Abstract
Age carries a detrimental impact on T cell function. In the past decade, analyses of the genome-scale transcriptional changes of T cells during aging have yielded a large amount of data and provided a global view of gene expression changes in T cells from aged hosts as well as subsets of T cells accumulated with age. Here, we aim to review the changes of gene expression in thymocytes and peripheral mature T cells, as well as the subsets of T cells accumulated with age, and discuss the gene networks and signaling pathways that are altered with aging in T cells. We also discuss future direction for furthering the understanding of the molecular basis of gene expression alterations in aged T cells, which could potentially provide opportunities for gene-based clinical interventions.
Keywords: aging, thymocytes, T cells, naïve and memory T cells, CD28− T cells
Introduction
Age carries a detrimental impact on T cell function. Age-associated decline of T cell function is complex and occurs at multiple levels (Haynes and Swain, 2006). Alteration in transcription is arguably one of the most studied and identifiable age-associated change in T cells. As part of the adaptation to a changing microenvironment with age (Linton et al., 2005), T cells undergo substantial changes in gene expression, resulting in enhanced or reduced aspects of their functions (Pawelec et al., 2005; Weng, 2006). Reported age-associated alterations in T cell functions include: increased cytotoxicity, enhanced secretion of inflammatory cytokines, reduced antigen-induced proliferation, and a decrease in cell division. The precise details of age-associated changes in transcription are beginning to be revealed, but the functional significance of these transcriptional alterations and their contribution to the age-associated changes in T cell function have not been fully characterized. A better understanding of the transcriptional changes in T cells that are associated with aging is an essential first step toward further functional assessment and eventual clinical interventions for older adults.
The development of methods, such as microarrays, allow the genome-scale assessment of gene expression and provide a means to understand the scope and magnitude of gene expression changes during aging (Prolla, 2005). The current commercially available whole genome arrays (Agilent, Affymetrix, and Illumina) contain the information of over 30,000 transcripts with most of the annotated protein-coding genes of human and mouse, and readily assess RNAs isolated from a few thousand to millions of cells. With sufficient biological repeats and proper statistical tools (Hyatt et al., 2006), the global gene expression pattern and significantly enhanced or reduced expressed genes are promptly identified; this provides sufficient transcriptional details to match the complexity of the age-associated changes. However, connecting the individual altered expressed genes to the altered functional networks and signaling pathways with aging is a monumental task.
In the past a few years, studies of global gene expression in T cells with aging have evolved from analyzing whole organ/tissue/whole populations of T cells to comparing well-defined subsets of T cells from different lymphoid tissues or organs, and from identification of significantly altered genes to connecting the gene networks and signaling pathways that are associated with these altered genes. The cumulative evidence shows that age-related alterations of transcription in T cells can be found in thymocytes (Lustig et al., 2007, 2009; Griffith et al., 2012), in peripheral mature T cells (Remondini et al., 2010), and T cell subsets (Fann et al., 2005; Mirza et al., 2011). Although neither the causes nor the consequences of all the transcriptional changes in T cells with age are currently known, the identification and validation of these age-associated changes pave a new avenue for a better understanding of cellular and functional alterations associated with aging. This information will be essential for developing strategies to slow down or even reverse the course of T cell aging. The purpose of this review is to summarize the age-associated gene expression changes on a global level that have been observed in recent studies of T cells and their subsets. We also discuss a potential link between the transcriptional changes and functional changes in aged T cells.
Transcriptional Changes in Aged Thymus and Peripheral Blood T Cells
Thymocytes
Initial microarray experiments were focused on comparing gene expression of the total organ/tissue/T cells between young and old hosts (mouse and human). Global gene expression analysis using total thymus from mice revealed that the largest number of gene expression changes with age, within total thymus, occurred in the categories of transcription regulators and enzymes, and were mostly involved with cell growth, proliferation, and death (Lustig et al., 2007). One of the more notable transcription regulators that decreased expression with age was the glucocorticoid receptor, which has been implicated in T cell development and survival within the thymus. Also detected was an increased expression of several heat shock proteins, which are necessary for cell survival.
A subsequent study focused specifically on thymocytes, demonstrated age-associated changes in the expression of genes involved in TCR signaling, antigen presentation, and lymphocyte development and function (Lustig et al., 2009). These age-related changes included increased expression of CXCR4 and CXCR6, as well as a decrease in CCL25. Another key change was an increase in Vav1, which is important for T cell development and activation. Genes involved with cell function included changes in various ribosomal proteins, which could indicate changes in translational activity with age, and a decrease in S100A, a calcium-binding protein involved in various cell functions. Interestingly, increased gene expression was consistent in genes associated with cancer.
An unexpected and significant difference was found in gene expression analyses of the immunoglobulin gene family from the thymus of old and young mice (Lustig et al., 2007). This observation has been noted in various gene expression studies (Schumacher et al., 2008; Swindell, 2009), even though some of these studies focused specifically on thymocytes or splenic T cells. Further analysis revealed the apparent presence of immunoglobulin on the cell surfaces of thymocytes as well as splenic T cells, but not intracellularly, indicating this might be an autoreactive phenomenon occurring with age (Lustig et al., 2009).
Not surprisingly, the age-associated change of the environment in which each T cell is located has an effect on the gene expression profile of thymocytes. Griffith et al. (2012) demonstrated that the most profound age-associated changes in gene expression (up to 30% of the genes) were found in the thymic stroma, and such changes affected gene expression changes in all subsets of thymocytes, which only had expression changes in about 3–4% of their total genes. However, the small proportion of changes within the developing thymocytes could be the most critical ones involved in T cell aging.
Mature T cells in the periphery
Changes in gene expression with age, specifically within the murine spleen, mirrored the overall trends of other organs; these included an increase in immune response, stress, and apoptosis related genes, along with a decrease in genes involved in cell growth, energy utilization, and lipid and carbohydrate metabolism (Schumacher et al., 2008). An interesting observation was that many of these expression patterns were similar between mouse strains with shortened, normal, or longer life spans, but the changes occurred at a pace relative to the expected life span for each strain.
Sole focus on gene expression analyses of T cells purified from mouse spleens showed critical gene expression changes in the old mice that may be responsible for their alterations in T cell function (Han et al., 2006). The authors found that old T cells express higher Socs3 and lower growth factor independent 1 (Gfi-1) compared to young T cells, which could contribute to the age-associated decline in proliferation. They also found the increase of apoptosis in old T cells could be explained by higher Gadd45 and lower Bcl2 in old T cells compared to young T cells. In addition, the authors observed an age-associated increase in many immunoglobulin-associated genes, as well as genes associated with innate immunity, such as lysozyme M and myeloperoxidase; moreover, this study showed a decrease in TCR-associated genes such as TCR-beta and various isoforms of Cd3, indicating that both the innate and adaptive arms of the immune system are involved in aging trends. Other categories of genes affected included structural proteins, calcium-binding proteins, and genes involved in electron transport.
Three basic patterns of changes in gene expression emerged from a study of T cells in peripheral blood of various aged adults: first, some genes increased in middle age then decreased; second, genes that decreased in middle age then increased, and lastly, some genes decreased with age (Remondini et al., 2010). Interestingly, they found no group of genes that only increased with age. Global clustering analysis revealed that the gene expression pattern in “successfully aged” adults (older than 90 years) more closely matched the patterns of the younger groups. Most of the genes involved in T cell signaling were in the group which increased in middle age, then decreased. Although it is difficult to strictly compare the patterns between the human and mouse studies with age, some common genes are evident. Not surprisingly, these genes are associated with TCR signaling, cytokines and their receptors, and cancer-related genes. Other common genes between human and mouse that changed expression with age included those from more general signaling pathways, such as Jak/Stat, PPAR, and mTOR signaling. This agrees with the observation that changes in gene expression occurring within aging T cells mirror changes occurring in other cell types throughout the body.
Genome-wide analyses of transcription alterations identified the change associated with aging as a whole of either tissue or of total T cells. As lymphoid tissue consists of different types of cells (T and B cells, stromal cells, etc.), total T cells also consist of different subsets (naïve, effector, and memory), it is unknown whether the identified age-associated changes are a result of aging in all types of cells or in a specific type of cell or a subset of a cell type. Therefore, it is necessary to further analyze the gene expression changes with age in defined a defined cell type, such as T cells, and their subsets.
Gene Expression Changes of T Cell Subsets in Aged Subjects
Human CD4 T cells
Although a global comparison of gene expression in naïve CD4 T cells with age is not available, a study comparing global gene expression of memory CD4 T cells (CD45RA−) between young (20–30 years old) and old (70–75 years old) human subjects show that the overall gene expression profiles of CD4 memory T cells are similar (Czesnikiewicz-Guzik et al., 2008). Based on the fold change (> or <1.25), 536 genes were identified as age-related, including up-regulated genes such as HLA region genes, CCR4, CCR8, CD26, CD58, IL17Rb, and LAIT1, and down-regulated genes such as CD28, CCR6, KLRB1, and KLRC2.
Another recent study compared gene expression of CD4 T cells between young and old donors after in vitro stimulation (Bektas et al., 2013). This study focused on examining changes in TCR-inducible gene expression via the NF-κB pathway, and found that most NF-κB target genes are not induced in a sustained manner in CD4 T cells from older compared to younger donors. Those genes that failed to exhibit sustained expression include CXCL10, TNFAIP2, CCL2, and CXCL5. On the other hand, a subset of NF-κB target genes such as CXCL1, CXCL11, IL1a, and IL6, continue to be up-regulated even in the absence of NF-κB induction. IL1 and IL6 are associated with a chronic pro-inflammatory state in the elderly and are dysregulated in CD4 T cells from old donors. In addition, the authors identified some immune function-related genes that were highly induced after 2 h of stimulation in old donors, including cell surface receptor and signaling molecules (SAMD4A, CD83, KCTD12, SOCS1, and CTLA4) and effector molecules (GZMH). Interestingly, some of the most noteworthy age-associated changes in murine models, such as up-regulation of the chemokines CCL3 and CCL4 and the cytokine IFN-γ, were not found in CD4 T cells of old donors in this study (Chen et al., 2003; Han et al., 2006; Mirza et al., 2011). Further studies are needed to determine the similarities and differences between mouse and human in age-associated changes in T cells.
The finding of low expression of CD28 in memory CD4 T cells from aged donors leads to further comparison of gene expression between CD28+ and CD28− memory CD4 T cell subsets in selected old donors (65–75 years old) (Czesnikiewicz-Guzik et al., 2008). CD28 is a co-stimulatory receptor that offers a critical signal for complete T cell activation during T cell receptor initiation. Absence of the CD28 signal causes partial T cell activation or even anergy. Strikingly, it was found that the difference in gene expression between CD28+ and CD28− memory CD4 T cells was greater than that of memory CD4 T cells between young and old subjects. CD28− memory CD4 T cells express high levels of several KIR genes and some surface receptors genes (CD70, CD74, and CCR5), but low levels of TCRB, CD27, IL7R, IL9R, and ICAM2. Although loss of CD28 protein expression with aging is mainly found in CD8 T cells, this study shows that reduced expression of CD28 mRNA is also found in CD4 T cells with aging.
Human CD8 T cells
A global comparative analysis of gene expression in human CD8 T cells isolated from young (23–27 year old) and old (65–81 year old) adults reported the identification of a total of 754 genes (505 down-regulated and 258 up-regulated) using an expression fold change more than 1.5 (Cao et al., 2010). The functions of these genes are involved in the regulation of transcription, protein phosphorylation, ubiquitination, intracellular transport, immune response, and apoptosis. Intriguingly, the down-regulated genes in CD8 T cells from old donors affect multiple stages of gene transcription: chromatin structure, transcription initiation, elongation, RNA stabilization, and protein translation and translocation; this suggests the impaired regulation of these fundamental molecular events might be responsible for the declined immune function in the elderly. In addition, genes related to the signaling pathways are also found down-regulated in CD8 T cells from old donors, such as T cell receptor signaling, IL-2 signaling, IGF-1 signaling, insulin receptor signaling, JAK/Stat signaling, MAPK/JNK signaling, PI3K/AKT signaling, Wnt/β-catenin signaling, and ERK/MAPK signaling. On the other hand, oxidative phosphorylation and apoptosis signaling are up-regulated in CD8 T cells from old donors. These alterations of gene expression provide a mechanism for the functional decline of the immune response in the elderly.
CD28− CD8 T cells
Accumulation of CD28− CD8 T cells is one of the hallmarks of human aging. The CD28− population is considered as aged CD8 T cells in humans (Weng et al., 2009). The study of CD28+ and CD28− memory CD8 T cells (CD45RA−) from young and middle aged donors (25–45 years old) identified <100 altered genes/transcripts based on the criteria of the false discovery rate (FDR) <0.05 and fold change >2 (Fann et al., 2005). Among these 45 genes,CD28− CD8 T cells highly expressed natural killer cell receptors, GZMB, PRF1, FASLG, IL12A, IL12, CCL4, CX3CR1, and CMKLR1. In contrast, they had low expression of IL3, IL23A, IL7R, and IL12RB2.
A subsequent study comparing CD28+ and CD28− CD8 T cells from young and old donors found that gene expression patterns of CD28− CD8 T cells from young and old subjects were similar (Lazuardi et al., 2009). In contrast, there were noticeable differences in gene expression of CD28+ CD8 T cells between young and old subjects. From the gene expression similarities, the levels from CD28+ CD8 T cells in old donors were located between young CD28+ and CD28− (young and old) CD8 T cells. The highly expressed genes identified by Fann et al. (2005) in CD28− CD8 T cells were also found in this study. In addition, high expression of PIK3CD, MAL, IL6R, CD62L, and CCR7 were found in CD28+ CD8 T cells of young subjects whereas high expression of GATA3, BIRC3, FAS, RGS1, and MAP3K1 were found in CD28+ CD8 T cells of old subjects. This study reveals that CD28− CD8 T cells have a common pattern of gene expression regardless of whether they are isolated from young or old subjects. However, whether the difference in gene expression in CD28+ CD8 T cells between young and old subjects is reflecting the impact of aging or the heterogeneous nature (different percentages of naïve and memory T cells) of CD28+ CD8 T cells in young and old subjects remains to be determined.
Interestingly, similarly reduced expression of some co-stimulatory molecules including CD28, NK cell receptor, and chemokine receptors were found in both CD4 and CD8 CD28− T cells with some differences in the expression of CD40L, KLRD1, KLRG1, and KLRK1 (Czesnikiewicz-Guzik et al., 2008). However, loss of CD28 surface expression was only prominent in CD8 T cells with aging. Further studies will be needed to compare the functional alteration of CD4 and CD8 subsets with aging.
Mouse CD4 and CD8 T cells
Studies in mouse T cells suggest that aging has a more profound detrimental impact on naïve compared to memory T cells (Ponnappan and Ponnappan, 2011). Gene expression comparison of naïve CD4 and CD8 T cells between young (3–4 months) and old (20 months) mice identified over 2000 age-associated genes in CD4 and CD8 T cells using a twofold change as the cutoff (Mirza et al., 2011). The functions of these genes are broad and involved in multiple cellular functions such as cell growth, cell cycle, cell death, inflammatory response, and cell trafficking. Some of those identified genes exhibited similar changes in both CD4 and CD8 T cells from old mice, such as Anxa1, Ccl1, Ccl5, Ccr2, Il4, Havcr2, and Ltb4r1. The enhanced expression of chemokines and chemokine receptors in aged mouse T cells was also observed in an earlier study (Mo et al., 2003). Some gene alterations are specific to either CD4 or CD8 T cells, such as Jak3, Socs1, and Pi3kcd in CD4, and Penk, Nfc2, and Irak3 in CD8 T cells of old mice.
Gene expression analysis of in vitro stimulated naïve T cells between young and old mice further revealed some defects in T cell signaling, cytokine production, and differentiation into Th2 cells. Gata3 and c-Maf were found up-regulated post-activation in naïve aged CD4 T cells, which may be responsible for the imbalanced Th2 immune response in the elderly. Ccl5 and Tlt4 are up-regulated in aged CD8 T cells from pre- to post-activation, while Ccl1, Ccl9, Il7r, and other genes are only up-regulated post-activation. Genes such as Tnfsf14 and S100a9 are up-regulated in aged CD8 T cells only in pre- and 12 h post-activation. In CD4 aged T cells, Ccl5 and Tlr4 are up-regulated at all timepoints. Rorc is not differentially expressed before activation, however its expression was decreased in aged CD4 T cells compared with young ones after TCR activation. In the pattern of lower expression in the elderly at both pre- and post-activation, many genes associated with microtubules, cell cycle replication, migration, and other functions were found in both CD4 and CD8 naïve T cells. Taken together, the highly expressed and up-regulated specific cytokines, chemokines, and their receptors in aged naïve T cells indicate that naïve CD4 and CD8 T cells in old mice have a pro-inflammatory status.
Gene Networks and Pathways Altered in Aged T Cells
A global view of gene expression profiles provides a means for examining the gene networks and signaling pathways of a defined biological process and function. Among the genes that were transcriptionally altered in aged T cells, substantial numbers of them are associated with basic cellular and molecular biological processes such as cell growth and proliferation, cell death and apoptosis, energy utilization and metabolism, and transcription regulation, which were also reported in other types of cells with aging (Kuilman et al., 2010). In Table 1, we combine the altered expressed genes from the literature based on their original selection and listed seven functional categories. We will focus on the molecular basis of three immune function-related alterations in T cells with aging in this review.
Table 1.
List of genes that are expressed either lower or higher in aged than in young T cells.
| Down-regulated |
Up-regulated |
||||
|---|---|---|---|---|---|
| Genes | Subsets | Reference | Genes | Subsets | Reference |
| T cell receptor signaling pathway | Cytokine/chemokine network | ||||
| AKT3 | h CD8 | Cao et al. (2010) | ACVR2A, IL15, TNFSF14 | h CD8 | Cao et al. (2010) |
| CD28 | h CD4N, CD8CD28- | Weng et al. (2009), Czesnikiewicz-Guzik et al. (2008) | Bmpr1a | m CD4N, CD8N | Mirza et al. (2011) |
| CD3G | h CD8 | Cao et al. (2010) | Ccl1, Il5, Il9, Il3, Il21 | m s-CD4N & CD8N | Mirza et al. (2011) |
| DLG1 | h CD8 | Cao et al. (2010) | Ccl17, Ccl20, Ccl22, Ccl24 | m s-CD4N | Mirza et al. (2011) |
| GRAP2 | h CD8 | Cao et al. (2010) | Ccl21c, Ccl8, Csf2 | m CD8N | Mirza et al. (2011) |
| GRB2 | h CD8CD28- | Lazuardi et al. (2009) | CCL3/Ccl3 | h CD4, m CD4N | Mirza et al. (2011), Chen et al. (2003) |
| ITK | h CD8 | Cao et al. (2010) | CCL4 | h CD8CD28- | Fann et al. (2005) |
| LCK | h CD8 | Cao et al. (2010) | CCL5/Ccl5 | h CD8, m CD4N, CD8N | Cao et al. (2010), Mirza et al. (2011) |
| MAPK1, MAPK14 | h CD8 | Cao et al. (2010) | CCR1 | h CD4 | Mo et al. (2003), Yung and Mo (2003) |
| NCK1 | h CD8 | Cao et al. (2010) | CCR2/Ccr2 | h CD4, m CD4N, CD8N | Mirza et al. (2011), Mo et al. (2003) |
| PDK1 | h CD8CD28- | Lazuardi et al. (2009) | CCR3 | h CD4 | Yung and Mo (2003) |
| PIK3CD | h CD8CD28- | Lazuardi et al. (2009) | CCR4/Ccr4 | h CD4, CD4N, m CD4N, s-CD8N | Czesnikiewicz-Guzik et al. (2008), Mirza et al. (2011), Mo et al. (2003), Yung and Mo (2003) |
| SOS2 | h CD8 | Cao et al. (2010) | CCR5/Ccr5 | h CD4, CD4N, m CD4N/CD8N | Czesnikiewicz-Guzik et al. (2008), Mirza et al. (2011), Mo et al. (2003), Yung and Mo (2003) |
| CCR6, CXCR2, CXCR5 | h CD4 | Mo et al. (2003) | |||
| Cytokine/chemokine network | CCR8 | h CD4N | Czesnikiewicz-Guzik et al. (2008), Mo et al. (2003) | ||
| Ccl19 | m s-CD4N | Mirza et al. (2011) | CD70, IL17RB | h CD4N | Czesnikiewicz-Guzik et al. (2008) |
| Ccl22 | m CD8N | Mirza et al. (2011) | CLCF1 | h CD8CD28- | Lazuardi et al. (2009) |
| Ccl24 | m s-CD8N | Mirza et al. (2011) | Csd2, Cxcl11 | m s-CD4N | Mirza et al. (2011) |
| Ccr9 | m CD4N, CD8N | Mirza et al. (2011) | Csf1, Cxcl10 | m CD4N | Mirza et al. (2011) |
| Cxcl1 | m CD4N | Mirza et al. (2011) | Csf2ra, Cxcl2 | m CD8N | Mirza et al. (2011) |
| Cxcl10, Cxcl12 | m CD8N | Mirza et al. (2011) | Csf3r | m s-CD8N | Mirza et al. (2011) |
| CXCR7, CXCR9 | h CD4, CD8 | Mo et al. (2003) | CX3CR1 | h CD8CD28- | Lazuardi et al. (2009), Fann et al. (2005) |
| Flt1 | m s-CD8N | Mirza et al. (2011) | CXCL12 | h CD4 | Hernandez-Lopez et al. (2010), Cane et al. (2012) |
| Ifna1 | m CD8N, s-CD4N | Mirza et al. (2011) | Cxcl9 | m CD8N, s-CD4N | Mirza et al. (2011) |
| Ifna13 | m CD8N | Mirza et al. (2011) | CXCR3/Cxcr3 | h CD4, CD8, m CD4N, CD8N | Cao et al. (2010), Mirza et al. (2011), Mo et al. (2003) |
| Ifnab | m CD4N | Mirza et al. (2011) | CXCR4 | h CD4 | Mo et al. (2003), Hernandez-Lopez et al. (2010), Cane et al. (2012) |
| Il12b, Kit, Pdgfb | m s-CD4N | Mirza et al. (2011) | FAS, IFNGR1 | h CD8CD28- | Lazuardi et al. (2009) |
| Cytokine/chemokine network | Cytokine/chemokine network | ||||
| Il12rb2 | m s-CD8N | Mirza et al. (2011) | Ifng, Il10, Il4 | m CD4N, CD8N | Mirza et al. (2011) |
| Il17a, Il6 | m s-CD8N | Mirza et al. (2011) | Il12rb1, Il13, Il5ra | m CD4N | Mirza et al. (2011) |
| Il6ra | m CD4N | Mirza et al. (2011) | Il18r1, Il20ra, Il23r, Il7r | m s-CD8N | Mirza et al. (2011) |
| Kit | m s-CD8N | Mirza et al. (2011) | Il18rap, Il1b | m CD8N | Mirza et al. (2011) |
| Ppbp | m CD4N | Mirza et al. (2011) | Il19, Il24, Il2ra, Inhba | m s-CD4N | Mirza et al. (2011) |
| Tnfrsf21 | m s-CD8N | Mirza et al. (2011) | Il1r2 | m CD4N, s-CD4N, CD8N | Mirza et al. (2011) |
| Tnfsf11a | m CD8N | Mirza et al. (2011) | Il2, Il22, Il23a | m s-CD8N | Mirza et al. (2011) |
| Tpo | m s-CD8N | Mirza et al. (2011) | Ltbr | m CD4N, CD8N | Mirza et al. (2011) |
| Xcr1 | m CD4N, s-CD8N | Mirza et al. (2011) | Tnfrsf13c | m CD4N | Mirza et al. (2011) |
| Natural killer cell-mediated cytotoxicity | Tnfsf13b, Tnfsf4, Tnfrsf17 | m CD4N, CD8N | Mirza et al. (2011) | ||
| Ifna1 | m s-CD4N | Mirza et al. (2011) | |||
| Klrd1, Klrk1 | m s-CD4N | Mirza et al. (2011) | Natural killer cell-mediated cytotoxicity | ||
| Nfatc4 | m s-CD4N | Mirza et al. (2011) | CD244 | h CD8 | Cao et al. (2010) |
| FAS | h CD8CD28- | Lazuardi et al. (2009) | |||
| Toll like receptor signaling pathway | GZMB | h CD8CD28- | Lazuardi et al. (2009) | ||
| Cd86 | m CD8N | Mirza et al. (2011) | IFNGR1 | h CD8CD28- | Lazuardi et al. (2009) |
| Cxcl10 | m CD8N | Mirza et al. (2011) | ITGB2 | h CD8 | Cao et al. (2010) |
| Ifna1, Ifna13 | m CD8N | Mirza et al. (2011) | KIR2DL3, KIR2DL4 | h CD4N | Czesnikiewicz-Guzik et al. (2008) |
| Ifnab | m CD4N | Mirza et al. (2011) | KIR2DL5A | h CD8CD28- | Lazuardi et al. (2009) |
| Il6 | m s-CD8N | Mirza et al. (2011) | KIR2DS5 | h CD8CD28- | Lazuardi et al. (2009) |
| Irf7 | m CD4N, s-CD8N | Mirza et al. (2011) | KIR3DL1 | h CD8CD28- | Lazuardi et al. (2009) |
| Lbp | m CD4N | Mirza et al. (2011) | KIR3DL2 | h CD4N, CD8CD28- | Czesnikiewicz-Guzik et al. (2008), Lazuardi et al. (2009) |
| Pik3cd | m CD4N | Mirza et al. (2011) | KLRC3 | h CD8CD28- | Lazuardi et al. (2009) |
| Rela | m CD4N | Mirza et al. (2011) | KLRD1 | h CD8 | Cao et al. (2010) |
| Tlr5, Tlr7 | m s-CD8N | Mirza et al. (2011) | KLRG1 | h CD8 | Henson and Akbar (2009) |
| MAPK1 | h CD8CD28- | Lazuardi et al. (2009) | |||
| Cell cycle regulator | NKG2D | h CD4 | Alonso-Arias et al. (2011) | ||
| CCND2, CCNH | h CD8 | Cao et al. (2010) | PRF1 | h CD8CD28- | Lazuardi et al. (2009) |
| FZR1 | h CD8 | Cao et al. (2010) | PTPN6 | h CD8 | Cao et al. (2010) |
| MAD1L1 | h CD8 | Cao et al. (2010) | VAV3 | h CD8CD28- | Lazuardi et al. (2009) |
| MYC | h CD8 | Cao et al. (2010) | |||
| RAD21 | h CD8 | Cao et al. (2010) | MAPK signaling pathway | ||
| RB1, RBL2 | h CD8 | Cao et al. (2010) | DUSP2, DUSP5,DUSP6, DUSP10 | h CD8 | Cao et al. (2010) |
| SMAD2, SMAD4 | h CD8 | Cao et al. (2010) | DUSP4 | h CD8CD28- | Lazuardi et al. (2009) |
| SMC3 | h CD8 | Cao et al. (2010) | FAS | h CD8CD28- | Lazuardi et al. (2009) |
| STAG1 | h CD8 | Cao et al. (2010) | FGFR1 | h CD8 | Cao et al. (2010) |
| TFDP2 | h CD8 | Cao et al. (2010) | FLNA | h CD8 | Cao et al. (2010) |
| YWHAE, YWHAZ | h CD8 | Cao et al. (2010) | GNA12 | h CD8 | Cao et al. (2010) |
| MAP3K1 | h CD8CD28- | Lazuardi et al. (2009) | |||
| Insulin signaling pathway | MAP3K5 | h CD8 | Cao et al. (2010) | ||
| AKT3 | h CD8 | Cao et al. (2010) | MAP3K8 | h CD8 | Cao et al. (2010) |
| BRAF | h CD8 | Cao et al. (2010) | MAP4K1 | h CD8 | Cao et al. (2010) |
| FLOT2 | h CD8 | Cao et al. (2010) | MAPK1 | h CD8CD28- | Lazuardi et al. (2009) |
| FOXO1 | h CD8 | Cao et al. (2010) | NR4A1 | h CD8 | Cao et al. (2010) |
| GRB2 | h CD8CD28- | Lazuardi et al. (2009) | RPS6KA5 | h CD8 | Cao et al. (2010) |
| Insulin signaling pathway | MAPK signaling pathway | ||||
| IRS1, IRS2 | h CD8 | Cao et al. (2010) | RRAS2 | h CD8CD28- | Lazuardi et al. (2009) |
| MAPK1 | h CD8 | Cao et al. (2010) | |||
| PDE3B | h CD8 | Cao et al. (2010) | Toll like receptor signaling pathway | ||
| PIK3CD | h CD8CD28- | Lazuardi et al. (2009) | Ccl3 | m CD4N | Mirza et al. (2011) |
| PIK3R2 | h CD8 | Cao et al. (2010) | Ccl5 | m CD4N | Mirza et al. (2011) |
| PRKACB, PRKAR1A | h CD8 | Cao et al. (2010) | Cd86 | m CD4N | Mirza et al. (2011) |
| RPS6KB1 | h CD8 | Cao et al. (2010) | Cxcl10 | m CD4N | Mirza et al. (2011) |
| SOS2 | h CD8 | Cao et al. (2010) | Tlr4 | m CD4N | Mirza et al. (2011) |
h, Human; m, mouse; CD4N, CD4 naïve T cells; CD8N, CD8 naïve T cells; s-, TCR activated.
Reduced expression of TCR and co-stimulation signaling associated genes
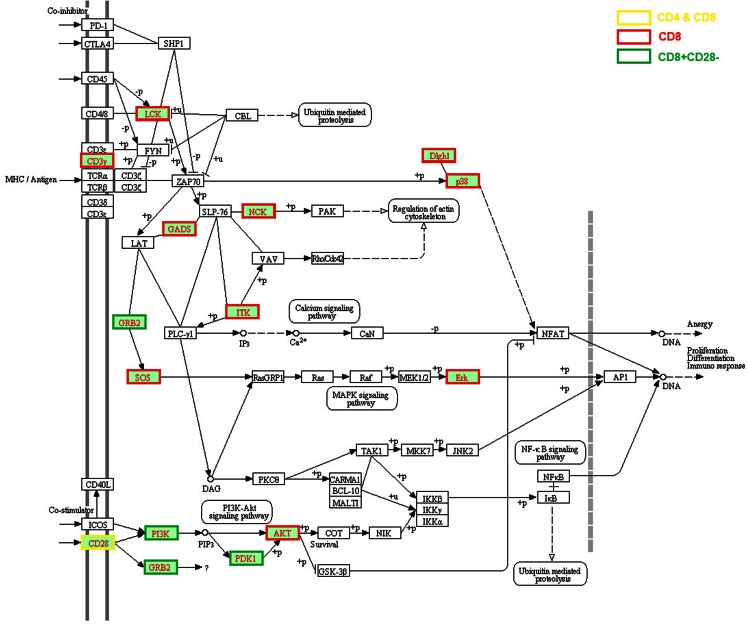
Impaired TCR signal transduction was observed in aged mice and humans two decades ago (Utsuyama et al., 1993; Whisler et al., 1996). Genes associated with several TCR signaling pathways were found to be expressed lower in aged hosts on both genome-scale (Table 1; Figure 1) and individual studies. First, CD3G expression was lower in human CD8 T cells (Cao et al., 2010), and phosphorylated Cd3z was decreased in aged mouse CD4 T cells, and almost absent in 18-month old mice (Garcia and Miller, 1997). Second, significantly reduced expression of CD28, particularly in CD8 T cells, was widely reported in humans with aging (Weng et al., 2009). Lower expression of CD28 was also found in human CD4 memory cells but to a much lower degree compared to CD8 T cells (Czesnikiewicz-Guzik et al., 2008). Intriguingly, several age-associated defects in TCR signaling occur at the post-translational level without obvious changes in the level of gene expression, such as: reduced phosphorylation of phospholipase Cγ1 (PLCγ1), JNK, second messengers such as inositol trisphosphate (IP3) and diacylglycerol (DAG), reduced influx of Ca2+ (Utsuyama et al., 1997; Kirk et al., 1999) in T cells of aged mice, decreased LCK activity (Fulop et al., 1999), and ERK phosphorylation in T cells of aged humans (Li et al., 2012). The consequence of these age-associated changes in TCR signaling is observed at the transcriptional levels of their downstream targets, such as the decreased expression of SOS, GADS, NKC, ITK, PI3K, PDK2, AKT, Erk, Dlgh1, and p38 (Table 1) (Utsuyama et al., 1993; Whisler et al., 1996). Certainly, more studies are needed to dissect the primary causes of the impaired expression of these age-related genes and their functional consequences.
Figure 1.
Age-related alteration of gene expression in the TCR signaling pathway. The age-related genes were assembled from the literature based on the significant changes reported by the original articles which were applied for all figures (Chen et al., 2003; Mo et al., 2003; Yung and Mo, 2003; Fann et al., 2005; Czesnikiewicz-Guzik et al., 2008; Henson and Akbar, 2009; Lazuardi et al., 2009; Weng et al., 2009; Cao et al., 2010; Hernandez-Lopez et al., 2010; Alonso-Arias et al., 2011; Mirza et al., 2011; Cane et al., 2012). Three-hundred five up-regulated and 472 down-regulated genes in aged human T cells including CD4 and CD8 T cells were analyzed using KEGG analysis in the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt). The entire TCR signaling pathway is included and the molecules with lower gene expression compared with young are highlighted in red text and different border color indicating specific T cell subsets. The altered expressed genes in the TCR signaling pathway include CD3G, LCK, GRAP2 (encode GADS), GRB2, ITK, NCK, MAPK signaling pathway molecules SOS2 and MAPK1 (encode Erk), CD28, PI3K-AKT signaling pathway molecules PI3K, PDK1, and AKT, DLG1 (encode DLGH1) and MAPK14 (encode p38).
Alteration of cytokines/chemokines network
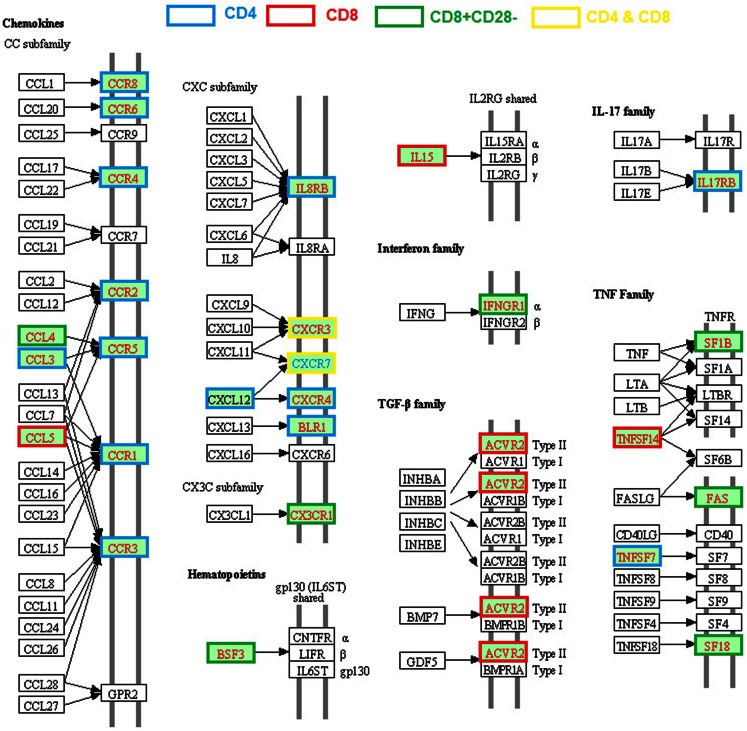
Alteration of expression of various cytokines, chemokines, and their receptors in CD4 or CD8 T cells from old humans and mice are well documented (Table 1; Figure 2). Unlike the age-associated TCR signaling pathway, enhanced expression of various cytokines of the TNF and TGF-β families, chemokines (CCL3, CCL4, CCL5, CXCL12), and chemokine receptors (CCR1, CCR2, CCR3, CCR4, CCR5, CCR6, CCR8, CXCR2, CXCR3, CXC4, CXCR5) were reported in CD4 or CD8 T cells from old humans and mice (Mo et al., 2003; Yung and Mo, 2003; Fann et al., 2005; Lazuardi et al., 2009; Cane et al., 2012).
Figure 2.
Age-related alteration of gene expression regarding cytokine–cytokine receptor interactions. The gene families of chemokines, interferon, TGFβ, IL17, and TNF are included and the molecules with higher or lower gene expression compared with young are highlighted in red and blue text, respectively. The different border color indicates specific T cell subsets.
Age-related enhanced expression of cytokines and their receptors include IL15, IL17RB, TNF family members (TNFSF7 and TNFSF14), and receptors (FAS, TNFRSF18, and TNFSF1B), IFN family receptor IFNGR1, and TGF-β family receptors ACVR2A. Most of these genes are involved in the inflammatory response, which could be the result of an increased inflammatory state during some chronic infections. Chemokine CXCL12 and its receptor CXCR4 are involved in regulating thymocyte development and differentiation from DN (CD44−CD25+, CD44−CD25−) to DP stages (Ara et al., 2003), but their roles in aged T cells have not been determined. In general, the high levels of expression of chemokines/chemokine receptors lead to an enhanced T cell chemotactic response and different patterns of tissue migration/residency of T cells in the old subjects (Cane et al., 2012). It is unclear whether such changes are a result of an increased inflammatory state and/or some chronic infection. Regardless, it is apparent this age-related change in T cell chemokine expression has an important functional consequence.
Enhanced gene expression related to NK cells function
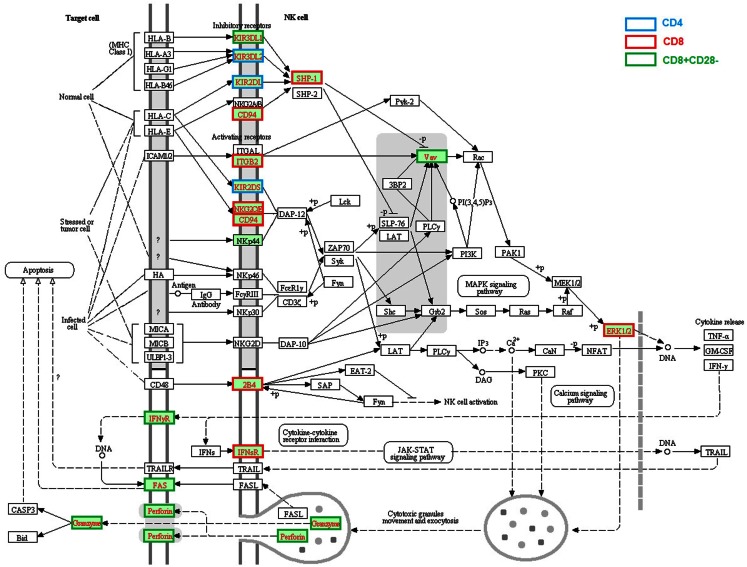
Enhanced expression of a cluster of genes associated with NK cell function is one of the most profound age-related changes in T cells, especially in human CD28− CD8 T cells (Table 1; Figure 3); it includes killer cell lectin-like receptors (KLR, KLRC3, KLRD1, and KLRG1), killer cell immunoglobulin-like receptors (KIR2DL3, KIR2DL4, KIRDL5A, KIR2DS5, KIR3DL1, and KIR3DL2), and CD244. Although these genes were primarily expressed in NK cells, aged memory, or CD28− T cells appear to increase their expression. Since CD28− cells are a unique subset of T cells in older humans, the acquired expression of NK cell receptors and some cytotoxic molecules (GZMB and PRF1) might reflect chronic infection and an increased inflammatory state (Fann et al., 2005). Another possibility is that gaining expression of NK receptors, especially KIRs, might be a compensation for a shrinking TCR repertoire in aged hosts (Abedin et al., 2005; Vallejo, 2006) since KIR recognizes MHC-I/Ag complex with lower affinity compared with that of TCR and was found only in the later stage of proliferative lifespan of oligoclonal T cells (Snyder et al., 2004). If this is true, aged T cells with a high level of NK cell receptors can function in both an MHC-restricted and non-restricted manner.
Figure 3.
Age-related alteration of gene expression regarding NK cell-mediated cytotoxicity. NK cell-mediated cytotoxicity was present and the molecules with higher gene expression compared with young were highlighted in red text. The different border color indicates specific T cell subsets. The altered expressed genes associated with NK mediated cytotoxicity in aged T cells include receptors KIR3DL1, KIR3DL2, KIR2DL, KIR2DS, KLRD1 (encode CD94), ITGB2, NKG2C/E, CD244 (encode 2B4), IFNGR, IFNSR, and FAS, signaling molecules PTPN6 (SHP-1), VAV and MARK1 (ERK1), and effector molecules GZMB and PRF1 (encode Perforin).
Conclusion
In the past decade, we have learned enormously from the global gene expression analysis of aged T cells. Alteration of several gene networks and pathways that are associated with aged T cells have now been identified in humans and mice, including T cell receptor and activation-related molecules, alteration of chemokine/chemokine receptor expression, gain of NK cell receptors and functions, and etc. Whether these identified alterations of gene expressions occur in all cells or in subsets of defined T cell populations remains to be determined. It is sufficient to say these alterations contribute to the overall decline of T cell function.
It is worth mentioning that not all microarray data, especially for some of the early studies, have the highest quality. The shortcomings include: incomplete gene list of the array used, insufficient number of biological repeats, and lack of standardized selection criteria of significantly altered genes. It is necessary in future studies to use the whole genome microarray chips with better designed experiments, such as using defined cell populations, standardized stimulation conditions and time, sufficient numbers of the biological repeats, and proper statistical tools for selection of significantly altered genes. In addition, future studies will need to dissect which specific stage of T cell development and differentiation these changes occurred. The precise relationship of gene expression change and function is essential for a better understanding these age-associated alterations in T cell functions.
The current commercial whole genome microarray chips offered by Agilent, Affymetrix, and Illumina have greatly improved from a few years ago, and become a standard method for global gene expression analysis. However, some shortcomings are also emerging with recent developments. Most of the microarray chips for gene expression analysis do not include miRNA and lncRNA, both of which serve critical biological functions and in which some alterations with aging and cell differentiation were recently reviewed (Gorospe and Abdelmohsen, 2011; Guttman et al., 2011). These microarray chips also do not provide information on different splicing isoforms of transcribed genes; thus, global transcriptional assessment by the traditional microarray method does not offer complete transcriptional information. RNA-Seq, a relatively new method based on next-generation sequencing, can readily address these shortcomings and provide more complete transcriptional analysis.
Identification of the alteration of several gene networks and pathways that are associated with aged T cells paves a way for further functional assessment and potential clinical interventions. Although the general approach such as supplementation of Vitamin E in mice showed some promising results, such as increased expression of genes involved in cell cycle regulation (Ccnb2, Cdc2, and Cdc6) and higher up-regulation of expression in old T cells following stimulation, a more focused approach targeting specific genes and gene networks that are altered in aged T cells may offer a more direct and specific treatment. With the advancement of the tools that can be used in manipulating gene expression in cells, experimental determination and verification of these specific genes/gene networks based therapeutic approach is feasible and will lead to the development of novel drugs that could postpone or even reverse the changes associated with aging in T cells.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Christina Slota for proof reading the manuscript. This research work was supported by the Intramural Research Program of the National Institute on Aging, NIH.
References
- Abedin S., Michel J. J., Lemster B., Vallejo A. N. (2005). Diversity of NKR expression in aging T cells and in T cells of the aged: the new frontier into the exploration of protective immunity in the elderly. Exp. Gerontol. 40, 537–548 10.1016/j.exger.2005.04.012 [DOI] [PubMed] [Google Scholar]
- Alonso-Arias R., Moro-Garcia M. A., Lopez-Vazquez A., Rodrigo L., Baltar J., Garcia F. M., et al. (2011). NKG2D expression in CD4+ T lymphocytes as a marker of senescence in the aged immune system. Age (Dordr.) 33, 591–605 10.1007/s11357-010-9200-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ara T., Itoi M., Kawabata K., Egawa T., Tokoyoda K., Sugiyama T., et al. (2003). A role of CXC chemokine ligand 12/stromal cell-derived factor-1/pre-B cell growth stimulating factor and its receptor CXCR4 in fetal and adult T cell development in vivo. J. Immunol. 170, 4649–4655 [DOI] [PubMed] [Google Scholar]
- Bektas A., Zhang Y., Wood W. H., III, Becker K. G., Madara K., Ferrucci L., et al. (2013). Age-associated alterations in inducible gene transcription in human CD4+ T lymphocytes. Aging (Albany NY) 5, 18–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane S., Ponnappan S., Ponnappan U. (2012). Altered regulation of CXCR4 expression during aging contributes to increased CXCL12-dependent chemotactic migration of CD4(+) T cells. Aging Cell 11, 651–658 10.1111/j.1474-9726.2012.00830.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J. N., Gollapudi S., Sharman E. H., Jia Z., Gupta S. (2010). Age-related alterations of gene expression patterns in human CD8+ T cells. Aging Cell 9, 19–31 10.1111/j.1474-9726.2009.00534.x [DOI] [PubMed] [Google Scholar]
- Chen J., Mo R., Lescure P. A., Misek D. E., Hanash S., Rochford R., et al. (2003). Aging is associated with increased T-cell chemokine expression in C57BL/6 mice. J. Gerontol. A Biol. Sci. Med. Sci. 58, 975–983 10.1093/gerona/58.11.B975 [DOI] [PubMed] [Google Scholar]
- Czesnikiewicz-Guzik M., Lee W. W., Cui D., Hiruma Y., Lamar D. L., Yang Z. Z., et al. (2008). T cell subset-specific susceptibility to aging. Clin. Immunol. 127, 107–118 10.1016/j.clim.2008.03.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fann M., Chiu W. K., Wood W. H., III, Levine B. L., Becker K. G., Weng N. P. (2005). Gene expression characteristics of CD28null memory phenotype CD8+ T cells and its implication in T-cell aging. Immunol. Rev. 205, 190–206 10.1111/j.0105-2896.2005.00262.x [DOI] [PubMed] [Google Scholar]
- Fulop T., Jr., Gagne D., Goulet A. C., Desgeorges S., Lacombe G., Arcand M., et al. (1999). Age-related impairment of p56lck and ZAP-70 activities in human T lymphocytes activated through the TcR/CD3 complex. Exp. Gerontol. 34, 197–216 10.1016/S0531-5565(98)00061-8 [DOI] [PubMed] [Google Scholar]
- Garcia G. G., Miller R. A. (1997). Differential tyrosine phosphorylation of zeta chain dimers in mouse CD4 T lymphocytes: effect of age. Cell. Immunol. 175, 51–57 10.1006/cimm.1996.1040 [DOI] [PubMed] [Google Scholar]
- Gorospe M., Abdelmohsen K. (2011). MicroRegulators come of age in senescence. Trends Genet. 27, 233–241 10.1016/j.tig.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith A. V., Fallahi M., Venables T., Petrie H. T. (2012). Persistent degenerative changes in thymic organ function revealed by an inducible model of organ regrowth. Aging Cell 11, 169–177 10.1111/j.1474-9726.2011.00773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Donaghey J., Carey B. W., Garber M., Grenier J. K., Munson G., et al. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 10.1038/nature10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. N., Adolfsson O., Lee C. K., Prolla T. A., Ordovas J., Meydani S. N. (2006). Age and vitamin E-induced changes in gene expression profiles of T cells. J. Immunol. 177, 6052–6061 [DOI] [PubMed] [Google Scholar]
- Haynes L., Swain S. L. (2006). Why aging T cells fail: implications for vaccination. Immunity 24, 663–666 10.1016/j.immuni.2006.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson S. M., Akbar A. N. (2009). KLRG1 – more than a marker for T cell senescence. Age (Dordr.) 31, 285–291 10.1007/s11357-009-9100-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Lopez C., Varas A., Sacedon R., Martinez V. G., Hidalgo L., Valencia J., et al. (2010). The CXCL12/CXCR4 pair in aged human thymus. Neuroimmunomodulation 17, 217–220 10.1159/000258728 [DOI] [PubMed] [Google Scholar]
- Hyatt G., Melamed R., Park R., Seguritan R., Laplace C., Poirot L., et al. (2006). Gene expression microarrays: glimpses of the immunological genome. Nat. Immunol. 7, 686–691 10.1038/ni0706-686 [DOI] [PubMed] [Google Scholar]
- Kirk C. J., Freilich A. M., Miller R. A. (1999). Age-related decline in activation of JNK by TCR- and CD28-mediated signals in murine T-lymphocytes. Cell. Immunol. 197, 75–82 10.1006/cimm.1999.1568 [DOI] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Mooi W. J., Peeper D. S. (2010). The essence of senescence. Genes Dev. 24, 2463–2479 10.1101/gad.1971610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazuardi L., Herndler-Brandstetter D., Brunner S., Laschober G. T., Lepperdinger G., Grubeck-Loebenstein B. (2009). Microarray analysis reveals similarity between CD8+CD28- T cells from young and elderly persons, but not of CD8+CD28+ T cells. Biogerontology 10, 191–202 10.1007/s10522-008-9167-1 [DOI] [PubMed] [Google Scholar]
- Li G., Yu M., Lee W. W., Tsang M., Krishnan E., Weyand C. M., et al. (2012). Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat. Med. 10, 1518–1524 10.1038/nm.2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton P. J., Li S. P., Zhang Y., Bautista B., Huynh Q., Trinh T. (2005). Intrinsic versus environmental influences on T-cell responses in aging. Immunol. Rev. 205, 207–219 10.1111/j.0105-2896.2005.00266.x [DOI] [PubMed] [Google Scholar]
- Lustig A., Carter A., Bertak D., Enika D., Vandanmagsar B., Wood W., et al. (2009). Transcriptome analysis of murine thymocytes reveals age-associated changes in thymic gene expression. Int. J. Med. Sci. 6, 51–64 10.7150/ijms.6.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig A., Weeraratna A. T., Wood W. W., III, Teichberg D., Bertak D., Carter A., et al. (2007). Transcriptome analysis of age-, gender- and diet-associated changes in murine thymus. Cell. Immunol. 245, 42–61 10.1016/j.cellimm.2007.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza N., Pollock K., Hoelzinger D. B., Dominguez A. L., Lustgarten J. (2011). Comparative kinetic analyses of gene profiles of naive CD4+ and CD8+ T cells from young and old animals reveal novel age-related alterations. Aging Cell 10, 853–867 10.1111/j.1474-9726.2011.00730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo R., Chen J., Han Y., Bueno-Cannizares C., Misek D. E., Lescure P. A., et al. (2003). T cell chemokine receptor expression in aging. J. Immunol. 170, 895–904 [DOI] [PubMed] [Google Scholar]
- Pawelec G., Akbar A., Caruso C., Solana R., Grubeck-Loebenstein B., Wikby A. (2005). Human immunosenescence: is it infectious? Immunol. Rev. 205, 257–268 10.1111/j.0105-2896.2005.00271.x [DOI] [PubMed] [Google Scholar]
- Ponnappan S., Ponnappan U. (2011). Aging and immune function: molecular mechanisms to interventions. Antioxid. Redox Signal. 14, 1551–1585 10.1089/ars.2010.3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla T. A. (2005). Multiple roads to the aging phenotype: insights from the molecular dissection of progerias through DNA microarray analysis. Mech. Ageing Dev. 126, 461–465 10.1016/j.mad.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Remondini D., Salvioli S., Francesconi M., Pierini M., Mazzatti D. J., Powell J. R., et al. (2010). Complex patterns of gene expression in human T cells during in vivo aging. Mol. Biosyst. 6, 1983–1992 10.1039/c004635c [DOI] [PubMed] [Google Scholar]
- Schumacher B., Van D. P., I, Moorhouse M. J., Kosteas T., Robinson A. R., Suh Y., et al. (2008). Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 4:e1000161. 10.1371/journal.pgen.1000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. R., Weyand C. M., Goronzy J. J. (2004). The double life of NK receptors: stimulation or co-stimulation? Trends Immunol. 25, 25–32 10.1016/j.it.2003.10.011 [DOI] [PubMed] [Google Scholar]
- Swindell W. R. (2009). Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics 10:585. 10.1186/1471-2164-10-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsuyama M., Varga Z., Fukami K., Homma Y., Takenawa T., Hirokawa K. (1993). Influence of age on the signal transduction of T cells in mice. Int. Immunol. 5, 1177–1182 10.1093/intimm/5.9.1177 [DOI] [PubMed] [Google Scholar]
- Utsuyama M., Wakikawa A., Tamura T., Nariuchi H., Hirokawa K. (1997). Impairment of signal transduction in T cells from old mice. Mech. Ageing Dev. 93, 131–144 10.1016/S0047-6374(96)01837-4 [DOI] [PubMed] [Google Scholar]
- Vallejo A. N. (2006). Age-dependent alterations of the T cell repertoire and functional diversity of T cells of the aged. Immunol. Res. 36, 221–228 10.1385/IR:36:1:221 [DOI] [PubMed] [Google Scholar]
- Weng N. P. (2006). Aging of the immune system: how much can the adaptive immune system adapt? Immunity 24, 495–499 10.1016/j.immuni.2006.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng N. P., Akbar A. N., Goronzy J. (2009). CD28(-) T cells: their role in the age-associated decline of immune function. Trends Immunol. 30, 306–312 10.1016/j.it.2009.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisler R. L., Newhouse Y. G., Bagenstose S. E. (1996). Age-related reductions in the activation of mitogen-activated protein kinases p44mapk/ERK1 and p42mapk/ERK2 in human T cells stimulated via ligation of the T cell receptor complex. Cell. Immunol. 168, 201–210 10.1006/cimm.1996.0067 [DOI] [PubMed] [Google Scholar]
- Yung R. L., Mo R. (2003). Aging is associated with increased human T cell CC chemokine receptor gene expression. J. Interferon Cytokine Res. 23, 575–582 10.1089/107999003322485071 [DOI] [PubMed] [Google Scholar]