antibodies (Molecular Probes) were applied to the samples at 1:200 dilution and incubated at room temperature for 1 hr. Stained dishes were mounted with Vectashield solution (Vector Laboratories) and coverslipped. Images were obtained with a Leica confocal microscope using appropriate laser settings. Leica colocalization software was used to quantitate double-stained samples.
Inhibition of Exocytosis. PC12 cells were transfected with plasmid constructs encoding GFP or GFP-LC fusion proteins and were selected with G418 for 2-3 days. The day before the assay, cells were plated at 2 × 105 cells per well in 24-well collagen-coated plates. Cells were loaded with [3H]norepinephrine (Amersham Pharmacia Biosciences) at 2 μCi/ml (1 μCi = 37 kBq) in culture medium at 37°C for 4 hr and then were washed twice with Ca2+- and Mg2+-free PBS. Exocytosis was induced by incubating the cells at 37°C for 15 min in release buffer (51 mM NaCl/100 mM KCl/0.5 mM MgCl2/2.2 mM CaCl2/5.6 mM d-glucose/15 mM Hepes, pH 7.4). A calcium-free buffer was used to measure basal release. After release, 500 μl of the buffer was added to 5 ml of Ready Protein Scintillation mixture (Beckman). Unreleased [3H]norepinephrine remaining in the cells was quantified by removing the cells with trypsin and adding them to fresh tubes containing the scintillation mixture. Samples were measured and data were expressed as percent release (dpm released divided by total dpm).
Production of Recombinant Proteins and Activity Assay Toward SNAP25. Recombinant BoNT/A-LC constructs (wild-type and mutants) were transformed into BL21-CodonPlus (DE3)-RIL Escherichia coli cells (Stratagene). Cultures were grown to an OD600 of 0.6-0.7 and then induced with 0.5 mM isopropyl β-d-thiogalactoside (IPTG) for 2 hr. Recombinant proteins were purified by affinity chromatography with BD Talon resin (BD Biosciences Clontech, Palo Alto, CA)). Purity of the proteins was assessed by SDS/PAGE. Activity of the rLC/A, rLC/A(AA), rLC/A(ΔC22), and rLC/A(ΔN8/ΔC22) was measured with an ELISA. Briefly, a recombinantly produced biotinylated substrate corresponding to SNAP25(134-206) was immobilized on a streptavidin-coated microtiter plate (Pierce). The appropriate rLC/A constructs and 900-kDa BoNT/A complex as a control were added to substrate-coated plates. Protease activity was determined by quantitating the formation of SNAP25(134-197) with a polyclonal antibody specific for the cleaved product (Allergan). Statistical modeling and parameter comparisons between data sets were carried out by using an extra-sum-of-squares F test (21, 22).
Results
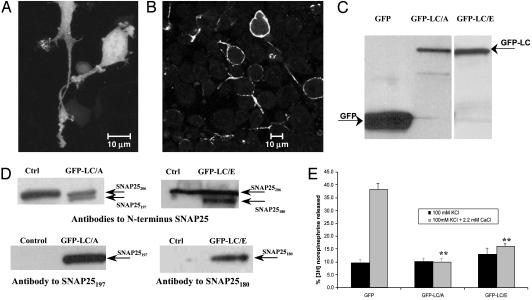
BoNT/A-LC Colocalizes with SNAP25 at the Plasma Membrane. To study the subcellular localization of BoNT-LCs, PC12 cells were transfected with plasmids encoding either GFP or a GFP-LC fusion. Imaging of GFP-expressing cells by using confocal microscopy demonstrated a homogeneous distribution of fluorescence throughout the cell (Fig. 1A). In contrast, the fluorescence of GFP-LC/A was localized in a punctate fashion along the plasma membrane of the cell body and neurites (Fig. 1B). Expression levels of GFP-LC/A were 10- to 20-fold lower than GFP alone, probably because of the unfavorable codon bias of clostridial genes (23) expressed in mammalian cells; however, protein analysis demonstrated single bands of the correct molecular weight for GFP and GFP-LC/A (Fig. 1C). Western blot analysis of cell lysates of nonselected cells probed with antibodies specific to SNAP25197 or the N terminus of SNAP25 demonstrated that GFP-LC/A is proteolytically active (Fig. 1D). Expressed GFP-LC/A in G418-selected cells inhibited [3H]norepinephrine release significantly (Fig. 1E), similar to cells electroporated with 0.5 nM BoNT/A (Fig. 5, which is published as supporting information on the PNAS web site). The punctate plasma membrane distribution of GFP-LC/A is similar to that of its substrate, SNAP25, in neuronal cells (24). In cells transfected with GFP-LC/A there was no intact SNAP25206 visible by immunocytochemistry (Fig. 6, which is published as supporting information on the PNAS web site). Staining of cells with a monoclonal antibody specific for SNAP25197, and polyclonal antibodies to GFP, and merging the fluorescent signals from confocal microscopy, revealed colocalization of SNAP25197 and GFP-LC/A (Fig. 2A). Analysis of several fields (n = 8) demonstrated that 76 ± 9% of GFP-LC/A colocalizes with SNAP25197 and 60 ± 10% of SNAP25197 colocalizes with GFP-LC/A. This result is in contrast to the data obtained staining for GFP and SNAP25206, (Fig. 2B) in which 19 ± 5% of GFP colocalized with SNAP25206 and 26 ± 7% of SNAP25206 colocalized with GFP. These data demonstrate the localization of both BoNT/A-LC and SNAP25197 to a common plasma membrane compartment.
Fig. 1.
Subcellular localization and activity of GFP-LC/A and GFP-LC/E in differentiated PC12 cells analyzed by using confocal microscopy. (A) GFP displayed a dispersed localization throughout the cell. (B) GFP-LC/A localized in a punctate manner in specific areas at the plasma membrane of the cell body and neurites, with no fluorescence in the cytoplasm of cells. (C) GFP-LC fusion proteins are expressed as a single band of 80 kDa. (D) GFP-LC/A and GFP-LC/E are active proteases and cleaved SNAP25206 into SNAP25197 and SNAP25180, respectively. (E) Cells transfected with GFP-LC/A or GFP-LC/E and selected with G418, showed a significant inhibition of [3H]norepinephrine release when compared with GFP-expressing cells (LC/A, P = 0.001; LC/E, P = 0.001; t test compared with GFP control).
Fig. 2.
(A) SNAP25197 and BoNT/A-LC colocalize in the same compartment at the plasma membrane. Differentiated cells expressing GFP-LC/A were immunostained with antibodies to GFP (green) and SNAP25197 (red). Separate images were taken for green and red fluorescence and digitally merged, and regions of colocalization were colored white. (B) GFP expressing PC12 cells were stained with antibodies to GFP (green) and SNAP25206 (red). (C) SNAP25180 and BoNT/E-LC are cytoplasmic proteins. GFP-LC/E showed cytoplasmic localization with nuclear exclusion. Cells displayed rounded morphology and lack of neurites even in differentiation media. PC12 cells were immunostained with antibodies to GFP (green) and SNAP25180 (red).
Despite sharing SNAP25 as a common substrate with BoNT/A-LC, BoNT/E-LC displayed a completely different subcellular distribution (Fig. 2C). GFP-LC/E was visualized in the cytoplasm, with few cells displaying plasma membrane fluorescence, and was mainly absent from nuclei. In addition, cells expressing GFP-LC/E in differentiation media were rounded with very few, short, and thin neurites compared with untransfected cells in the same dish. These results are consistent with reports in the literature of PC12 cells transfected with BoNT/E-LC and differentiated with staurosporine (24). The proteolytic activity of GFP-LC/E was confirmed by Western blot analysis of SNAP25206 and SNAP25180 (Fig. 1D) and inhibition of neurotransmitter release from PC12 cells (Fig. 1E). Immunostaining with an antibody specific for the BoNT/E-LC cleavage product, SNAP25180, revealed a substantially different localization pattern. Fluorescence was observed primarily in the cytoplasm, to a lesser extent at the plasma membrane, and was absent from nuclei (Fig. 2C). It has previously been hypothesized that cleavage of 26 aa from SNAP25 by BoNT/E-LC hastens the removal of nonfunctional SNAP25180 from the plasma membrane, allowing the incorporation of newly synthesized SNAP25206 (6). Our work provides experimental evidence of a shift in localization of SNAP25 from the plasma membrane to the cytosol after being cleaved to SNAP25180. Examination of more than 15 fields demonstrated that 73 ± 8% of GFP-LC/E colocalized with SNAP25180, and 51 ± 15% of SNAP25180 colocalized with GFP-LC/E. The cellular distribution of GFP-LC/E and SNAP25180 is in marked contrast to the observed distribution for either GFP-LC/A or SNAP25197.
Localization Signals Are Present in BoNT/A-LC Sequence. In an attempt to identify the presence of localization signals in BoNT/A-LC, several truncated variants were prepared. A truncated form of BoNT/A-LC, comprising residues Tyr-9 to Leu-415 (ΔN8/ΔC22) was reported as the minimal essential domain of the endoprotease with a structure (PDB structure ID code 1E1H) similar to the BoNT/A-LC structure in the holotoxin (8), and retaining a diminished catalytic activity toward SNAP25 (25). Deleted amino acids are depicted in yellow in the ribbon structure shown in Fig. 3A and Fig. 7, which is published as supporting information on the PNAS web site. GFP-LC/A (ΔN8/ΔC22) no longer localized to the plasma membrane; instead, fluorescence appeared in perinuclear structures as well as throughout the cytoplasm (Fig. 3B). The protein was not degraded as shown by Western blot analysis (Fig. 3C) and retained minimal activity with respect to cleavage of SNAP25 (Fig. 3D). These data suggest that signals involved in directing BoNT/A-LC to the plasma membrane may reside within the deleted N and/or C termini (Fig. 3A). Further analysis of the sequence revealed the presence of a putative dileucine motif (D/EXXXLL) at the C terminus (Fig. 3A, in red) that was present only in the BoNT/A serotype (FEFYKLL) (Table 1 and Fig. 8, which is published as supporting information on the PNAS web site). Targeting and trafficking of proteins to different compartments depend on sorting signals contained within the protein (26, 27). These signals interact with specific recognition molecules, which are components of membrane-bound transport intermediates (28). Direct or indirect interaction of dileucine motifs with adaptor protein (AP) complexes, such as AP1, AP2, and AP3, has been demonstrated for several proteins. Most reported proteins containing identified dileucine motifs are transmembrane proteins (Table 1), with the Nef protein of primate lentiviruses being the only nontransmembrane protein known to traffic by means of a dileucine motif (26, 29-31).
Fig. 3.
Mutations and truncations of BoNT/A-LC affect subcellular localization. (A) Ribbon model of the BoNT/A-LC crystal structure extracted from the BoNT/A holotoxin (150 kDa) x-ray structure (8) residues 1-430. The dileucine motif at the C terminus is shown in red, truncations are shown in yellow. (B) GFP-LC/A(ΔN8/ΔC22) has no plasma membrane localization. Fluorescence can be detected in a structure in the middle of the cell (70% of cells) along with some cytoplasmic protein (30% of cells). (C and F) GFP-LC/A mutant proteins are expressed as a single 80-kDa band. Expression levels are GFP-LC/A(AA) = GFP-LC/A(ΔC22) ≥ GFP-LC/A > GFP-LC/A(ΔN8) >> GFP-LC/A(ΔN8/ΔC22). (D and I) Mutant BoNT/A-LC proteins retain enzymatic activity toward the cleavage of SNAP25. Levels of activity are GFP-LC/A > GFP-LC/A(AA) = GFP-LC/A(ΔC22) > GFP-LC/A(ΔN8). GFP-LC/A(ΔN8/ΔC22) had only residual activity. LCAG, LCAAG, and tLCAG in D represent the constructs encoding LC/A, LC/A(AA), and LC/A(ΔN8/ΔC22), respectively, with GFP fused at the C terminus. (E) The C terminus of LC/A contains an active dileucine motif. GFP-LC/A(AA) localizes at the plasma membrane but it is more diffuse within the membrane, a noticeable percentage of cells displayed fluorescence in the cytoplasm. (G) GFP-LC/A(ΔC22) causes a change in localization similar to GFP-LC/A(AA). (H) Signals involved in plasma membrane localization are present at the N terminus of BoNT/A-LC. GFP-LC/A(ΔN8) localized in the cytoplasm with nuclear exclusion. Approximately 5% of the cells displayed localization similar to the double-truncated protein GFP-LC/A(ΔN8/ΔC22).
Table 1. Proteins containing active dileucine motifs.
| Protein | Sequence | Refs. |
|---|---|---|
| Rat VMAT2 | EEKMAIL | 40, 41 |
| Rat VMAT1 | EEKRAIL | 42 |
| Rat VAChT | SERDVLL | 40, 41 |
| Human CD3γ | SDKQTLL | 27 |
| Human CD4 | SQIKRLL | 43 |
| M6P/IGF2R | DSDEDLL | 44 |
| GLUT4 | RRTPSLL | 45 |
| TGF-β RII | SEHCAIIL | 46 |
| IRAP | ESSAKLL | 37 |
| Human LIMP II | DERAPLI | 47 |
| Human tyrosinase | EEKQPLL | 48 |
| Lentivirus Nef | GENNSLL | 29, 30, 31 |
| gp 130 | ESTQPLL | 49 |
| Shal K+ channel | FETQHHHLLHC | 50 |
| Na 1.2 (Na+ channel) | IHCLDIL | 51 |
| Yeast Vam3p (syntaxin) | NEQSPLL | 39 |
| VAMP4 (synaptobrevin) | SERRNLL | 32 |
| Synaptotagmin I | EEVDAML | 48 |
| BoNT/A | FEFYKLL | This paper |
| BoNT/B-G | Not present |
Several proteins have been shown to contain active C-terminal dileucine motifs (boldface). Of special interest are the SNARE proteins VAMP4, yeast Vam3p (syntaxin), and synaptotagmin I. BoNT/A is the only clostridial neurotoxin containing a dileucine motif at the C terminus of the LC. VMAT, vesicular monoamine transporter; VAChT, vesicular acetylcholine transporter; IRAP, insulin-regulated aminopeptidase. Phosphorylation of −5 Ser in hCD4, M6P/IGF2R, and gp 130 apparently suffices for the absence of D/E.
BoNT/A-LC Contains an Active Dileucine Motif. Mutation of the leucines into alanines has been shown to disrupt the motif and thereby affect interaction with AP adaptors, protein internalization, and/or intracellular localization. For example, mutation of the motif present in VAMP4 affects its steady-state distribution, with a large proportion of VAMP4 being found in peripheral structures in addition to the normal trans-Golgi network (TGN) distribution (32). A dileucine to dialanine GFP-LC/A(AA) mutant (L427A and L428A) was expressed in PC12 cells (Fig. 3C), revealing that disruption of the dileucine motif caused changes in the steady-state distribution at the plasma membrane. GFP-LC/A(AA) displayed a periplasmalemmal distribution with some protein in the cytoplasm, suggesting that the protein can reach the target membrane but may be loosely anchored (Fig. 3E). A similar pattern of localization (Fig. 3G) was observed when GFP-LC/A(ΔC22), lacking 22 C-terminal amino acids, was expressed, suggesting that the primary localization signal present at the C terminus is the dileucine motif. Localization changes for both LC/A C-terminal mutants are consistent with reports demonstrating that mutations of dileucine motifs yield proteins that have a loose association with the membrane due to disrupted interactions with complexing proteins (32). Both GFP-LC/A(ΔC22) and GFP-LC/A(AA) mutants were expressed in PC12 cells at higher levels than GFP-LC/A (Fig. 3 C and F). Proteolysis of SNAP25 could be detected in the transfected cells at slightly lower levels than seen with wild-type LC/A (Fig. 3 D and I).
Because neither truncation (ΔC22) nor mutation (LL → AA) of the C terminus produced the same effect as the double-truncation mutant (ΔN8/ΔC22), we postulated that signals important for plasma membrane targeting were also present at the N terminus. A GFP-LC/A(ΔN8) with eight N-terminal amino acids removed was expressed in differentiated PC12 cells (Fig. 3F). A complete loss of plasma membrane localization was observed (Fig. 3H), with fluorescence dispersed throughout the cytoplasm and nuclear exclusion. In ≈5% of these cells, localization was similar to the GFP-LC/A (ΔN8/ΔC22) mutant. The GFP-LC/A(ΔN8) mutant was expressed at lower levels than GFP-LC/A (Fig. 3F) and appeared less efficient at cleaving SNAP25 (Fig. 3I). The sequence of the first eight amino acids at the N terminus is different between serotype A and E, as shown in the sequence alignment in Fig. 8. These data support the presence of signals or structural elements important for plasma membrane localization in the N terminus of LC/A.
BoNT/A-LC Mutants Have Reduced Catalytic Activity. To assess the effects of LC/A mutations on functional exocytosis, we measured the release of [3H]norepinephrine. GFP-LC/A(ΔC22) and GFP-LC/A(AA) inhibited exocytosis to the same extent as GFP-LC/A. However, in cells expressing either GFP-LC/A(ΔN8) or GFP-LC/A(ΔN8/ΔC22) inhibition of [3H]norepinephrine exocytosis was minimal or absent (Fig. 4A). Cellular inhibition of neurotransmitter release correlated well with the intracellular SNAP25197 Western blot data; however, similarities and differences in activity are not accurately quantifiable in these cellular analyses, because of variable expression levels in cells transfected with different plasmid constructs. To further characterize their proteolytic activities, BoNT/A-LC mutants were expressed in E. coli as polyhistidine-tagged fusion proteins (Fig. 4B). Protease activity was analyzed in an ELISA using a substrate corresponding to SNAP25(134-206) and SNAP25197-specific antibodies (Fig. 4C). Statistical modeling and parameter comparisons between data sets were carried out by using an extra-sum-of-squares F test. No differences were observed for the activity of BoNT/A-LC with affinity tags at the N or C terminus and data are presented as a combination. Surprisingly, analysis of the various recombinant LC/A constructs revealed that the dialanine mutant (rLC/A(AA)) was 26-fold less active than the wild-type LC/A construct. Upon truncation of the 22-aa C-terminal tail containing the dileucine motif [rLC/A(ΔC22)], the catalytic activity was significantly reduced 3-fold compared with the dialanine mutant. These results are interesting because removal of the C-terminal tail has minimal impact beyond the substitution of two amino acids. When both the N and C termini were deleted, rLC/A(ΔN8/ΔC22), the catalytic activity was further reduced (greater than 600-fold reduction compared with wild-type LC/A). These results demonstrate that the diminished cellular LC activity can be attributed in part to decreased catalytic activity. In addition, a decrease in cellular SNAP25 cleavage by LC/A mutants with reduced membrane association may also be due, in part, to a lack of localization with SNAP25 at the plasma membrane. BoNT/A-LC catalytic activity is not needed for plasma membrane localization because an inactive LC/A [GFP-LC/A(H227Y)] with intact N and C termini localizes to the plasma membrane. Reduced activity of the rLC mutants may be due to changes in the protein structure resulting in changes in the catalytic activity, and these findings warrant further protein structure/function analysis.
Fig. 4.
Effect of mutations on the BoNT/A-LC catalytic activity. (A) GFP-LC/A, GFP-LCA(AA), and GFP-LC/A(ΔC22) completely inhibited exocytosis, GFP-LC/A(ΔN8) inhibited exocytosis to a lesser extent, and GFP-LC/A(ΔN8/ΔC22) failed to inhibit exocytosis (t test). (B) Schematic of the constructs prepared in pET30 plasmids containing wild-type LC/A and LC/A mutants. (C) Activity of purified wild-type rLC/A and rLC/A mutants in the SNAP25 ELISA. Wild-type rLC/A catalytic activity was unaffected by the position of the His6 tag. Data presented and statistical analyses are from at least three independent ELISAs per construct with three replicates per point assayed. Wild-type LC/A has a significantly lower (right-shifted) EC50 and lower response maximum compared with BoNT/A complex (F3,42 = 28.8, P < 0.05). Comparison of the activities of LC/A mutants to wild-type LC/A reveals similar response maxima but significantly lower EC50 values (LL → AA, F3,53 = 41.2; ΔC22, F3,53 = 65.3;ΔN8ΔC22, F3,53 = 122.2; all P < 0.05). Further comparisons of the mutant LC/A forms reveal that both ΔC22 (F2,26 = 3.6, P < 0.05) and ΔN8ΔC22 (F2,26 = 31.0, P < 0.05) have lower EC50s than the LL → AA form, and that ΔN8ΔC22 also has a lower EC50 than the ΔC22 mutant LC/A (F2,16 = 58.2, P < 0.05).
Discussion
This study demonstrates that BoNT/A-LC associates with the plasma membrane of neuronal cells through signals present in the primary sequence of the protein. It also demonstrates that BoNT/A-LC resides in close proximity to SNAP25197 at the plasma membrane, whereas BoNT/E-LC and SNAP25180 reside in the cytoplasm. This difference is striking because the duration of action of BoNT serotypes in animals (33, 34) and cell culture models (17, 18) is generally accepted to be BoNT/A >> BoNT/E. Direct comparisons of the duration of action of BoNT/A and BoNT/E in human extensor digitorum brevis muscles showed that BoNT/E-treated muscles were fully recovered after 30 days, whereas BoNT/A muscles remained fully inhibited by 90 days (35). This distinct subcellular localization could make BoNT/E-LC more accessible to degradation in the neuron, whereas BoNT/A-LC may be partially protected from degradation by localizing to a slow recycling compartment at the plasma membrane.
The data presented also demonstrate that a signal for plasma membrane localization resides within the first eight amino acids at the N terminus of LC/A; alternatively, these amino acids may constitute an important structural element within the protein. Moreover, an active dileucine motif present at the C terminus of BoNT/A-LC has been identified. The change in localization due to disruption or deletion of the dileucine motif may be more relevant when exogenous toxin is administered to neurons because of the role of this motif in protein trafficking, since BoNT/A is trafficked intracellularly through acidic endosomes. Acidification of the vesicle is responsible for a conformational change of the translocation domain facilitating delivery of BoNT/A-LC to the cytoplasm (3, 4, 36). It is hypothesized that the C terminus of BoNT/A-LC may be accessible for interaction with adaptor proteins and BoNT/A-LC may be directed to the plasma membrane in this manner. Examples of the role of dileucine motifs in the retargeting process from acidic endosomes have been reported. For example, the dileucine motif of the insulin-regulated aminopeptidase (IRAP) plays a role in retargeting the protein from the acidic endosomes of the endocytic recycling compartment to the plasma membrane (37). The dileucine motif is specific to serotype A and may play a role in trafficking of BoNT/A-LC from the acidic endosomal compartment to the plasma membrane. Interestingly, BoNT/C1 neurotoxin also causes a long inhibition of transmitter release (18) but lacks a dileucine motif at the C terminus and differs from serotype A in the first 8 aa at the N terminus (Fig. 8). In a previous report, expression of a pEGFP/BoNT/C1-LC construct in HIT-T15 cells appeared to yield GFP fluorescence dispersed throughout the cell, including the nucleus (38), similar to that what we have previously reported for BoNT/B-LC.† Although shorter than serotype A (18), B and C1 do have a long duration of action in neuronal cells, supporting an alternative mechanism for these serotypes other than plasma membrane localization for the persistence of the endopeptidase activity. BoNT/A-LC is unique in being a protein of bacterial origin containing a functional dileucine motif. These data support sequence-specific signals as determinants of intracellular localization and as a basis for the duration and toxicity differences between the two different botulinum neurotoxin serotypes. Elucidating the mechanism of the subcellular localization and trafficking of BoNTs within neurons will aid in understanding the long and variable duration of action of BoNT activity in neurons.
Supplementary Material
Acknowledgments
We thank Li Zhang for providing the clostridial genomic DNA used for cloning, and Dr. George Sachs (University of California, Los Angeles) for assistance with confocal microscopy and scientific direction. We are also indebted to Todd Herrington for early studies on the motifs.
Abbreviations: BoNT, botulinum neurotoxin; LC, light chain; SNAP25, synaptosome-associated protein of 25 kDa.
Footnotes
Fernández-Salas, E., Steward, L. E., Ho, H. & Aoki, K. R. (2002) Naunyn-Schmiedeberg's Arch. Pharmacol. 365, Suppl. 2, 44 (abstr.).
References
- 1.Simpson, L. L. (2000) Biochimie 82, 943-953. [DOI] [PubMed] [Google Scholar]
- 2.Brin, M. F., Binder, W., Blitzer, A., Schenrock, L. & Pogoda, J. M. (2002) in Scientific and Therapeutic Aspects of Botulinum Toxin, eds. Brin, M. F., Hallett, M. & Jankovic, J. (Lippincott Williams & Wilkins, Philadelphia), pp. 233-250.
- 3.Chaddock, J. A. & Melling, J. (2001) in Molecular Medical Microbiology, ed. Sussman, M. (Academic, San Diego), pp. 1141-1152.
- 4.Schiavo, G., Matteoli, M. & Montecucco, C. (2000) Physiol. Rev. 80, 717-766. [DOI] [PubMed] [Google Scholar]
- 5.Verheyden, J., Blitzer, A. & Brin, M. F. (2001) Semin. Cutan. Med. Surg. 20, 121-126. [DOI] [PubMed] [Google Scholar]
- 6.Dolly, J. O., Lisk, G., Foran, P. G., Meunier, F., Mohammed, N., O'Sullivan, G. & de Paiva, A. (2002) in Scientific and Therapeutic Aspects of Botulinum Toxin, eds. Brin, M. F., Hallett, M. & Jankovic, J. (Lippincott Williams & Wilkins, Philadelphia), pp. 91-102.
- 7.Lacy, D. B. & Stevens, R. C. (1999) J. Mol. Biol. 291, 1091-1104. [DOI] [PubMed] [Google Scholar]
- 8.Lacy, D. B., Tepp, W., Cohen, A. C., DasGupta, B. R. & Stevens, R. C. (1998) Nat. Struct. Biol. 5, 898-902. [DOI] [PubMed] [Google Scholar]
- 9.Swaminathan, S. & Eswaramoorthy, S. (2000) Nat. Struct. Biol. 7, 693-699. [DOI] [PubMed] [Google Scholar]
- 10.Humeau, Y., Doussau, F., Grant, N. J. & Poulain, B. (2000) Biochimie 82, 427-446. [DOI] [PubMed] [Google Scholar]
- 11.Rizo, J. & Südhof, T. C. (2002) Nat Rev. Neurosci. 3, 641-653. [DOI] [PubMed] [Google Scholar]
- 12.Blasi, J., Chapman, E. R., Link, E., Binz, T., Yamasaki, S., De Camilli, P., Südhof, T. C., Niemann, H. & Jahn, R. (1993) Nature 365, 160-163. [DOI] [PubMed] [Google Scholar]
- 13.Binz, T., Blasi, J., Yamasaki, S., Baumeister, A., Link, E., Südhof, T. C., Jahn, R. & Niemann, H. (1994) J. Biol. Chem. 269, 1617-1620. [PubMed] [Google Scholar]
- 14.Jurasinski, C. V., Lieth, E., Dang Do, A. N. & Schengrund, C. L. (2001) Toxicon 39, 1309-1315. [DOI] [PubMed] [Google Scholar]
- 15.Meunier, F. A., Lisk, G., Sesardic, D. & Dolly, J. O. (2003) Mol. Cell. Neurosci. 22, 454-466. [DOI] [PubMed] [Google Scholar]
- 16.Naumann, M. (2002) in Scientific and Therapeutic Aspects of Botulinum Toxin, eds. Brin, M. F., Hallett, M. & Jankovic, J. (Lippincott Williams & Wilkins, Philadelphia), pp. 303-308.
- 17.Keller, J. E., Neale, E. A., Oyler, G. & Adler, M. (1999) FEBS Lett. 456, 137-142. [DOI] [PubMed] [Google Scholar]
- 18.Foran, P. G., Mohammed, N., Lisk, G. O., Nagwaney, S., Lawrence, G. W., Johnson, E., Smith, L., Aoki, K. R. & Dolly, J. O. (2003) J. Biol. Chem. 278, 1363-1371. [DOI] [PubMed] [Google Scholar]
- 19.Varshavsky, A. (1996) Proc. Natl. Acad. Sci. USA 93, 12142-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, F., Foran, P., Shone, C. C., Foster, K. A., Melling, J. & Dolly, J. O. (1997) Biochemistry 36, 5719-5728. [DOI] [PubMed] [Google Scholar]
- 21.DeLean, A., Munson, P. J. & Rodbard, D. (1978) Am. J. Physiol. 235, 97-102. [DOI] [PubMed] [Google Scholar]
- 22.DeLean, A., Munson, P. J., Guardabasso, V. & Rodbard, D. (1988) A User's Guide to allfit (Laboratory of Theoretical and Physical Biology, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD), Version 2.6.
- 23.Johnson, E. A. & Bradshaw, M. (2001) Toxicon 39, 1703-1722. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Arca, S., Alberts, P., Zahraoui, A., Louvard, D. & Galli, T. (2000) J. Cell Biol. 149, 889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadkhodayan, S., Knapp, M. S., Schmidt, J. J., Fabes, S. E., Rupp, B. & Balhorn, R. (2000) Protein Expression Purif. 19, 125-130. [DOI] [PubMed] [Google Scholar]
- 26.Kirchhausen, T. (1999) Annu. Rev. Cell Dev. Biol. 15, 705-732. [DOI] [PubMed] [Google Scholar]
- 27.Letourneur, F. & Klausner, R. D. (1992) Cell 69, 1143-1157. [DOI] [PubMed] [Google Scholar]
- 28.Boehm, M. & Bonifacino, J. S. (2002) Gene 286, 175-186. [DOI] [PubMed] [Google Scholar]
- 29.Bresnahan, P. A., Yonemoto, W., Ferrell, S., Williams-Herman, D., Geleziunas, R. & Greene, W. C. (1998) Curr. Biol. 8, 1235-1238. [DOI] [PubMed] [Google Scholar]
- 30.Craig, H. M., Pandori, M. W. & Guatelli, J. C. (1998) Proc. Natl. Acad. Sci. USA 95, 11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenberg, M., DeTulleo, L., Rapoport, I., Skowronski, J. & Kirchhausen, T. (1998) Curr. Biol. 8, 1239-1242. [DOI] [PubMed] [Google Scholar]
- 32.Peden, A. A., Park, G. Y. & Scheller, R. H. (2001) J. Biol. Chem. 276, 49183-49187. [DOI] [PubMed] [Google Scholar]
- 33.Adler, M., Keller, J. E., Sheridan, R. E. & Deshpande, S. S. (2001) Toxicon 39, 233-243. [DOI] [PubMed] [Google Scholar]
- 34.Aoki, K. R. (2002) Toxicon 40, 923-928. [DOI] [PubMed] [Google Scholar]
- 35.Eleopra, R., Tugnoli, V., Rossetto, O., De Grandis, D. & Montecucco, C. (1998) Neurosci. Lett. 256, 135-138. [DOI] [PubMed] [Google Scholar]
- 36.Koriazova, L. K. & Montal, M. (2003) Nat. Struct. Biol. 10, 13-18. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, A. O., Subtil, A., Petrush, R., Kobylarz, K., Keller, S. R. & McGraw, T. E. (1998) J. Biol. Chem. 273, 17968-17977. [DOI] [PubMed] [Google Scholar]
- 38.Lang, J., Zhang, H., Vaidyanathan, V. V., Sadoul, K., Niemann, H. & Wollheim, C. B. (1997) FEBS Lett. 419, 13-17. [DOI] [PubMed] [Google Scholar]
- 39.Darsow, T., Burd, C. G. & Emr, S. D. (1998) J. Cell Biol. 142, 913-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varoqui, H. & Erickson, J. D. (1998) J. Biol. Chem. 273, 9094-9098. [DOI] [PubMed] [Google Scholar]
- 41.Tan, P. K., Waites, C., Liu, Y., Krantz, D. E. & Edwards, R. H. (1998) J. Biol. Chem. 273, 17351-17360. [DOI] [PubMed] [Google Scholar]
- 42.Liu, Y., Krantz, D. E., Waites, C. & Edwards, R. H. (1999) Trends Cell Biol. 9, 356-363. [DOI] [PubMed] [Google Scholar]
- 43.Aiken, C., Konner, J., Landau, N. R., Lenburg, M. E. & Trono, D. (1994) Cell 76, 853-864. [DOI] [PubMed] [Google Scholar]
- 44.Chen, H. J., Yuan, J. & Lobel, P. (1997) J. Biol. Chem. 272, 7003-7012. [DOI] [PubMed] [Google Scholar]
- 45.Holman, G. D. & Sandoval, I. V. (2001) Trends Cell Biol. 11, 173-179. [DOI] [PubMed] [Google Scholar]
- 46.Ehrlich, M., Shmuely, A. & Henis, Y. I. (2001) J. Cell Sci. 114, 1777-1786. [DOI] [PubMed] [Google Scholar]
- 47.Sandoval, I. V., Arredondo, J. J., Alcalde, J., Gonzalez Noriega, A., Vandekerckhove, J., Jimenez, M. A. & Rico, M. (1994) J. Biol. Chem. 269, 6622-6631. [PubMed] [Google Scholar]
- 48.Blagoveshchenskaya, A. D., Hewitt, E. W. & Cutler, D. F. (1999) Mol. Biol. Cell 10, 3979-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibson, R. M., Schiemann, W. P., Prichard, L. B., Reno, J. M., Ericsson, L. H. & Nathanson, N. M. (2000) J. Biol. Chem. 275, 22574-22582. [DOI] [PubMed] [Google Scholar]
- 50.Rivera, J. F., Ahmad, S., Quick, M. W., Liman, E. R. & Arnold, D. B. (2003) Nat. Neurosci. 6, 243-250. [DOI] [PubMed] [Google Scholar]
- 51.Garrido, J. J., Fernandes, F., Giraud, P., Mouret, I., Pasqualini, E., Fache, M. P., Jullien, F. & Dargent, B. (2001) EMBO J. 20, 5950-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.