Abstract
Huntington's disease is an autosomal dominant neurodegenerative disorder caused by expansion of a polyglutamine tract in the huntingtin protein that results in intracellular aggregate formation and neurodegeneration. Pathways leading from polyglutamine tract expansion to disease pathogenesis remain obscure. To elucidate how polyglutamine expansion causes neuronal dysfunction, we generated Drosophila transgenic strains expressing human huntingtin cDNAs encoding pathogenic (Htt-Q128) or nonpathogenic proteins (Htt-Q0). Whereas expression of Htt-Q0 has no discernible effect on behavior, lifespan, or neuronal morphology, pan-neuronal expression of Htt-Q128 leads to progressive loss of motor coordination, decreased lifespan, and time-dependent formation of huntingtin aggregates specifically in the cytoplasm and neurites. Huntingtin aggregates sequester other expanded polyglutamine proteins in the cytoplasm and lead to disruption of axonal transport and accumulation of aggregates at synapses. In contrast, Drosophila expressing an expanded polyglutamine tract alone, or an expanded polyglutamine tract in the context of the spinocerebellar ataxia type 3 protein, display only nuclear aggregates and do not disrupt axonal trafficking. Our findings indicate that nonnuclear events induced by cytoplasmic huntingtin aggregation play a central role in the progressive neurodegeneration observed in Huntington's disease.
Huntington's disease (HD) is characterized by neurodegeneration and formation of neuronal intracellular inclusions secondary to abnormal expansion of a CAG tract (encoding polyglutamine, polyQ) in exon 1 of the huntingtin gene (Htt) (1). Expansion of the CAG tract past the pathological threshold of ≈35-40 repeats ensures disease manifestation, presumably due to promotion of an abnormal protein conformation (2). Although intracellular aggregates are a prominent hallmark of polyQ disease, their role in disease pathogenesis is debated (3). In the case of HD, Htt-immunopositive aggregates have been observed in both the nucleus and the cytoplasm of affected neurons, although their primary subcellular localization remains controversial (4). One difficulty in characterizing the subcellular localization of Htt aggregates has been the widespread use of exon 1 models, which encode only the first 81 aa of the Htt protein with the polyQ tract. These models widely demonstrate nuclear localization of Htt-immunopositive aggregates, although it is unclear whether exon 1 faithfully mimics the localization of the full-length endogenous protein (3,135 aa).
Although some evidence links intranuclear aggregates and disease manifestation (5), recent findings suggest that the presence of aggregates in the nucleus may not be central to neuropathology (6, 7). HD disease symptoms have been reported to arise from neuronal cell loss before nuclear aggregates are detectable but after neuritic aggregates have appeared (6, 8). In transgenic mice, the presence of aggregates in neurites correlates with development of neuropathological symptoms (9), and expression of the abnormal protein alters synaptic function (10, 11). To explore the mechanisms by which polyQ aggregates may cause neuronal dysfunction, we generated transgenic Drosophila that express the first 548 aa of the human Htt gene with either a pathogenic polyQ tract of 128 repeats (Htt-Q128) or a nonpathogenic tract of 0 repeats (Htt-Q0). This N-terminal motif contains regions of strong homology between Htt isoforms from Drosophila to humans and is more likely to faithfully mimic endogenous Htt localization than constructs containing only the first 81 aa, which display no sequence conservation between Drosophila and human Htt homologs. Our findings indicate that cytoplasmic Htt aggregates sequester other cytoplasmic polyQ-containing proteins and block axonal transport.
Materials and Methods
Drosophila Genetics and Generation of Htt Constructs. Drosophila melanogaster were maintained on standard medium at 22°C. cDNAs for Htt-Q0 and Htt-Q128 were generously provided by M. R. Hayden (Department of Medical Genetics, University of British Columbia, Vancouver) and subsequently subcloned into pHS or pUAST vectors. Transgenic Drosophila encoding Q127 were obtained from Parsi Kazemi-Esfarjani (12), SCA3-Q78 from Nancy Bonini (13), and Dishevelled Q108 from Leslie Thompson (14).
Western Blot Analysis. To obtain samples for Western analysis of pHS-Htt lines, 20 Drosophila of each indicated genotype were collected and heat shocked at 37°C for 40 min, allowed to recover at room temperature for 1 h, heat shocked again for 40 min, and incubated at 28°C overnight before processing. Drosophila were frozen in liquid nitrogen, heads were isolated and homogenized in sample buffer, and proteins were separated on 10% SDS/PAGE gels. The gels were immunoblotted with mouse anti-Htt MAb2166 (Chemicon) at 1:1,000, and immunoreactive bands were visualized by using ECL (Pierce).
Electrophysiological Analysis. Electroretinograms and extracellular recordings from the dorsal longitudinal flight muscles were performed as described (15). Developmental temperature-shift experiments were performed by exposing Drosophila larvae to two 1-h 37°C heat shocks each day.
Morphological Analysis. Immunostaining was performed on wandering third instar larvae reared at 25°C as described (15). Primary antibodies used were mouse anti-Htt MAb2166 (Chemicon) at 1:500, rabbit anti-GFP sc8334 (Santa Cruz Biotechnology) at 1:500, anti-hemagglutinin to label Q127 and SCA3-Q78 at 1:1,000, rabbit anti-Dishevelled at 1:500, and rabbit anti-synaptotagmin at 1:500. FITC-horseradish peroxidase was used at 1:10,000 to label neuronal membranes. Visualization and quantification were performed on a Pascal confocal microscope (Zeiss) by using Cy2-conjugated and rhodamine red-conjugated secondary antibodies (Molecular Probes, Chemicon, and The Jackson Laboratory). Htt-Q128 aggregate diameter was determined by digitally capturing images at ×40 magnification and computing diameters (in μm) by using lsm 5 pascal analysis software. All error measurements are SD. Corneal pseudopupil analysis of adult ommatidia was performed as described (16).
Larval Locomotion and Adult Behavioral Analysis. Wandering third-instar larvae grown at room temperature were collected, washed gently with distilled water, and placed individually on the center of a flat layer of 0.7% agar on an evenly illuminated light box. Larval locomotion was recorded with a digital video camera (Canon XL1S) attached to a ×16 zoom lens. Recording stopped at 2.5 min or when the larva reached the edge of the agar layer, with documentation of the recording time. Distance traveled by each larva was quantified by measuring centimeters traversed by a crawling larva on the agar surface during the time recorded. Speed was calculated by dividing the distance traveled by the seconds of recording time. Drosophila viability assays were performed on elav-GAL4, Htt-Q128/white, Htt-Q128/elav-GAL4, and Htt-Q0/elav-GAL4 flies by daily quantification of lethality for 200 females of each genotype. Flies were aged at 25°C, with 20 flies per food vial, and transferred every 2 days.
Results and Discussion
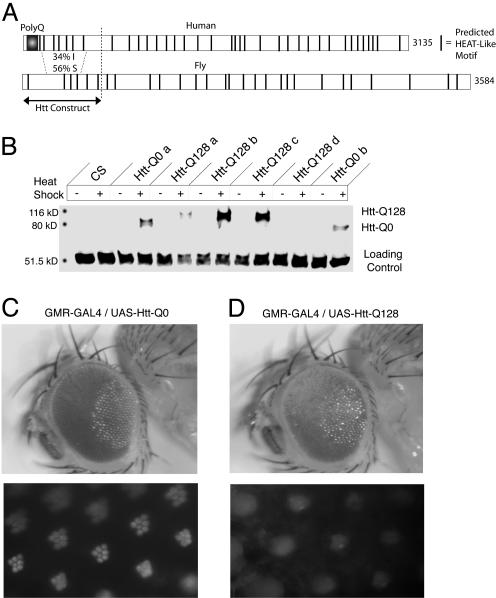
To characterize neuronal defects that result from an expanded polyQ tract within the Htt gene, we generated transgenic Drosophila expressing N-terminal fragments of human Htt containing 0 (Htt-Q0) or 128 (Htt-Q128) glutamines. The Htt constructs were engineered to include the first 548 aa of the human Htt protein, which include and extend well beyond the 81-aa product encoded by the first exon of the gene. The 548-aa fragment is truncated close to the site of cleavage by caspase-3, thought to be a crucial step in the generation of aggregate-forming Htt fragments (17, 18). This region also encompasses the highest stretch of homology between the Drosophila and human Htt proteins (Fig. 1A). We expressed Htt-Q0 and Htt-Q128 fragments by using UAS/GAL4 or a heat-shock promoter. To confirm transgene expression, Htt protein was compared between pHS-Htt Drosophila maintained at room temperature and after exposure to a heat-shock paradigm (Fig. 1B). Western blotting with anti-human Htt antibodies detected no Htt protein in control Canton S or in pHS-Htt lines maintained at room temperature. In contrast, Htt-Q0 and Htt-Q128 lines showed abundant Htt expression after heat shock. Transgene expression was also established in pUAST-Htt strains that were crossed to a neuronal GAL4 driver (data not shown).
Fig. 1.
Generation of a Drosophila transgenic model of HD. (A) Domain structure of the human and Drosophila Htt homologs with predicted Huntington elongation factor 3 protein phosphatase 2A Tor1 (HEAT)-like motifs indicated. The 548-aa N terminus of human Htt used for transgenic construction is indicated. (B) Heat-shock induction of Htt in Q0 and Q128 pHS strains. Western blotting was performed with an antibody generated to the N terminus of human Htt that recognizes both the Q0 and Q128 variants. (C and D) External morphology and pseudopupil analysis of transgenic Drosophila expressing either Htt-Q0 (C) or Htt-Q128 (D) driven by the eye-specific GMR-GAL4 driver. Flies were aged for 2-4 days at 25°C before analysis. Htt-Q128 causes a rough-eye phenotype with loss of pigmentation, abnormal bristle pattern, and photoreceptor degeneration.
To determine the functional consequences of Htt-Q128 expression on neuronal activity and morphology, we examined effects in the visual system. Previous Drosophila models of polyQ diseases have demonstrated that eye-specific expression of expanded polyQ proteins leads to a rough-eye phenotype and photoreceptor degeneration (12, 14, 19, 20). To determine whether the 548-aa Htt transgene caused similar effects, Htt-Q0 and Htt-Q128 were expressed by using the eye-specific GMR-GAL4 driver, and the resulting eye phenotypes were observed by external morphology and the corneal pseudopupil method. Whereas expression of Htt-Q0 did not perturb external-eye appearance or ommatidial morphology (Fig. 1C), expression of Htt-Q128 caused a rough-eye phenotype with corresponding photoreceptor degeneration (Fig. 1D). Thus, polyQ expansion in the context of a larger Htt fragment results in neurodegeneration, as observed in other polyQ disease models.
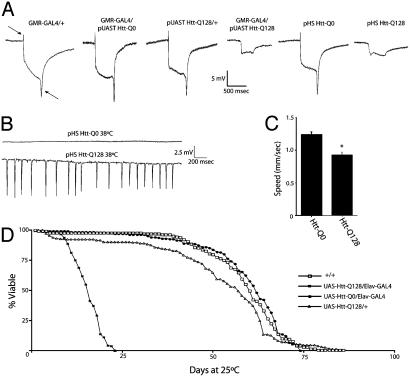
To characterize the physiological effects of mutant Htt expression, we recorded electroretinograms from transgenic animals (Fig. 2A). A normal electrical response to light was seen in Drosophila expressing the GMR-GAL4 driver alone, Htt-Q0 with GMR-GAL4, or Htt-Q128 without the GMR-GAL4 driver. In contrast, Drosophila expressing Htt-Q128 with the GMR-GAL4 driver showed reduced photoreceptor depolarization and complete abolishment of synaptic transmission in response to light. Similar abnormal electroretinograms were observed in heat-shocked pHS-Htt-Q128 lines after a developmental heat shock paradigm but not with control pHS-Htt-Q0 strains (Fig. 2 A). We also assayed synaptic activity in the giant fiber flight circuit, a pathway important in escape responses and flight initiation. Wild-type Drosophila display little to no spontaneous activity when the temperature is raised to 38°C. In contrast, robust spontaneous seizure activity was recorded in Htt-Q128 flies at 38°C after a developmental heat-shock paradigm (Fig. 2B). No seizure activity was recorded in Htt-Q0 flies at 38°C (Fig. 2B). Together, these results indicate that Htt-Q128 expression results in neurodegeneration, accompanied by widespread defects in membrane excitability and brain activity.
Fig. 2.
Physiological and behavioral analysis of Htt-Q128-expressing transgenic Drosophila. (A) Both UAS-Htt-Q128/GMR-GAL4 and pHS-Htt-Q128 exhibit reduced photoreceptor depolarization and abolished synaptic transmission, as indicated by loss of the on/off transients (arrows) in electroretinogram recordings. (B) Extracellular recordings from the DLM flight muscles of Htt-Q0 and Htt-Q128 Drosophila are shown. A developmental heat-shock paradigm that induces Htt-Q128 expression results in abnormal seizure activity in the flight circuit at 38°C. (A and B) Drosophila were aged for 2-4 days at 25°C. (C) Quantitative analysis of wandering third-instar larval crawling behavior indicates that pan-neuronal expression of Htt-Q128 disrupts motor pattern generation, resulting in a significant decrease in locomotor speed. Thirty larvae were analyzed for each genotype. (D) Viability analysis of Drosophila maintained at 25°C indicates that pan-neuronal expression of Htt-Q128 with a second chromosome elav-GAL4 driver results in decreased lifespan compared with control strains. Expression of Htt-Q128 with the stronger X-chromosome elav-GAL4 driver C155 leads to 100% pharate adult lethality.
To establish whether neuronal Htt-Q128 transgene expression causes defects at earlier stages of Drosophila development, we performed quantitative locomotion assays to examine the function of the motor central pattern generator in third-instar larvae. When Htt transgenes were expressed with the pan-neuronal elav-GAL4 driver C155, Htt-Q128 larvae showed a significant reduction in locomotor speed of >25%, from 1.23 mm/s in control Htt-Q0 animals to 0.92 mm/s in Htt-Q128 larvae (Student's t test, P < 0.001; Fig. 2C). Adult transgenic flies also displayed abnormal motor behavior caused by pan-neuronal expression of the Htt-Q128 protein. Whereas expression of Htt-Q128 with the C155 neuronal GAL4 driver causes pharate adult lethality with no viable adult escapers, Htt-Q128 driven by a weaker second chromosome elav-GAL4 driver results in fully viable adults. Several days after eclosion, flies expressing Htt-Q128, but not Htt-Q0, began to exhibit uncoordinated movement and abnormal grooming behaviors. The behavioral defects worsened with age, resulting in premature death. To quantify the reduction in viability, lifespan curves were generated for control adults, Htt-Q128 adults without elav-GAL4, or adults expressing Htt-Q0 or Htt-Q128 with elav-GAL4 (Fig. 2D). Compared to controls, Htt-Q128/elav-GAL4 animals showed a dramatic reduction in lifespan, with a decrease in the T50 (age at which 50% of the culture has died) by 70%, indicating a highly significant effect of Htt-Q128 expression on viability in Drosophila (Fig. 2D).
A hallmark of HD is the formation of Htt-immunopositive intracellular aggregates in neurons. To determine whether intracellular aggregates are formed in transgenic Htt Drosophila, both Htt-Q128 and Htt-Q0 strains were crossed to flies containing C155 elav-GAL4 to direct expression of Htt within the nervous system. Htt-immunopositive staining was visualized in both central and peripheral neurons of dissected third-instar larvae. Whereas Htt staining remained diffuse throughout the cytoplasm of neurons in Drosophila expressing Htt-Q0 (Fig. 3A), distinct aggregates were observed in the cytoplasm and processes of neurons in lines expressing Htt-Q128 (Fig. 3B). Contrary to what has been observed in exon 1 HD models, we found no evidence of nuclear aggregate localization. To verify that Htt aggregation is based on the length of the polyQ tract and not on protein concentration, Htt levels were quantitated for several Htt-Q0 and Htt-Q128 transgenic lines crossed to elav-GAL4. Levels of Htt protein were generally higher in Htt-Q128 lines than in Htt-Q0 lines, likely because of sequestration of the mutant protein into stable aggregates. However, low-expressing Htt-Q128 lines that produced transgenic protein at a level comparable with that in Htt-Q0 strains still exhibited aggregates, whereas Htt-Q0 lines did not (data not shown), indicating that polyQ tract expansion and not protein concentration alone is necessary for formation of aggregates. Aggregate formation was also time-dependent. Although Htt levels were visibly high in the central and peripheral nervous system of Htt-Q128/elav-GAL4 embryos, the protein remained largely diffuse in the cytoplasm with rare occurrence of aggregates (data not shown). By the third instar larval stage, essentially all Htt was observed in aggregates, with relatively little nonaggregate staining (Fig. 3B). We conclude that Htt-Q128 forms cytoplasmic neuronal aggregates in a time-dependent manner.
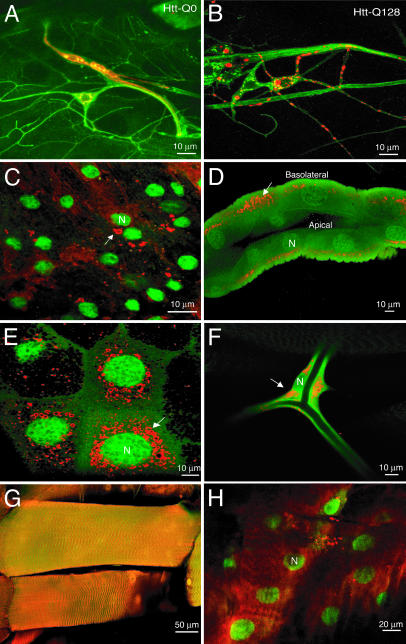
Fig. 3.
Cytoplasmic aggregation of Htt-Q128 in neuronal and nonneuronal tissues. (A) Immunocytochemical detection of Htt (red) and neuronal membranes by anti-horseradish peroxidase (green) in multidendritic neurons of Htt-Q0-expressing third-instar larva. Htt-Q0 is found diffusely throughout the cytoplasm. (B) Immunocytochemical detection of Htt (red) and horseradish peroxidase (green) in multidendritic neurons of Htt-Q128-expressing third-instar larva. Unlike Htt-Q0, Htt-Q128 is found in cytoplasmic aggregates throughout the cell body and neurites. (C-E) Expression of UAS-Htt-Q128 (red) and UAS-GFP-nls (green) by tubP-GAL4 in the CNS (C), gut (D), salivary gland (E), trachea (F), muscle (G), and epidermis (H) of third-instar larvae. In all cases, cytoplasmic aggregates are observed. However, muscle and epidermis form far fewer aggregates than other tissues. In polarized cells like the gut, basolateral transport of Htt aggregates is observed, with an absence of aggregates in the apical domain. The nucleus is indicated by N, and Htt aggregates are indicated by arrows.
Although the causative proteins for many of the polyQ repeat diseases are expressed widely or ubiquitously, aggregate formation and cell death occur in subsets of neurons that differ between the diseases. To examine the effect of cellular context on aggregate formation, the Htt-Q128 protein was expressed in different cell types by using a tubulin GAL4 driver. We generated transgenic lines containing both the UAS-Htt-Q128 construct and UAS-GFP fused to a nuclear localization signal, allowing for covisualization of Htt-immunopositive aggregates and GFP-stained cell nuclei in expressing cells. Immunocytochemical analysis demonstrated the formation of cytoplasmic aggregates in both neuronal and nonneuronal tissues, including CNS neurons (Fig. 3C), gut (Fig. 3D), salivary glands (Fig. 3E), and trachea (Fig. 3F). Interestingly, Htt aggregates were differentially distributed in polarized cells such as the gut, with transport of Htt aggregates to the basolateral domain and exclusion from the apical surface (Fig. 3D). Similar aggregate transport was found in neurons (see below), indicating that Htt aggregates undergo a cytoskeletal association that allows for directed transport. The Htt-Q128 protein was found in a more diffuse, nonaggregated state in certain cell types, including muscle and epidermis (Fig. 3 G and H), suggesting that some tissues may be more resistant to Htt aggregation. In cell types in which aggregate formation occurred, only cytoplasmic aggregates (as opposed to nuclear aggregates) were observed, suggesting differences between the larger Htt fragments used in our study compared with exon 1 HD models.
Although the polyQ repeat diseases share a similar CAG repeat expansion in the causative gene, the pattern of neurodegeneration and behavioral dysfunction is distinct for each, indicating that protein context for expanded polyQ tracts is critical to disease manifestation. To examine the importance of protein context in the subcellular localization of polyQ-containing proteins, immunocytochemical analysis was performed on larvae expressing an expanded polyQ tract alone (Q127) (12), the mutant polyQ protein responsible for Machado-Joseph disease (SCA3-Q78) (13), or an expanded polyQ tract (Q108) previously engineered into the nonpathogenic dishevelled gene (14). In contrast to the cytoplasmic localization of Htt aggregates, both Q127 and SCA3-Q78 aggregates localized exclusively to the nucleus (Fig. 4 B and C). Very few Dishevelled-immunopositive aggregates were observed (Fig. 4D), and the protein was present diffusely in the cytoplasm. These results demonstrate that the protein context in which the polyQ tract is found exquisitely controls both aggregate localization and aggregate formation.
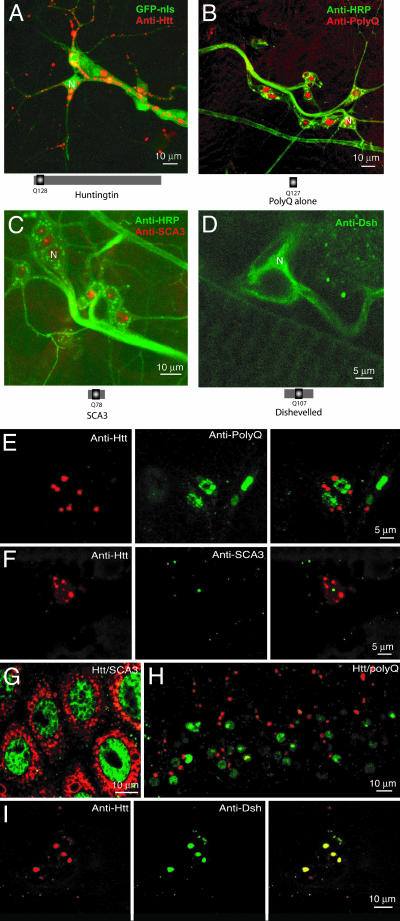
Fig. 4.
Protein context is important for polyQ-mediated aggregation and aggregate localization. (A-D) Immunolocalization of aggregates in third-instar larvae expressing Htt-Q128 (A), Q127 (B), SCA3-Q78 (C), and Dsh-Q108 (D) in multidendritic neurons with the C155 elav-GAL4 driver. Htt-Q128 aggregates are exclusive to the cytoplasm, whereas Q127 and SCA3-Q78 aggregates are found only in the nucleus. Dsh-Q108 is mostly diffuse in the cytoplasm, with few aggregates visible. The nucleus is represented by N. (E and F) Double transgenic third-instar larvae expressing Htt-Q128 and Q127 (E) or Htt-Q128 and SCA3-Q78 (F) with the C155 elav-GAL4 driver were dissected and immunostained for Htt (red) and SCA3/Q127 (green). Multidendritic sensory neurons were identified as in A-D and assayed for aggregate colocalization. No colocalization of aggregates is observed, indicating distinct nuclear and cytoplasmic aggregation pathways. Similar results are observed in the CNS (H) and in nonneuronal tissues such as the salivary glands (G). (I) In contrast, in double transgenic larvae expressing Htt-Q128 and Dsh-Q108 with C155 elav-GAL4, Htt-Q128 (red) is able to completely sequester Dsh-Q108 (green) into aggregates, as indicated by colocalization (yellow) in Right.
To test whether Htt-Q128 can interact and coaggregate with other polyQ repeat proteins, we generated double transgenic Htt-Q128; Q127 and Htt-Q128; SCA3-Q78 strains. When the Htt-Q128 and Q127 proteins were coexpressed, both central and peripheral neurons showed localization of Htt-Q128 aggregates to the cytoplasm, whereas Q127 aggregates were restricted to the nucleus (Fig. 4 E and H). Likewise, in strains expressing Htt-Q128 and SCA3-Q78, aggregates were segregated independently in the cytoplasm and nucleus (respectively) of both neuronal (Fig. 4F) and nonneuronal (Fig. 4G) cells. These results suggest that the trafficking and aggregation of nuclear and cytoplasmic aggregates are independently regulated.
To determine whether Htt-Q128 might interact with cytoplasmic proteins containing an expanded polyQ tract, double transgenic strains were made containing Htt-Q128 and Dishevelled-Q108 (Dsh-Q108). As demonstrated (Fig. 4D), Dsh-Q108 formed few aggregates when expressed alone; however, when coexpressed with Htt-Q128, the subcellular distribution of Dsh-Q108 shifted from a diffuse cytoplasmic pattern to a complete sequestration into aggregates that colocalized with Htt-Q128 (Fig. 4I). These findings indicate that Htt aggregates are able to trap and sequester Dsh-Q108 into cytoplasmic aggregates. Similar interactions between Htt aggregates and endogenous cytoplasmic polyQ-containing proteins might be predicted to play an important role in disease pathology.
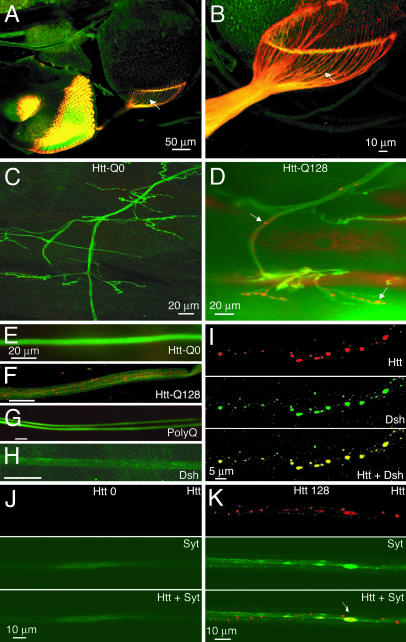
As described (Fig. 3D), Htt-Q128 forms cytoplasmic aggregates that are associated with cytoskeletal transport systems. When Htt-Q128 was expressed with the eye-specific driver GMR-GAL4, Htt aggregates were abundantly transported along axons entering the CNS of the developing visual system and accumulated in pathfinding photoreceptor growth cones (Fig. 5 A and B). Similarly, when Htt-Q128 was expressed with the C155 driver, aggregates were transported in larval motor axons and accumulated at presynaptic neuromuscular junction terminals (Fig. 5 D and F). No aggregates were observed in axons from animals expressing Htt-Q0 (Fig. 5 C and E). Axonal transport of aggregates was not observed in transgenic animals that produce exclusively nuclear aggregates (Q127 and SCA3-Q78) (Fig. 5G), suggesting that axonal and synaptic defects that may occur downstream of cytoplasmic aggregate formation are likely specific to HD. Additionally, no Dsh-Q108 aggregates were observed in axon bundles (Fig. 5H). However, when Dsh-Q108 was coexpressed with Htt-Q128, Dsh-Q108 protein trapped by Htt-Q128 aggregates was also transported along axons (Fig. 5I). Similar trapping of endogenous cytoplasmic polyQ proteins may sequester them away from their natural cellular locations and contribute to neuronal dysfunction.
Fig. 5.
Htt-Q128 aggregates block axonal transport. (A and B) Expression of Htt-Q128 by GMR-GAL4 results in axonal transport of Htt aggregates (arrows) in the developing visual system. (C and D) Expression of Htt-Q128 (D), but not Htt-Q0 (C), by C155 elav-GAL4 results in axonal transport and synaptic accumulation of Htt aggregates at neuromuscular junctions in third-instar larvae. Axonal transport of aggregates is abundant in Htt-Q128-expressing third-instar larvae (F) but absent in animals expressing Htt-Q0 (E), Q127 (G), or Dsh-Q108 (H). Nerves are stained green by anti-horseradish peroxidase, and polyQ proteins are visualized in red. (I) Consistent with the trapping of Dsh-Q108 by Htt-Q128 aggregates in the cell bodies of multidendritic sensory neurons, Htt-Q128 also traps and transports Dsh-Q108 in peripheral nerves. (J and K) Expression of Htt-Q128 (K), but not Htt-Q0 (J), results in an accumulation of the synaptic protein synaptotagmin I in sites of axonal swelling, colocalizing with large accumulations of Htt-Q128 aggregates. Synaptotagmin I is not trapped in Htt-Q128 aggregates, as indicated by the lack of protein colocalization in smaller Htt-Q128 aggregates, but rather concentrates specifically at sites where larger aggregate accumulations result in swollen axons, indicating blockage of axonal transport.
In observing axonal aggregates in Htt-Q128-expressing animals, it was noted that the diameter of Htt aggregates often exceeded that of normal larval axons. This suggested that large Htt aggregates might physically block axonal transport, as would be manifested by axonal swellings at the sites of blockage. We tested this hypothesis in Htt-Q128-expressing animals by observing the localization of synaptotagmin I, a synaptic vesicle protein localized to synapses in Drosophila. Normal transport of the synaptotagmin protein along axons is below the threshold for immunocytochemical detection. This pattern of trafficking, as observed in Htt-Q0-expressing animals (Fig. 5J), is abruptly altered in Htt-Q128-expressing Drosophila. Instead of the normal diffuse localization along axonal tracts, synaptotagmin became concentrated at specific points along axons that corresponded to large areas of Htt-immunopositive aggregate accumulation, suggesting sites of axonal blockage (Fig. 5K). These synaptotagmin-rich areas of Htt aggregate accumulation were quantified in 100-μm segments along peripheral nerves and were observed at a density of 6.1 ± 2.6 sites per 100 μm. In contrast, synaptotagmin-immunopositive accumulations alone without Htt aggregate colocalization were only observed at a density of 1.3 ± 0.9 per 100 μm. The average diameter of Htt-Q128 aggregate accumulations at putative axonal blockage sites was 2.4 ± 0.6 μm. These findings suggest that axonal segments can be obstructed by Htt aggregates. Over time, the cumulative blockage of axons and synaptic terminals in postmitotic neurons is likely to contribute to the progressive physiological defects and neuronal dysfunction we have documented in Htt-Q128-expressing Drosophila, as well as to late-onset neurodegeneration in HD patients.
Many human neurodegenerative diseases have been successfully modeled in Drosophila with replication of key neuropathological features, including late onset and progressive neurodegeneration. Existing Drosophila models of HD (for review, see ref. 21) target expression of the first exon of the mutant Htt protein to the fly retina, either by means of the use of an eye-specific promoter (19) or the GAL4/UAS system (22). In both HD models, expression of Htt with a pathogenic number of glutamine repeats results in nuclear accumulation of aggregates and progressive neurodegeneration of photoreceptor cells, suggesting a role for nuclear aggregates in disease pathology. Indeed, nuclear aggregate-mediated impairment of transcription has become a favored hypothesis to explain polyQ-mediated neurodegeneration (23). Our findings using a larger Htt transgene suggest that nonnuclear pathology associated with cytoplasmic and neuritic aggregates is likely to play an essential role in disease progression as well. Given that Drosophila polyQ disease models with either nuclear-restricted (expanded polyQ alone, mutant SCA-3, or Htt exon 1) or cytoplasm-restricted (Htt-Q128) aggregates both exhibit neurodegeneration, it is likely that multiple pathways for polyQ-mediated dysfunction exist. Indeed, we have been unable to rescue adult lethality in Htt-Q128-expressing Drosophila with any of the previously published genetic suppressors of Drosophila transgenic polyQ models (12, 20, 24, 25). These negative results likely reflect distinct modes of toxicity between nuclear and cytoplasmic aggregates and suggest that preventing polyQ-mediated Htt toxicity may require more research on the role of nonnuclear aggregates. We have demonstrated a potential role for cytoplasmic and neuritic aggregates in the sequestration of cytoplasmic polyQ proteins and in the blockage of axonal transport. Consistent with our hypothesis that Htt-Q128 expression causes neurodegeneration secondary to impairment of axonal transport, recent studies have found that neurodegeneration is a primary consequence of axonal transport defects in non-polyQ diseases as well, including Alzheimer's disease (26-28). During review of our manuscript, two reports have been published that document similar axonal transport defects in HD models (29, 30). Determining the precise mechanisms by which Htt aggregates physically attach to the axonal cytoskeleton will likely provide important insights into the mechanisms of axonal transport blockage in HD. In summary, our results indicate that cytoplasmic aggregate formation in HD sequesters endogenous polyglutamine proteins and blocks axonal transport, contributing to neurophysiological dysfunction and neurodegeneration.
Acknowledgments
We thank Leslie Thompson, Nancy Bonini, and Parsi Kazemi-Esfarjani for Drosophila strains; Michael Hayden for Htt cDNAs; Paul Garrity for help with pseudopupil analysis; Barry Ganetzky, in whose laboratory this project was initiated; and Ling-Ling Ho and Jessica Slind for help with generation of transgenic Drosophila. This work was supported by the National Institutes of Health, the David and Lucile Packard Foundation, and the Merck/Massachusetts Institute of Technology Collaboration Program.
Abbreviations: HD, Huntington's disease; polyQ, polyglutamine.
References
- 1.The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971-983. [DOI] [PubMed] [Google Scholar]
- 2.Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R., Bates, G. P., Davies, S. W., Lehrach, H. & Wanker, E. E. (1997) Cell 90, 549-558. [DOI] [PubMed] [Google Scholar]
- 3.Kopito, R. R. (2000) Trends Cell Biol. 10, 524-530. [DOI] [PubMed] [Google Scholar]
- 4.DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P. & Aronin, N. (1997) Science 277, 1990-1993. [DOI] [PubMed] [Google Scholar]
- 5.Davies, S. W., Turmaine, M., Cozens, B. A., DiFiglia, M., Sharp, A. H., Ross, C. A., Scherzinger, E., Wanker, E. E., Mangiarini, L. & Bates, G. P. (1997) Cell 90, 537-548. [DOI] [PubMed] [Google Scholar]
- 6.Gutekunst, C. A., Li, S. H., Yi, H., Mulroy, J. S., Kuemmerle, S., Jones, R., Rye, D., Ferrante, R. J., Hersch, S. M. & Li, X. J. (1999) J. Neurosci. 19, 2522-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saudou, F., Finkbeiner, S., Devys, D. & Greenberg, M. E. (1998) Cell 95, 55-66. [DOI] [PubMed] [Google Scholar]
- 8.Sapp, E., Penney, J., Young, A., Aronin, N., Vonsattel, J. P. & DiFiglia, M. (1999) J. Neuropathol. Exp. Neurol. 58, 165-173. [DOI] [PubMed] [Google Scholar]
- 9.Li, H., Li, S. H., Cheng, A. L., Mangiarini, L., Bates, G. P. & Li, X. J. (1999) Hum. Mol. Genet. 8, 1227-1236. [DOI] [PubMed] [Google Scholar]
- 10.Li, H., Li, S. H., Johnston, H., Shelbourne, P. F. & Li, X. J. (2000) Nat. Genet. 25, 385-389. [DOI] [PubMed] [Google Scholar]
- 11.Usdin, M. T., Shelbourne, P. F., Myers, R. M. & Madison, D. V. (1999) Hum. Mol. Genet. 8, 839-846. [DOI] [PubMed] [Google Scholar]
- 12.Kazemi-Esfarjani, P. & Benzer, S. (2000) Science 287, 1837-1840. [DOI] [PubMed] [Google Scholar]
- 13.Warrick, J. M., Paulson, H. L., Gray-Board, G. L., Bui, Q. T., Fischbeck, K. H., Pittman, R. N. & Bonini, N. M. (1998) Cell 93, 939-949. [DOI] [PubMed] [Google Scholar]
- 14.Marsh, J. L., Walker, H., Theisen, H., Zhu, Y. Z., Fielder, T., Purcell, J. & Thompson, L. M. (2000) Hum. Mol. Genet. 9, 13-25. [DOI] [PubMed] [Google Scholar]
- 15.Reickhof, G. E., Yoshihara, M., Guan, Z. & Littleton, J. T. (2003) J. Biol. Chem. 278, 41099-41108. [DOI] [PubMed] [Google Scholar]
- 16.Van Vactor, D. L., Jr., Cagan, R. L., Kramer, H. & Zipursky, S. L. (1991) Cell 67, 1145-1155. [DOI] [PubMed] [Google Scholar]
- 17.Kim, Y. J., Yi, Y., Sapp, E., Wang, Y., Cuiffo, B., Kegel, K. B., Qin, Z. H., Aronin, N. & DiFiglia, M. (2001) Proc. Natl. Acad. Sci. USA 98, 12784-12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellington, C. L., Ellerby, L. M., Gutekunst, C. A., Rogers, D., Warby, S., Graham, R. K., Loubser, O., van Raamsdonk, J., Singaraja, R., Yang, Y. Z., et al. (2002) J. Neurosci. 22, 7862-7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson, G. R., Salecker, I., Dong, X., Yao, X., Arnheim, N., Faber, P. W., MacDonald, M. E. & Zipursky, S. L. (1998) Neuron 21, 633-642. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Funez, P., Nino-Rosales, M. L., de Gouyon, B., She, W. C., Luchak, J. M., Martinez, P., Turiegano, E., Benito, J., Capovilla, M., Skinner, P. J., et al. (2000) Nature 408, 101-106. [DOI] [PubMed] [Google Scholar]
- 21.Muqit, M. M. & Feany, M. B. (2002) Nat. Rev. Neurosci. 3, 237-243. [DOI] [PubMed] [Google Scholar]
- 22.Steffan, J. S., Bodai, L., Pallos, J., Poelman, M., McCampbell, A., Apostol, B. L., Kazantsev, A., Schmidt, E., Zhu, Y. Z., Greenwald, M., et al. (2001) Nature 413, 739-743. [DOI] [PubMed] [Google Scholar]
- 23.Ross, C. A. (2002) Neuron 35, 819-822. [DOI] [PubMed] [Google Scholar]
- 24.Chan, H. Y., Warrick, J. M., Gray-Board, G. L., Paulson, H. L. & Bonini, N. M. (2000) Hum. Mol. Genet. 9, 2811-2820. [DOI] [PubMed] [Google Scholar]
- 25.Kazantsev, A., Walker, H. A., Slepko, N., Bear, J. E., Preisinger, E., Steffan, J. S., Zhu, Y. Z., Gertler, F. B., Housman, D. E., Marsh, J. L., et al. (2002) Nat. Genet. 30, 367-376. [DOI] [PubMed] [Google Scholar]
- 26.Hafezparast, M., Klocke, R., Ruhrberg, C., Marquardt, A., Ahmad-Annuar, A., Bowen, S., Lalli, G., Witherden, A. S., Hummerich, H., Nicholson, S., et al. (2003) Science 300, 808-812. [DOI] [PubMed] [Google Scholar]
- 27.LaMonte, B. H., Wallace, K. E., Holloway, B. A., Shelly, S. S., Ascano, J., Tokito, M., Van Winkle, T., Howland, D. S. & Holzbaur, E. L. (2002) Neuron 34, 715-727. [DOI] [PubMed] [Google Scholar]
- 28.Gunawardena, S. & Goldstein, L. S. (2001) Neuron 32, 389-401. [DOI] [PubMed] [Google Scholar]
- 29.Gunawardena, S., Her, L. S., Brusch, R. G., Laymon, R. A., Niesman, I. R., Gordesky-Gold, B., Sintasath, L., Bonini, N. M. & Goldstein, L. S. (2003) Neuron 40, 25-40. [DOI] [PubMed] [Google Scholar]
- 30.Szebenyi, G., Morfini, G. A., Babcock, A., Gould, M., Selkoe, K., Stenoien, D. L., Young, M., Faber, P. W., MacDonald, M. E., McPhaul, M. J. & Brady, S. T. (2003) Neuron 40, 41-52. [DOI] [PubMed] [Google Scholar]