Abstract
Combined analyses of gene networks and DNA sequence variation can provide new insights into the aetiology of common diseases. Here, we used integrated genome-wide approaches across seven rat tissues to identify gene networks and the loci underlying their regulation. We defined an interferon regulatory factor 7 (IRF7)1-driven inflammatory network (iDIN) enriched for viral response genes, which represents a molecular biomarker for macrophages and was regulated in multiple tissues by a locus on rat chromosome 15q25. At this locus, Epstein-Barr virus induced gene 2 (Ebi2 or Gpr183), which we localised to macrophages and is known to control B lymphocyte migration2,3, regulated the iDIN. The human chromosome 13q32 locus, orthologous to rat 15q25, controlled the human equivalent of iDIN, which was conserved in monocytes. For the macrophage-associated autoimmune disease type 1 diabetes (T1D) iDIN genes were more likely to associate with T1D susceptibility than randomly selected immune response genes (P = 8.85 × 10−6). The human locus controlling the iDIN, was associated with the risk of T1D at SNP rs9585056 (P = 7.0 × 10−10, odds ratio = 1.15), which was one of five SNPs in this region associated with EBI2 expression. These data implicate IRF7 network genes and their regulatory locus in the pathogenesis of T1D.
While genome-wide association studies (GWAS) have uncovered many common genetic variants associated with human diseases, the molecular mechanisms by which DNA variation affects disease risk remain poorly characterised4. To translate genetic association into biological function DNA variation has been correlated with gene expression to identify the genetic drivers of gene networks, which are coordinately regulated by transcription factors (TFs), that represent important determinants of disease aetiology5,6,7. Here we used a panel of recombinant inbred (RI) rat strains8 to study TF-driven gene networks and their regulatory loci and integrated these data with human gene expression and GWAS data to identify genes, networks and pathways for human disease (Supplementary Fig. 1).
We combined expression quantitative trait loci (eQTLs) from fat, kidney and heart8,9 with new eQTL data in aorta, skeletal muscle, adrenal and liver to create genome-wide eQTL datasets across seven rat tissues. We used a two-step procedure to integrate eQTLs and TF-target genes to identify TF-driven gene networks (Supplementary Information). In the first step, we indentified 147 TFs whose expression mapped to 587 eQTLs across seven tissues, which were mostly (>90%) under trans-regulatory genetic control, in keeping with previous studies in yeast10,11. In the second step, we tested for enrichment of transcription factor binding sites (TFBSs)12 in the putative promoter sequences of genes whose expression mapped to trans-eQTLs. Out of the 13 TF-driven gene networks identified (Supplementary Table 1) we observed the strongest TFBS enrichment for interferon regulatory transcription factor Irf7 (P < 1 × 10−6, False Discovery Rate, FDR < 5 × 10−5). Irf7 TFBSs were predicted in the promoters of 23 genes, including Irf7 itself, that all mapped to a single trans-eQTL on rat chromosome 15q25 in adrenal, kidney, heart and liver. We confirmed experimentally a subset of the predicted Irf7-targets by chromatin immunoprecipitation and quantitative PCR that established direct interaction of Irf7 with the promoters of these genes (Fig. 1a-c). Taken together, this provides evidence for a TF-driven regulatory cascade in which genetic variation on chromosome 15q25 modulates the expression of Irf7 and Irf7-target genes.
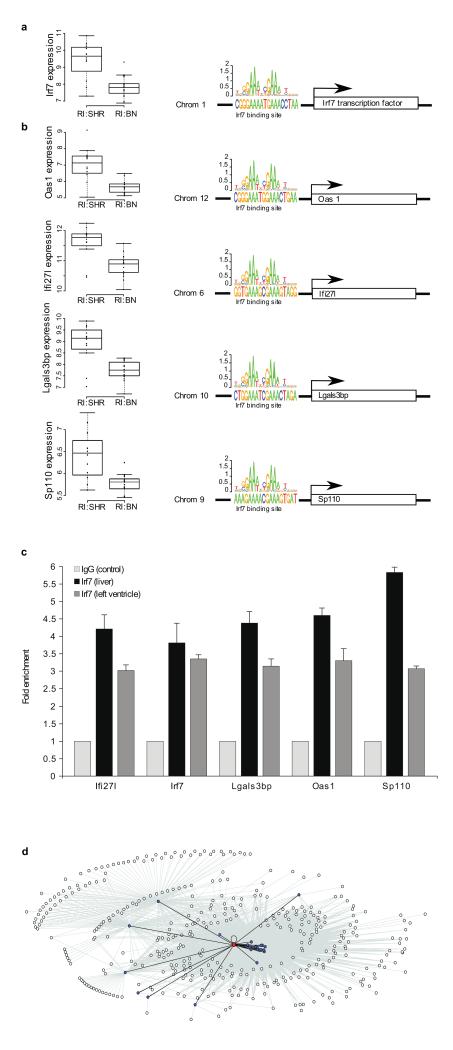
Figure 1. The Irf7-driven inflammatory gene network (iDIN).
Trans-regulated expression of a, Irf7 and b, genes containing Irf7 transcription factor binding sites by rat chromosome 15q25 at SNP J666808. Left panels, gene expression in the left ventricle is shown in the recombinant inbred (RI) rat strains grouped by SHR or BN genotype at SNP J666808 (SHR allele, RI.SHR; BN allele, RI.BN). Right panels, transcription factor binding site predictions are represented for the five (out of 23 predicted) Irf7-target genes. The chromosome (Chrom) encoding the Irf7-target is shown to the left of the predicted Irf7 binding sites. These data provide evidence for a regulatory cascade in which a locus on chromosome 15q25 regulates the expression of Irf7 on chromosome 1 in an allele-dependent manner with consequent effects on Irf7-target genes mediated through Irf7 transcription factor binding sites. c, Quantitative chromatin immuno-precipitation of predicted Irf7-target genes. Direct binding of Irf7 to the promoters of the predicted targets Ifi27l, Irf7, Lgals3bp, Oas1, and Sp110 was confirmed in liver and heart tissues. Fold enrichments are shown relative to non-immune IgG control. d, The expanded Irf7-driven inflammatory gene network (iDIN) comprising 305 genes. Nodes represent genes, the node representing Irf7 is coloured red and its predicted targets are coloured blue. Edges connect genes that are either predicted Irf7-targets (black) or show significant Pearson correlation (FDR < 0.1%) to one of the predicted targets (grey).
Irf7 is a master regulator of the type 1 interferon response1 and genes directly regulated by Irf7 may comprise the core components of a larger network, which we identified by genome-wide co-expression analysis of Irf7-target genes across tissues (Supplementary Information). This revealed a network of 247 genes across seven tissues, which was expanded to 305 genes in four of the seven tissues where additional gene expression data were available (FDR < 0.1%) (Supplementary Table 2). Gene ontology (GO) analysis of the network showed enrichment for specific biological processes, including “immune response” (P = 7.5 × 10−29), and “response to virus” (P = 9.6 × 10−7) (Supplementary Table 3). We designated the network the Irf7-driven inflammatory gene network (iDIN) (Fig. 1d), which was most enriched for expression in mouse bone marrow macrophages (P = 1.6 × 10−159) and human monocytes (P = 6.0 × 10−177) with high levels of expression in other immune cells, including B lymphocytes (Supplementary Fig. 2).
Whilst a core of 23 Irf7-target genes mapped to the same trans-eQTL on rat chromosome 15 the overall genetic control of the iDIN was unknown. We used sparse Bayesian regression models13 to determine the association between expression levels of iDIN genes across seven tissues with genome-wide single nucleotide polymorphisms (SNPs) and identify regulatory ‘hot-spots’14. The same rat 15q25 locus, which controlled Irf7 and its targets in trans, was associated with iDIN expression in all tissues (FDR < 1%) and showed the strongest evidence for common regulation in five out of seven tissues with increased iDIN genes expression associated with the Spontaneously Hypertensive Rat (SHR) allele (Fig. 2). The iDIN, which is highly expressed in immune cells, may represent a molecular signature of macrophages that are associated with risk of common inflammatory diseases15 and autoimmune disease type 1 diabetes (T1D)16. Hence, we characterized expression of Cd68, an established marker of macrophages17, in SHR and Brown Norway (BN) hearts and the RI strains. Cd68 mRNA levels were elevated in SHR as compared to BN heart (P = 0.01), which reflected increased numbers of macrophages (P = 2 × 10−22). In the RI strains, Cd68 was under trans-acting genetic control at the 15q25 locus that regulates the iDIN (Supplementary Fig. 3).
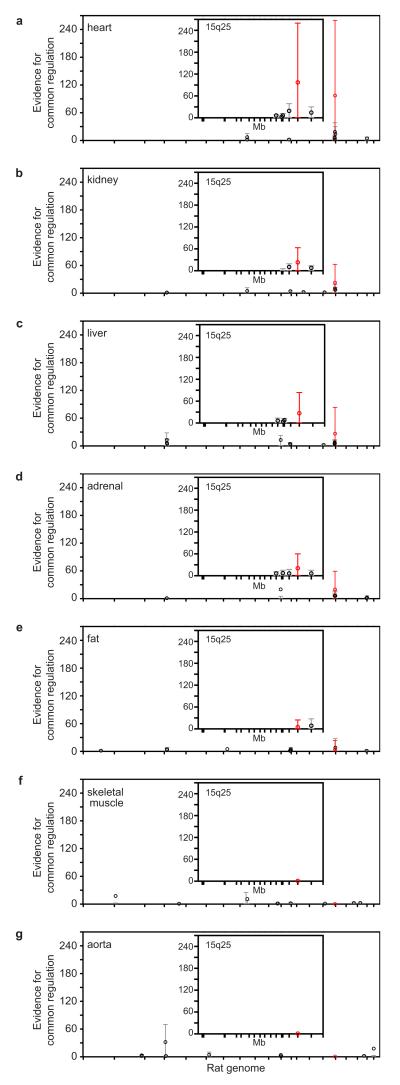
Figure 2. Genetic mapping of regulatory ‘hot-spots’ for the iDIN.
a-g, For each rat autosomal chromosome (x-axes), the strength of evidence for a SNP being a regulatory ‘hot-spot’ for controlling the network is measured by the average Bayes Factor (y-axes). Controlling the FDR at 1% level for each eQTL, the average Bayes Factor indicates the evidence in favour of common genetic regulation versus no genetic control, and is reported as a ratio between the strengths of these models (Supplementary Information). For the 10 largest regulatory hot-spots the average Bayes Factors (circles) and their 90% range (5th-95th percentiles) are reported; a single SNP (J666808) that is consistently and most strongly associated with the network in 5 out of 7 tissues is highlighted in red. Inserts, average Bayes Factors and 90% range for the SNPs on rat chromosome 15q25 (87,479,238 - 108,949,015 bp). SNP positions in the region are indicated by tick marks.
We then analyzed genetic variation in the RI strains using SNPs18 from the 15q25 region, which contains seven annotated protein-coding genes, and determined the expression of iDIN genes in seven inbred rat strains of known genotype that refined the locus to a 700kb region (Supplementary Fig. 4). Using the SHR genome sequence19, only Dock9, Ebi2 and Tm9sf2 exhibited DNA variation within the region, which was synonymous for Dock9, non-synonymous but not predicted to be functional for Tm9sf2 and a 5′UTR SNP for Ebi2 (Supplementary Table 4). Ebi2 was the only differentially expressed gene between parental strains within the region, was cis-regulated in heart and kidney and highly expressed in myeloid cell types (Supplementary Fig. 4 and 5). We assessed the effect of the Ebi2 5′UTR SNP by luciferase assay, the SHR allele resulted in reduced luciferase activity as compared to the BN allele (Supplementary Fig. 5).
Ebi2 (or Gpr183) encodes an orphan G protein-coupled receptor (GPCR) that controls B cell migration2,3 and is a candidate for the regulation of the iDIN at the chromosome 15q25 region. We localised Ebi2 expression to Cd68+ve macrophages within the rat heart (Supplementary Fig. 6), an observation that we confirmed and extended across tissues (pancreas, liver, kidney and heart) in the Ebi2GFP/+ mouse2 (Supplementary Fig. 7). SiRNA knockdown of Ebi2 in primary cultures of rat macrophages (Supplementary Fig. 8a) increased expression of Irf7, the central hub of the iDIN, and of iDIN genes (Supplementary Fig. 8b). This suggests that Ebi2 is a negative regulator of the innate immune response in macrophages, which would be consistent with lower Ebi2 expression in the SHR that has more macrophages than the BN rat (Supplementary Fig. 3).
To translate our findings to humans, we tested whether the iDIN was recapitulated in human immune cells using genome-wide expression data from monocytes isolated from 1,490 individuals from the Gutenberg Heart Study (GHS)20. We performed TFBS enrichment and co-expression analysis, analogous to that performed in the rat, and identified the human IRF7-driven network (Supplementary Table 5), which exhibited strong overlap with the rat iDIN (P = 9.1 × 10−20) and was most strongly annotated by the GO term “response to virus” (P = 1.9 × 10−13) (Supplementary Table 6). Using monocyte gene expression data from a distinct cohort of 758 subjects from the Cardiogenics Study (Supplementary Information), we found the same set of co-regulated IRF7-target genes (Supplementary Table 5) and significant overlap with the expanded IRF7-driven network indentified in the GHS (P = 8.3 × 10−23).
We determined whether the human chromosome 13q32 locus (spanning ~1 Mb, Supplementary Table 7), which is orthologous to the critical rat chromosome 15q25 region, was associated with expression of iDIN genes in humans. Multivariate analysis of the Cardiogenics monocyte expression and genotype data revealed that six SNPs in the 13q32 region (including rs9557217, P = 5.0 × 10−5; and rs9585056, P = 1.1 × 10−3) were associated with trans-regulated expression of IRF7 and IRF7-target genes (Supplementary Fig. 9). We did not, however, detect a signal for trans-regulation of IRF7 or IRF7-target genes at the 13q23 locus in the GHS cohort. This may reflect the differences between the monocyte selection protocols used in the two studies (Supplementary Information and data not shown).
In both the GHS and Cardiogenics cohorts, EBI2 expression in monocytes was cis-regulated at the 13q32 locus but the peak SNPs differed between the two cohorts (most associated SNPs: Cardiogenics, rs9585056, P = 2.2 × 10−8; GHS, rs9517725, P = 6.8 × 10−13) (Fig. 3). However, a formal hypothesis test21 of a common causal genetic variant was not rejected (P = 0.14). Two of the five SNPs, rs9557217 and rs9585056 contained in the model explaining EBI2 expression also exhibited a significant trans-effect on iDIN gene expression in the Cardiogenics cohort (Supplementary Fig. 9), suggesting common regulatory control by this locus on the IRF7 network and EBI2 expression.
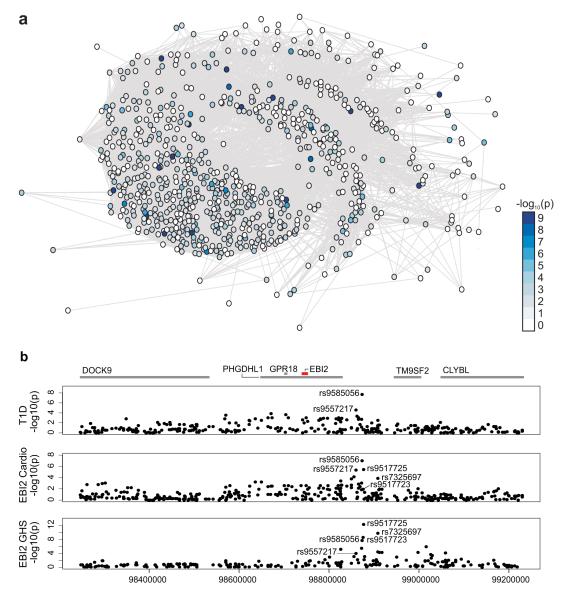
Figure 3. A gene network and locus for T1D risk.
a, Schematic representation of the union of IRF7-driven gene networks that was created using the set of human orthologues of rat iDIN genes and human iDIN genes. A Wilcoxon rank test based gene set enrichment analysis (modified from Holden et al.)30 showed SNPs close to iDIN genes to be significantly more likely to associate with T1D in large-scale GWAS than SNPs close to randomly selected genes (P = 2.5 × 10−10) and randomly selected immune response genes (P = 8.8 × 10−6). Nodes represent iDIN genes and the node colour indicates the P-values (-log10 scale) of the association with T1D (see Methods). b, Results of EBI2 eQTL analysis in GHS (top panel), Cardiogenics (middle panel) and T1D association (bottom panel) at the human chromosome 13 locus that is orthologous to the 700kb rat chromosome 15q25 region. The upper panel shows the nominal –log10 P-values of marker regression against gene expression of EBI2 for all SNPs in the region. We defined EBI2 eQTL models by selecting SNPs using lasso regression (Supplementary Information) in GHS (rs9585056, rs9517723, rs7325697). When adding imputed SNPs, rs9517725 explains most of the variation of the EBI2 expression (P = 6.8 × 10−13) at this locus. Lasso model selection in Cardiogenics yielded an overlapping set of three SNPs (rs9557217, rs9585056, rs9517725). The lower panel shows the –log10 P-values of T1D association with SNPs in the region. SNP rs9585056 showed the strongest association with T1D (P = 7.0 × 10−10) amongst the genotyped markers.
Monocyte-derived macrophages are critical determinants of inflammatory processes important for common disease15 including autoimmune T1D22. The iDIN is expressed in macrophages, enriched for immune response genes and contained IFIH1, a well-characterised T1D susceptibility gene23,24. We evaluated the association of the human orthologues of rat iDIN genes and genes in the human iDIN (Fig. 3) with T1D (Supplementary Information). SNPs close to (<=1Mb) any iDIN genes were significantly more likely to associate with T1D in large-scale GWAS than SNPs close to genes not in the network (P = 2.4 × 10−10) (Supplementary Table 8). We also tested the iDIN association with T1D against all genes annotated by the GO term “immune response” and established an overrepresentation of T1D associated genes (P = 8.85 × 10−6), indicating that the iDIN more specifically categorizes T1D genes than the GO term “immune response”.
In a GWAS meta-analysis of T1D in 7,514 cases and 9,045 controls25, we found evidence for association of the chromosome 13q32 region at SNP rs9585056 (P = 1.3 × 10−7) that had not been reported before (Fig. 3b). We genotyped this SNP in two independent large cohorts and increased the strength of the T1D association (combined P = 7.0 × 10−10, odds ratio (95% confidence interval) = 1.15 (1.09-1.21), Supplementary Table 9). The minor C allele of SNP rs9585056 was associated with T1D risk, lower EBI2 expression in both GHS and Cardiogenics cohorts and, on average, increased expression levels of iDIN genes in the Cardiogenics cohort. Although we cannot discriminate between single and multiple causal variants, overall, these results show an overlap of association signals in the same region on human chromosome 13q32 for iDIN genes, EBI2 cis-regulation and T1D. We also noted that the EBI1 (or CCR7) and EBI3 (or IL27B) genes are also associated with T1D susceptibility: EBI1 is in the confirmed T1D region 17q21.225, and EBI3 encodes the beta subunit of the IL-27 cytokine, for which alpha-subunit gene, IL27, is in the T1D region 16p11.225, suggesting a link between EBV infection and T1D.
The immunopathology of autoimmune T1D is characterised by infiltration of the pancreas with B and T lymphocytes and macrophages16. We have shown that iDIN genes contribute to T1D risk and implicate the innate viral response pathway and macrophages in the aetiology of T1D. Genetic control points that perturb biological networks can represent important loci for disease risk6 and the new T1D susceptibility locus that we identified may regulate innate immune response genes in macrophages, as we demonstrated in the rat. Ebi2, which controls Irf71, represents a candidate for trans-regulation of the human iDIN and for T1D risk. A role for IRF7 in the pathogenesis of T1D is supported by functional studies26 and by other T1D genes, namely TLR7, TLR827, and IFIH123,24, which are regulated by or act through IRF728. Our study shows that co-expression networks across species provide functional annotation of genes in biological processes that can be used to reveal the signal of common genetic variation of small effect that is not detected by GWAS.
Methods Summary
Genome-wide expression data in the rat were generated from seven tissues: adrenal, aorta, fat, kidney, left ventricle, liver and skeletal muscle using Affymetrix RAE 230a and RAE 230_2 chips. eQTL mapping was carried out using the genetic map of the BXH/HXB RI strains generated in a previous large scale effort by the STAR consortium18, as previously described8,9. In humans, expression data from isolated monocytes were obtained from 1,490 population-based individuals from the Gutenberg-Heart Study20 and from 758 individuals from the Cardiogenics Study. eQTL data were analyzed in conjunction with TFBS enrichment analysis using PASTAA12 to identify core gene networks centred on transcription factors. The core networks were expanded to include genes showing co-expression (FDR < 0.1%) with any of the core network genes in seven rat tissues and isolated human monocytes. Association between expression levels of the network genes and genome-wide SNPs in the rat was carried out using sparse Bayesian regression models13, and the major regulatory control points (‘hot-spots’)14 for the entire network were identified. Genes at the locus associated with the rat network were characterised by DNA sequencing, RNA-Sequencing, quantitative PCR analyses, luciferase assay and combined in situ hybridisation and immunohistochemistry. A combined network, comprised of the union or intersection of the rat and human networks, was constructed and analyzed for association with T1D by means of a stratified Wilcoxon rank test to compare SNPs genotyped in T1D GWAS25,29 close (<=1Mb) to any network gene or to those close to any gene not in the network (see www.t1dbase.org for all T1D SNP association data). SNPs across the human locus, that is orthologous to rat chromosome 15q25 controlling the network, were tested for association with T1D as described elsewhere25. Supplementary Fig. 1 provides an overview of the study design. Full Methods are provided in Supplementary Information.
Supplementary Material
Acknowledgements
We acknowledge funding from the German National Genome Research Network (NGFN-Plus ‘Genetics of Heart Failure’), the Helmholtz Association Alliance on Systems Biology (MSBN), EURATools (LSHG-CT-2005-019015), European Union FP6 (LSHM-CT-2006-037593), PHC ALLIANCE 2009 (19419PH), UK National Institute for Health Research Biomedical Research Unit (Royal Brompton and Harefield NHS Trusts, University Hospitals of Leicester NHS Trusts) and Biomedical Research Centre (Imperial College NHS Trust) awards, the British Heart Foundation, grant P301/10/0290 from the Grant Agency of the Czech Republic and 1M6837805002 from the Ministry of Education of the Czech Republic, the Fondation Leducq the Medical Research Council UK, Research Councils UK, the Juvenile Diabetes Research Foundation International, the Wellcome Trust. The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement N° HEALTH-F4-2010-241504 (EURATRANS).
Footnotes
Author Contributions S.A.C., N.H. and E.P. initiated the study. M.H., E.P., N.H. and S.A.C. participated in the conception, design and coordination of the study. H.L., Y.L., R.S., Y.A.L., S.P., C.R., K.S. and R.B. performed genetic, biochemical and functional analysis in the rat. E.E.G. and J.G.C. provided Ebi2GFP/+ mouse data. M.P. and T.J.A. contributed materials and discussion of the manuscript. M.H., E.P., C.W., D.J.S., D.C., A.B., S.R.L., L.B., M.R. and L.T. designed and applied the modelling methodology and statistical analyses. M.H., E.P. and H.S. performed expression QTL analysis in the rat. L.B. conceived and performed the Bayesian analysis. C.W., D.J.S. and D.C. performed association analyses in humans. M.H., O.H., H.R. and M.V. conceived and performed bioinformatics analyses in the rat. J.E., C.H., S.M., W.H.O., C.M.R., N.J.S., H.S., A.H.G., S.B., T.M., T.Z., S.S., A.Z., M.R., L.T. and F.C. provided the human monocyte expression data and contributed to the transcriptomic analyses in the Cardiogenics and the Gutenberg Heart Study cohorts. M.H., E.P., N.H. and S.A.C. wrote the paper with significant contributions from C.W. and J.A.T. All authors discussed the results and commented on the manuscript. M.H. and E.P. contributed equally to the paper and are listed in alphabetical order.
References
- 1.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 2.Pereira JP, Kelly LM, Xu Y, Cyster JG. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 2009;460:1122–1126. doi: 10.1038/nature08226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gatto D, Paus D, Basten A, Mackay CR, Brink R. Guidance of B cells by the orphan G protein-coupled receptor EBI2 shapes humoral immune responses. Immunity. 2009;31:259–269. doi: 10.1016/j.immuni.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 4.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubner N, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 9.Petretto E, et al. Integrated genomic approaches implicate osteoglycin (Ogn) in the regulation of left ventricular mass. Nat Genet. 2008;40:546–552. doi: 10.1038/ng.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 11.Yvert G, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 12.Roider HG, Manke T, O’Keeffe S, Vingron M, Haas SA. PASTAA: identifying transcription factors associated with sets of co-regulated genes. Bioinformatics. 2009;25:435–442. doi: 10.1093/bioinformatics/btn627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petretto E, et al. New insights into the genetic control of gene expression using a Bayesian multi-tissue approach. PLoS Computational Biology. 2010;6 doi: 10.1371/journal.pcbi.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breitling R, et al. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 2008;4:e1000232. doi: 10.1371/journal.pgen.1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140:871–882. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- 16.Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat Rev Endocrinol. 2009;5:219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- 17.Holness CL, Simmons DL. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood. 1993;81:1607–1613. [PubMed] [Google Scholar]
- 18.Saar K, et al. SNP and haplotype mapping for genetic analysis in the rat. Nat Genet. 2008;40:560–566. doi: 10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atanur SS, et al. The genome sequence of the spontaneously hypertensive rat: Analysis and functional significance. Genome Res. 2010;20:791–803. doi: 10.1101/gr.103499.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeller T, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plagnol V, Smyth DJ, Todd JA, Clayton DG. Statistical independence of the colocalized association signals for type 1 diabetes and RPS26 gene expression on chromosome 12q13. Biostatistics. 2009;10:327–334. doi: 10.1093/biostatistics/kxn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Herrath M. Diabetes: A virus-gene collaboration. Nature. 2009;459:518–519. doi: 10.1038/459518a. [DOI] [PubMed] [Google Scholar]
- 23.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth DJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 25.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009 doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, et al. Interferon-alpha initiates type 1 diabetes in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2008;105:12439–12444. doi: 10.1073/pnas.0806439105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper JD, et al. Follow-up of 1715 SNPs from the Wellcome Trust Case Control Consortium genome-wide association study in type I diabetes families. Genes Immun. 2009;10(Suppl 1):S85–94. doi: 10.1038/gene.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawai T, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 29.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holden M, Deng S, Wojnowski L, Kulle B. GSEA-SNP: applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics. 2008;24:2784–2785. doi: 10.1093/bioinformatics/btn516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.