SUMMARY
Extensive behavioral and biochemical characterization of cannabinoid-mediated effects on the central nervous system has revealed at least three lines of evidence supporting the role of a putative guanine nucleotide-binding protein-coupled cannabinoid receptor for cannabimimetic effects, (i) stereoselectivity, (ii) inhibition of the adenylate cyclase/cAMP second messenger system, and (iii) radioligand-binding studies with the synthetic cannabinoid [3H]CP-55,940 indicating a high degree of specific binding to brain tissue preparations. Based on recent findings from our laboratory demonstrating that Δ9-tetrahydrocannabinol markedly inhibited forskolin-stimulated cAMP accumulation in mouse spleen cells, the presence of a guanine nucleotide-binding protein-coupled cannabinoid receptor associated with mouse spleen cells and its functional role in immune modulation were investigated. In the present studies, stereoselective immune modulation was observed with the synthetic bicyclic cannabinoid (−)-CP-55,940 versus (+) CP-56,667 and with 11-OH-Δ8-tetrahydrocannabinol-dimethylheptyl, (−)-HU-210 versus (+)-HU-211. In both cases, the (−)-enantiomer demonstrated greater immunoinhibitory potency than the (+)-isomer, as measured by the in vitro sheep red blood cell antibody-forming cell response. Radioligand binding studies produced a saturation isotherm exhibiting approximately 45–65% specific binding to mouse spleen cells. Scatchard analysis demonstrated a single binding site on spleen cells, possessing a Kd of 910 pm and a Bmax of approximately 1000 receptors/spleen cell. RNA polymerase chain reaction of isolated splenic RNA using specific primers for the cannabinoid receptor resulted in the amplification of a 854-kilobase predicted product that hybridized with cannabinoid receptor cDNA, demonstrating the presence of cannabinoid receptor mRNA in mouse spleen. Together, these findings strongly support the role of a cannabinoid receptor in immune modulation by cannabimimetic agents.
Δ9-THC, the major psychoactive component of marijuana, and a number of structurally related cannabinoid compounds have been widely established as being immunosuppressive (1). Although the mechanisms responsible for the immunological effects of these agents are presently unknown, they have traditionally been attributed to the lipophilic properties of the cannabinoids, which were thought to produce nonspecific perturbations of the cell membrane. Interest in the development of cannabinoid therapeutic agents for a variety of applications, including analgesia, attenuating nausea and vomiting due to cancer chemotherapy, decreasing intraocular pressure in glaucoma, stimulating appetite, and decreasing bronchial constriction, has been a catalyst for elucidating the mechanism by which cannabinoids mediate their broad spectrum of physiological effects. Recent isolation and characterization of a cDNA from rat brain cortex are considered by many to be definitive evidence for the putative cannabinoid receptor (2). The translated sequence of this cDNA revealed a peptide product possessing the characteristics of the G protein-coupled receptor family. Although novel, the identification of a cannabinoid receptor was not completely unexpected, based on at least three lines of evidence that strongly implicated a receptor-associated mechanism for cannabimimetic effects, (i) Structure-activity relationship studies with a variety of cannabinoid compounds have demonstrated enantioselective effects in a number of experimental systems (2–7). (ii) Radioligand binding studies have indicated a high degree of specific binding of the synthetic cannabinoid [3H]CP-55,940 to brain (4, 6, 8, 9). (iii) Cannabinoids have been shown to inhibit adenylate cyclase in both membrane and whole-cell preparations of brain and in neuronal cell lines, further implicating the role of a G protein-coupled receptor (10–14).
Recently, a clone encoding a receptor protein possessing all the characteristics of a G protein-coupled receptor, HGMP08, was isolated from a human brain stem cDNA library (15). Interestingly, HGMP08 was found to share 97.3% homology with the rat cannabinoid receptor cloned by Matsuda et al. (2). This same probe also revealed detectable amounts of transcripts when utilized to screen a cDNA library from human testis (7). CHO-K1 cells transfected with the isolated construct from testis, BS/08, demonstrated stereoselective inhibition of forskolin-stimulated cAMP accumulation. What is most intriguing from these studies is the identification of the putative cannabinoid receptor in non-neuronal tissue. Recent studies from our laboratory have shown that Δ9-THC markedly inhibits adenylate cyclase in mouse spleen cells, suggesting the presence of the putative cannabinoid receptor associated with the immune system (16). Based on these findings, the objective of the present studies was to determine whether the cannabinoid receptor exists in mouse spleen, thus indirectly supporting the role of a cannabinoid receptor in immune suppression by cannabinoid compounds. The role of a cannabinoid receptor in cannabinoid-mediated immune modulation has not been addressed.
Materials and Methods
Chemicals
Δ9-THC was provided by the National Institute on Drug Abuse. CP-55,940 and CP-56,667 were gifts of Dr. Lawrence Melvin (Pfizer, Inc., Groton, CT). Dr. Raphael Mechoulom (Hebrew University, Israel) supplied HU-210 and HU-211.
Mice
Virus-free female B6C3F1 mice (5–6 weeks of age) were purchased from the Frederick Cancer Research Center. On arrival, mice were randomized, transferred to plastic cages containing a sawdust bedding (four mice/cage), and quarantined for 1 week. Mice were given food (Purina Certified Laboratory Chow) and water ad libitum and were not used for experimentation until their body weight was 17–20 g. Animal holding rooms were kept at 21–24° and 40–60% relative humidity with a 12-hr light/dark cycle.
In vitro antibody assays
Spleens from untreated mice were isolated aseptically and made into a single-spleen cell suspension. The spleen cell suspension was adjusted to 1.0 × 107 cells/ml in RPMI1640 supplemented with 5% fetal bovine serum (Hyclone, Logan, UT), 2 mm l-glutamine, antibiotic-antimycotics (100 units/ml penicillin, 100 µg/ ml streptomycin, and 0.25 µg/ml fungizone) (GIBCO, Grand Island, NY), and 5 × 10−5 m 2-mercaptoethanol and was transferred in 500-µl aliquots to a 48-well Costar culture plate (Cambridge, MA) set up in quadruplicate for each treatment group. Cannabinoid compounds were added directly, in 5 µl of vehicle (0.01% dimethylsulfoxide or 0.1% ethanol, final culture concentration), to the respective wells of the 48-well culture plate. Each well was sensitized with 6.5 × 106 sRBC and cultured for 5 days in a Bellco stainless steel tissue culture chamber pressurized to 6.0 psi with a gas mixture consisting of 10% O2, 7% CO2, and 83% N2. The culture chamber was continuously rocked for the duration of the culture period (i.e., 5 days). Enumeration of the AFC response was performed as previously described (16). Briefly, spleen cells were resuspended in each well of the 48-well culture plate. A 50-µl aliquot of cell suspension was taken from each well and added to a 12- × 75-mm heated culture tube containing 400 µl of 0.5% melted agar (DIFCO, Detroit, MI) solution in Earle’s balanced salt solution and 0.05% DEAE-dextran (Pharmacia, Piscataway, NJ). Additionally, each agar tube received 25 µl of guinea pig complement and 25 µl of indicator sRBC. The tubes were immediately vortex mixed, a 200-µl aliquot of the mixture was transferred to a 100- × 15-mm Petri dish, and the agar solution was covered with a 45- × 50-mm microscope coverslip. Once the agar had solidified, the Petri dishes were incubated at 37° for 3 hr. After the 3-hr incubation, the AFC were enumerated at 6.5× magnification using a Bellco plaque viewer. During the 3-hr incubation period the number of spleen cells per well and viability (described below in Pronase determination of viability) were determined using a Coulter counter. Results from quadruplicate cultures were expressed as the mean ± standard error of AFC/106 recovered splenocytes.
Pronase determination of viability
Aliquots of spleen cell suspensions were incubated with an equal volume (100 µl) of Pronase (5 mg/kg; Calbiochem-Behring Corp., San Diego, CA) for 10 min at 37°. After the incubation, the splenocyte solution was diluted with 10 ml of Isoton (Coulter, Addison, NJ), counted in a Coulter counter, and compared with a 100-µl aliquot of the same test sample of splenocytes without Pronase. The percent viability = (cell counts with Pronase/cell counts without Pronase) × 100.
Radioligand binding studies
The filtration procedure used for [3H]CP-55,940 binding (specific activity, 175 dpm/nmol) (9) was a modification of the centrifugation method described by Devane et al. (4). Spleens were isolated from untreated mice and made into a single-cell suspension. The cells were washed, centrifuged, and resuspended in Ca2+/Mg2+-free Hanks’ balanced salt solution (GIBCO) at a concentration of approximately 1.5 × 108 cells/ml. The binding assay was performed in AquaSil (Pierce, Rockford, IL) siliconized 13- × 100-mm disposable glass culture tubes to which was added a 1-ml volume of reaction buffer (606 mg of Tris · HC1, 36.8 mg of EDTA, and 500 mg of BSA in 100 ml of Ca2+/Mg2+-free Hanks’ balanced salt solution), radioligand [3H]CP-55,940 (ranging from 0.1 to 5 nm), 1 µm unlabeled CP-55,940, and 100 µl of intact spleen cells (1.5 × 107 cells). The reaction mixture was incubated at 30° for 60 min. After this incubation, the reaction was stopped by the addition of 2 ml of ice-cold washing buffer (3.63 g of Tris · HCl and 600 mg of BSA in 600 ml of distilled H2O), and the reaction mixture was washed twice with 4 ml of washing buffer during vacuum filtration through polyethylenimine-pretreated 2.4-cm GF/C glass microfiber filters (Whatman International, Maid-stone, England). The filters were transferred into 20-ml polyethylene scintillation vials containing 10 ml of scintillation fluid and 1 ml of H2O and were then placed on a shaker platform for 1 hr. The samples were assayed for 3H using a Beckman LS1801 scintillation counter. Cell binding of [3H]CP-55,940 in the presence and absence of unlabeled CP-55,940 (1 µm) was determined. Specific binding was calculated by subtracting nonspecific from total binding. A saturation binding isotherm was plotted to demonstrate the relationship between total, non-specific, and specific radioligand binding to mouse spleen cells. A Scatchard analysis was performed to determine the affinity (Kd) and the number of binding sites per spleen cell (Bmax). Scatchard and Hill analyses were performed with the EBDA/LIGAND computer program.
RNA preparation
RNA was prepared from isolated splenocytes and rat brain cerebellum using RNAzol B (Biotecx), a commercially available modification of the acid phenol extraction technique.
RNA analysis
Equivalent amounts of RNA (typically 10 µg for a minigel) were denatured by heating at 70° for 5 min in loading buffer and were loaded onto 1% formaldehyde-agarose horizontal gels. Loading buffer was 50% formamide, 6% formaldehyde, 20 mm boric acid, 10% glycerol, 0.2 mm EDTA, 0.25% bromphenol blue, 0.25% xylene cyanol. After denaturation, 1 µl of 1 mg/ml ethidium bromide was added to aid visualization of the RNA samples. The gels consisted of 1% agarose in 20 mm boric acid, pH 8.3, 0.2 mm EDTA, 3% formaldehyde, and were run in a buffer of the same composition.
RNA PCR
Total RNA from splenocytes and cerebellum was reverse transcribed and amplified for cannabinoid receptor cDNA using a Perkin Elmer RNA PCR kit, as follows: 1 µg of total RNA was added to a tube containing reverse transcription mixture (4 mm MgCl2, 50 mm KC1, 10 mm Tris · HCl, pH 8.3, 1 mm dGTP, 1 mm dATP, 1 mm TTP, 1 mm dCTP, 1 unit/µl placental RNase inhibitor, 2.5 units/µl Moloney murine leukemia virus reverse transcriptase, and 2.5 µm random hexamers), in a final volume of 20 µl. Reverse transcription was carried out at 42° for 1 hr, and the enzyme was inactivated by heating at 99° for 5 min and cooled to 4°. The entire reverse transcription mixture was diluted into 100 µl with MgCl2, KCL, and Tris · HCl, pH 8.3, added to final concentrations of 2 mm, 50 mm, and 10 mm, respectively. Taq polymerase (2.5 units) was added and PCR primers (0.15 µm final) based on bp 1–21 of the cannabinoid receptor and bp 822–843 (on the opposite strand) were used to amplify a 843-bp DNA. The thermocycler cycles used were 2 min at 95° for one cycle, 1 min at 95° and 4 min at 60° for 35 cycles, 7 min at 60° for one cycle, and 4° soak.
For the c-fos experiment, primers based on bp 232–252 and bp 322–342 (on the opposite strand) of rat c-fos cDNA were chosen. Intron 1 of the c-fos gene is between these primers, so any genomic DNA present would be seen as a 800-bp PCR product; otherwise, a 110-bp product would be produced from the c-fos mRNA.
For the DNA PCR, 1 µg of total RNA was amplified using the same conditions as for the RNA PCR, except that the reverse transcriptase and RNase inhibitors were omitted and only the PCR cycles were performed.
The gels were transferred to GeneScreen (NBN) nylon membranes in 50 mm NaOH, 1 m NaCl.
Hybridization
The hybridization buffer consisted of 50% form-amide, 6× standard saline citrate (20× standard saline citrate is 3 m NaCl, 0.3 m sodium citrate, pH 7.0), 0.1 mg/ml salmon sperm DNA, 50 mm Tris, pH 8.0, and 5× Denhardt’s (50× Denhardt’s is 1 g of Ficoll, 1 g of polyvinylpyrrolidone, and 1 g of BSA fraction V in 100 ml of H2O). Filters were hybridized at 42° using 107 dpm of radiolabeled probe (specific activity, >5 × 108 dpm/µg).
Probe preparation
The probe used in these experiments was a rat cannabinoid receptor cDNA that one of us (M.E.A.) isolated from a λZAP rat brain cDNA library using a probe based on sequences unique to the published cannabinoid receptor (2). Oligonucleotide probes based on bp 1–21 and bp 1410–1422 on the opposite strand were chosen for use in PCR to generate a 1389-bp probe specific for the cannabinoid receptor. Sequence analysis of the isolate used in these experiments indicated identity with the published sequence. A 1.6-kilobase insert containing the cannabinoid receptor sequence was radiolabeled by random priming with T7 DNA polymerase for use as a probe for the reverse transcription-PCR.
Statistics
The mean ± standard error was determined for each treatment group of a given experiment. The homogeneity of the results was determined using Bartlett’s test for homogeneity (17). Homogeneous data were evaluated by a parametric analysis of variance. When significant differences occurred, treatment groups were compared with the vehicle controls using Dunnett’s t test (18). Nonhomogeneous data were evaluated for significance using Wilcoxon’s rank test (19).
Results
Comparison of immunosuppressive potencies of enantiomeric pairs (−)-CP-55,940 versus (+)-CP-56,667 and (−)-HU-210 versus (+)-HU-211
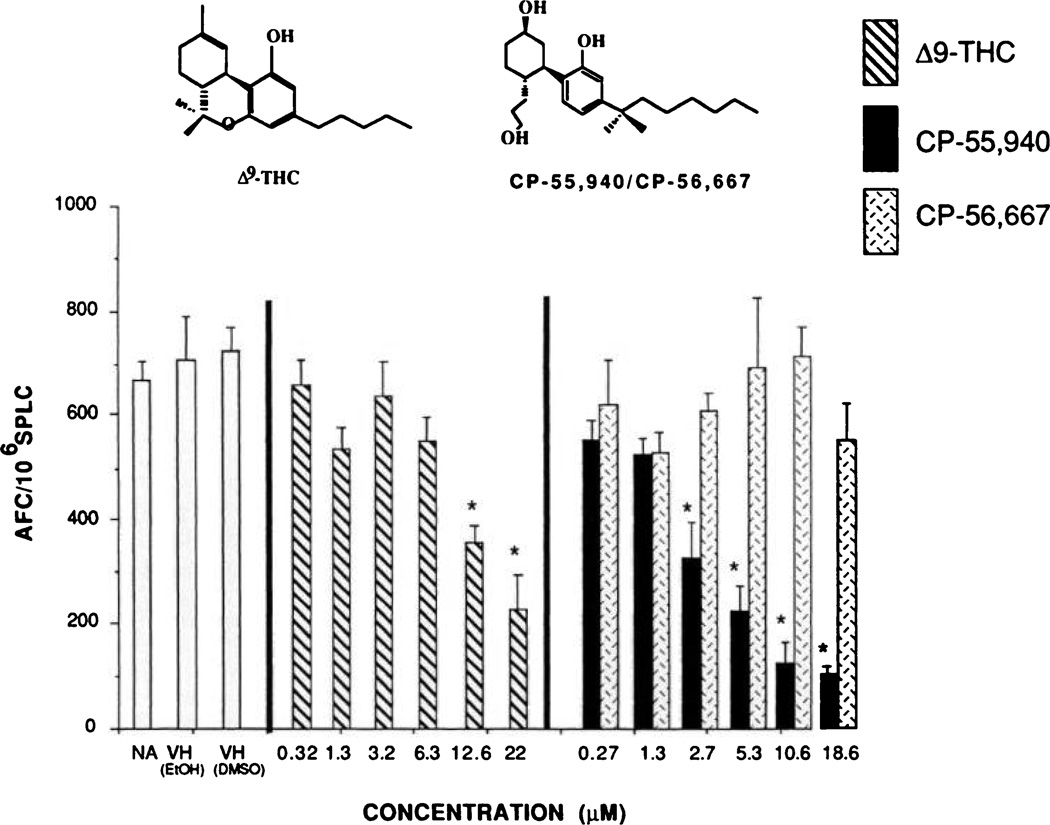
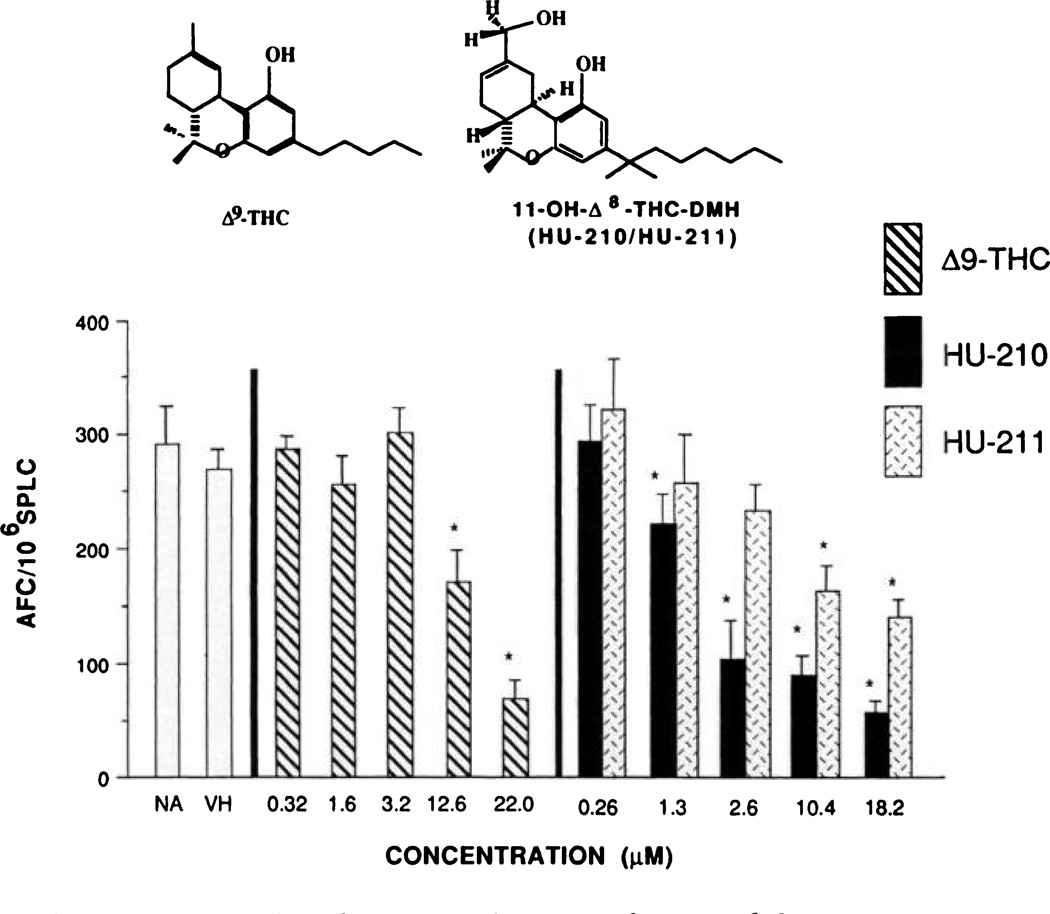
Enantioselective CNS-associated effects have been widely reported for a variety of structurally related cannabimimetic compounds. Historically, the (−)-enantiomers characteristically demonstrate significantly greater potency than the (+)-enantiomers. A second aspect of cannabinoid structure-activity is that synthetic cannabinoids possessing the dimethylheptyl aliphatic side chain consistently exhibit markedly greater potencies than Δ9-THC (5, 20). In light of this, the immunomodulatory potencies of two enantiomeric cannabinoid pairs possessing the dimethyl-heptyl side chain, CP-55,940 versus CP-56,667 and HU-210 versus HU-211, were compared with each other and with that of Δ9-THC, as measured by the in vitro sRBC AFC response. With respect to both of the enatiomeric pairs, the (−)-enantiomers demonstrated greater immunoinhibitory potency than the (+)-isomers. This was especially striking with CP-55,940 and CP-56,667, in which CP-56,667 was devoid of immunosuppressive activity at concentrations as high as 12 µm (Fig. 1). Conversely, 12 µm CP-55,940 produced approximately a 90% inhibition of the AFC response. Although not as striking as the CP-55,940 versus CP-56,667 comparison, HU-210 also demonstrated greater immunoinhibitory activity than did HU-211 (Fig. 2). However, the profile of activity for HU-210 and HU-211 in the AFC response did not show as great a separation in potency as reported for this enantiomeric pair in the CNS. Of equal importance as the enantioselective effects demonstrated with these cannabinoids was the finding that both CP-55,940 and HU-210 exhibited greater immunosuppressive potency than did Δ9-THC, which produced an approximately 67% inhibition of the AFC response at 22 µm. It is important to emphasize that there was no effect on cell number or viability at any of the concentrations tested.
Fig. 1.
Direct addition of stereoisomers CP-55,940 and CP-56,667 to naive spleen cell culture and their effect on the in vitro sRBC AFC response. Spleens from naive female B6C3F, mice were isolated aseptically and made into single-cell suspensions. The splenocytes were washed, adjusted to 1.0 × 107 cells/ml, and transferred in 500-µl aliquots to wells of a 48-wel culture plate. Quadruplicate cultures were prepared with vehicle [0.01% dimethylsulfoxide (DMSO) or 0.1% ethanol (EtOH) final concentration in culture], CP-55,940, or CP-56,667 and were sensitized with sRBC. Cultures were subsequently assayed, using sRBC, for their day 5 IgM antibody response by enumerating the number of AFC, spleen cell viability, and total recovered cells/culture. Bars, mean ± standard error as determined for each group. * p < 0.05, as determined by Dunnett’s t test, compared with the vehicle group.
Fig. 2.
Direct addition of stereoisomers HU-210 and HU-211 to naive spleen cell culture and their effect on the in vitro sRBC AFC response. Spleens from naive female B6C3F1 mice were isolated aseptically and made into single-cell suspensions. The splenocytes were washed, adjusted to 1.0 × 107 cells/ml, and transferred in 500-µl aliquots to wells of a 48-well culture plate. Quadruplicate cultures were prepared with vehicle (0.1 % ethanol, final concentration in culture). HU-210, or HU-211 and were sensitized with sRBC. Cultures were subsequently assayed, using sRBC, for their day 5 IgM antibody response by enumerating the number of AFC, spleen cell viability, and total recovered cells/culture. Bars, mean ± standard error, as determined for each group. *, p < 0.05, as determined by Dunnett’s t test, compared with the vehicle group.
[3H]CP-55,940 binding to spleen cells
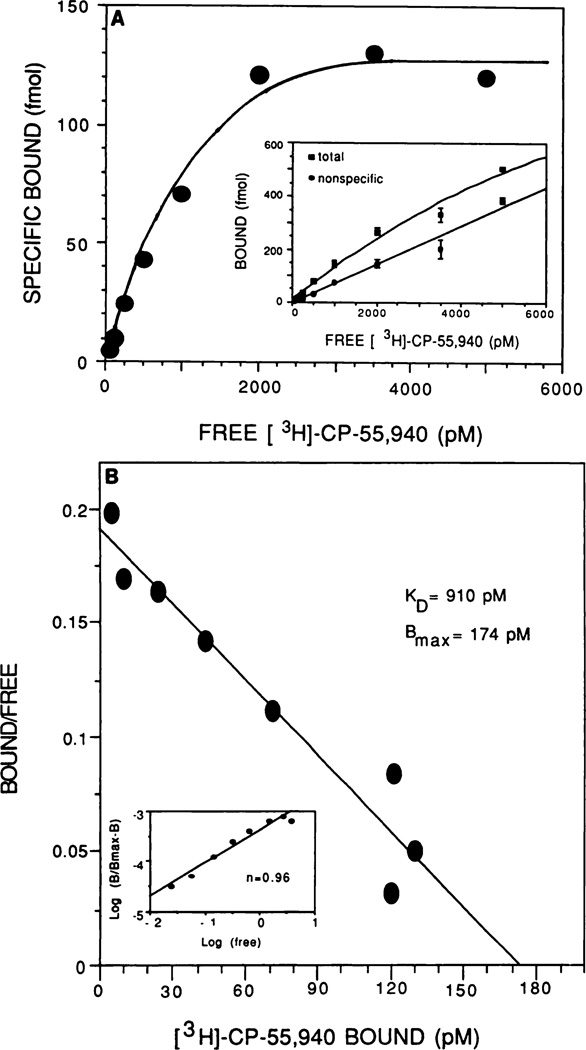
Radioligand-binding experiments revealed a high degree of specific binding of [3H]CP-55,940 to mouse spleen cells. As shown in the saturation isotherm (Fig. 3A), specific binding ranged from 45 to 65%. Scatchard analysis (Fig. 3B) demonstrated single-site binding on spleen cells, with a Kd of 910 pm and a Bmax of approximately 1000 receptors/spleen cell. Additionally, the Hill coefficient was approximately 1. Similar radioligand studies were performed using spleen cell lysates prepared by sonication (data not shown). No significant difference was observed in Kd or Bmax values between intact and disrupted spleen cells.
Fig. 3.
In vitro radioligand binding of [3H]CP-55,940 to mouse spleen cells. Spleens from naive female B6C3F1 mice were isolated aseptically and made into single-cell suspensions. The splenocytes were washed, adjusted to 2.0 × 106 cells/ml in buffer (50 mm Tris · HCl, pH 7.4, 1 mm EDTA, 3 mm MgCl2, 5 mg/ml BSA), and transferred in 1-ml aliquots to silanized glass culture tubes. Spleen cells were incubated in the presence of [3H]CP-55,940 at 30° for 1 hr. The reaction was terminated by addition of cold 50 mm Tris · HCl plus 5 mg/ml BSA (pH 7.4) and was then rapidly filtered through Whatman OF/C glass fiber filters. Specific binding was defined as the difference between the binding that occurred in the presence and in the absence of 1 µm unlabeled ligand. A, Saturation isotherm; B, Scatchard plot, for which EBDA analysis was used to determine Kd and Bmax values.
RNA PCR
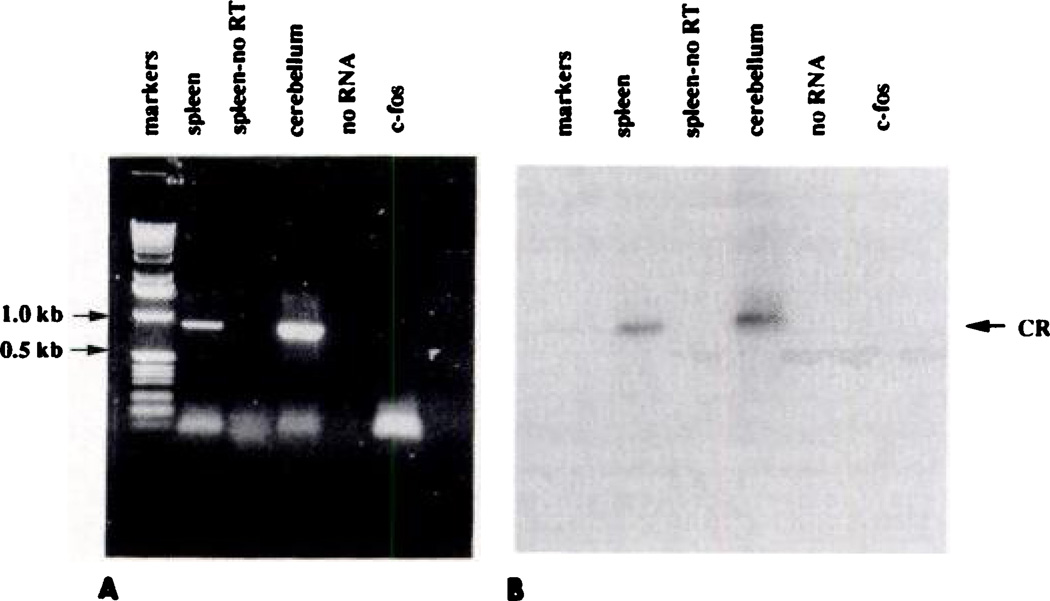
Enatioselective immune inhibition, specific binding of [3H] CP-55,940 to spleen cells, and our previously reported findings that Δ9-THC markedly inhibited forskolin-stimulated accumulation of intracellular cAMP in spleen cells (16) strongly implicate the presence of a G protein-coupled cannabinoid receptor in mouse spleen. However, Northern analysis of mouse spleen showed no detectable quantity of mRNA for the putative cannabinoid receptor (data not shown). This observation is consistent with similar findings reported for rat (2) and dog (7) spleen. In light of these negative results, RNA PCR was attempted, based on the possibility that message for the cannabinoid receptor is present in spleen but in quantities insufficient to be detected by Northern analysis. Total RNA was reverse transcribed into cDNA and then primers specific for the cDNA of interest were used to selectively amplify the desired product using PCR. In Fig. 4A, lane 1, the 835-bp predicted product using the cannabinoid receptor primers is shown. The gel was transferred to nylon membranes and hybridized with cannabinoid receptor cDNA probe. Fig. 4B shows the corresponding autoradiogram from this gel, indicating the presence of cannabinoid receptor mRNA in mouse spleen. Because of the extreme sensitivity of PCR and the possibility that contaminating genomic DNA was responsible for the identified splenic product, a series of controls were included in this study in order to rule out the possibility that the product identified was due to amplification of genomic DNA. In the first control, shown in Fig. 4B, lane 2, no product was observed when DNA PCR was performed on the splenic RNA preparation using the same primers as those used for RNA PCR. A second control, which consisted of another set of primers based on bp 234–254 and 324–344 (on the opposite strand) of rat c-fos cDNA, was also utilized. Intron 1 of the c-fos gene is between these primers, so any genomic DNA present would be seen as a 800-bp PCR product; otherwise, a 110-bp product would be produced from the c-fos mRNA. Only a 110-bp product was seen (Fig. 4B, lane 4).
Fig. 4.
Reverse transcription-PCR of mouse spleen RNA. RNA from splenocytes and rat brain cerebellum was amplified as described in Materials and Methods. A, Agarose gel Lanes 1–4, PCR products using the cannabinoid receptor primers (bp 1–21 and 822–843 on the opposite strand). Lane 1,2 µl from the RNA PCR reaction using splenocyte RNA, which is the predicted size of 835 bp. Lane 2, 2 µl from the DNA PCR reaction using splenocyte RNA, indicating the absence of DNA contamination of the RNA. Lane 3, 2 µl from the RNA PCR reaction using rat cerebellar RNA. Lane 4, product from the DNA PCR reaction with no added RNA. Lane 5, 2 µl of the PCR product from the RNA PCR reaction using the c-fos primers. These primers were based on bp 232–252 and 322–342 on the opposite strand of the c-fos gene. The predicted size of 110 bp, as seen, indicates the absence of DNA contamination (which would be seen as a 800-bp product). The gel was transferred to nylon membranes and hybridized with the cannabinoid cDNA probe. B, Resulting autoradiogram.
Discussion
Indirect evidence supporting the functional role of a G protein-coupled cannabinoid receptor in CNS-associated effects has been emerging over the past decade. These findings have provided a mechanistic framework to explain selective cannabinoid-mediated effects inconsistent with the notion that this class of agents act indirectly through disruption of cell membrane processes after intercalation into the lipid bilayer. Most compelling have been three lines of evidence, (i) enantioselective CNS-associated effects, (ii) inhibition of forskolin-stimulated adenylate cyclase, and (iii) a high degree of specific binding of the synthetic cannabinoid [3H]CP-55,940 to brain tissue. Direct evidence for the existence of a cannabinoid receptor has been recently forthcoming from two independent laboratories in which a G protein-coupled receptor has been isolated and cloned from human (15) and rat brain (2). Although cannabimimetic agents are widely established as being immunomodulatory, the mechanism for this inhibition has been elusive. A recent finding by our laboratory, i.e., immunoinhibitory but noncytotoxic concentrations of Δ9-THC dose-dependently inhibited forskolin stimulation of adenylate cyclase in mouse spleen cells (16), was the rationale for attempting to identify a cannabinoid receptor in mouse spleen.
One of the most compelling observations arguing against cannabinoid-mediated immunomodulation occurring via non-specific disruption of cell membrane processes is the marked difference observed in the potencies of the (−)- and (+)-cannabinoid stereoisomers. This was most apparent with (−)-CP-55,940 versus (+)-CP-56,667, as measured by inhibition of the sRBC AFC response. At concentrations at which the (−)-enantiomer, CP-55,950, produced approximately a 90% inhibition of the AFC response, the (+)-enantiomer, CP-56,667, was completely devoid of activity. These differences in potencies cannot be attributed to differences in lipophilicity between enantiomers, because both have identical chemical structures and lipophilic properties. It is also highly unlikely that this phenomenon is due to differences in drug metabolism, because cannabinoids are readily metabolized via cytochrome P-450 (21), a family of enzymes found in extremely low abundance in lymphoid cells. The observation that cannabinoid analogs possessing the dimethylheptyl aliphatic side chains have greater immunosuppressive potency than does Δ9-THC is also significant because it further underscores the similarities in activity of these compounds between the immune system and neuronal tissue and suggests that, structurally, the receptor may have little tissue to tissue variability.
Radioligand-binding studies with mouse spleen cells revealed a relatively high degree of CP-55,940 specific binding. On the other hand, the average number of receptors per spleen cell in this heterogeneous spleen cell pool was relatively small (~1000/spleen cell). Because a heterogeneous spleen cell preparation was utilized for these studies, it is presently unclear whether this receptor is present on all spleen cells in relatively low abundance or whether the receptor is present in higher abundance than predicted by these studies but expressed only on select spleen cell subpopulations. It is important to emphasize that recent characterization of β-adrenergic receptors on splenic B cells, also a G protein-coupled receptor, similarly revealed approximately 750–1500 receptors/cell, suggesting that this is not an unusually low number of receptors in association with lymphoid cells (22). Studies are presently underway in our laboratory to characterize the relative number of putative cannabinoid receptors expressed on purified splenic B cells, T cells, and macrophages. The Kd value observed in these studies, 910 pm, is slightly higher than those previously reported by Devane et al. (4) in rat brain P2 membrane preparations (Kd, 133 pm) using the same radioligand, [3H]CP-55,940; however, these differences may be partially due to assay differences (filtration versus centrifugation). Radioligand binding of [3H]HU-210 to rat brain P2 membrane preparations under conditions identical to those utilized for intact spleen cells resulted in a Kd of 1.2 nm (9). Similarly, Herkenham et al. (6) has reported a Kd of ~1 nm for [3H]CP-55,940 in rat brain slices. One intriguing aspect pertaining to these binding analyses, including our own studies, is that, in spite of the fact that cannabinoids demonstrate relatively high affinity of binding to the cannabinoid receptor, relatively high concentration of cannabinoids are required to produce functional effects in biological systems. This perplexing relationship for the cannabinoid receptor has been demonstrated in a number of tissue and cell preparations, including N18TG2 neuroblastoma cells (4, 11), rat brain preparations (6, 23), Sertoli cells (24), and mouse spleen cells (16). This phenomenon, as well as whether cannabinoid receptor subtypes exist, is currently being investigated.
Identification and quantitation of mRNA for the putative cannabinoid receptor have been elusive in splenic tissue. Northern analyses by several groups have been unable to identify mRNA for the cannabinoid receptor in rat (2) and dog spleen (7). Similar results were obtain in our own studies using mouse spleen. However, based on indirect evidence discussed above, which supported the presence of a cannabinoid receptor in spleen cells but in relatively low abundance, reverse PCR was performed in an attempt to amplify potentially low levels of mRNA for the putative receptor. Interestingly, the predicted product for the cannabinoid receptor was in fact amplified from splenic RNA using this approach. It is important to emphasize that a number of controls were included in these studies, which ruled out the possibility that the product was amplified from genomic DNA. These results were subsequently confirmed using RNase protection, a technique markedly more sensitive than Northern analysis for detecting and quantitating the presence of mRNA. These findings indicate the presence of mRNA for the cannabinoid receptor in mouse spleen and suggest that the reason why Northern analysis has proven to be an unsuitable technique for detecting the presence of this mRNA may be due to either its very low abundance in spleen, as predicted by our binding data, and/or the possibility that the mRNA for the cannabinoid receptor in spleen is highly unstable.
Many questions remain unanswered pertaining to the putative cannabinoid receptor, including (i) its endogenous ligand, (ii) its role in the CNS, and (iii) its role in non-neural tissues, to mention several. However, it must be emphasized that the cannabinoid receptor has been found in two separate non-neural tissues, human testis (7) and, now, mouse spleen. The receptor as identified in both rat (2) and human brain (15) has >97% homology between these two species. In our studies, a rat cDNA for the receptor isolated from rat brain was used successfully to identify the presence of the cannabinoid receptor in mouse spleen, indirectly suggesting homology with the rat and human receptor. These findings, as well as the fact that the cannabinoid receptor is highly conserved between species, suggest a more holistic functional role for the receptor than would have initially been predicted if it were present solely in association with the CNS.
In summary, this series of studies indicate the presence of the putative cannabinoid receptor on spleen cells and implicate its role in immune modulation by cannabimimetic agents. Its presence is supported by the stereoselective immunomodulatory effects produced by cannabimimetic agents, by the high degree of specific binding demonstrated with [3H]CP,55,940 with mouse spleen cells, and by the presence of mRNA for the cannabinoid receptor in mouse spleen. Additionally, previous studies from our laboratory demonstrated that Δ9-THC markedly inhibited forskolin-stimulated cAMP accumulation in mouse spleen cells (16), results similar to those reported using brain-derived tissue preparations and cell lines (10–14). These findings are significant because they provide a mechanism by which this class of agents interact with the immune system, as well as providing a potentially relevant model for further characterizing the functional role of the cannabinoid receptor, not only in immune modulation but also in other tissues, including the CNS.
Acknowledgments
This work was supported in part by funds from Grants DA07908, DA03672, and DA05274 and Thomas and Kate Miller Jeffress Memorial Trust Grant J-183.
ABBREVIATIONS
- Δ9-THC
Δ9-tetrahydrocannabinol
- sRBC
sheep red blood cells
- G protein
guanine nucleotide-binding protein
- CNS
central nervous system
- AFC
antibody-forming cell
- PCR
polymerase chain reaction
- HU-210/HU-211
11-OH-Δ8-tetrahydrocannabinol-dimethylheptyl
- CP-55,940/CP-56,667
(cis)-3-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-(trans)-4-(3-hydroxypropyl)cyclohexanol
- bp
base pair(s)
- BsA
bovine serum albumin
References
- 1.Munson AE, Fehr KO. Immunological effects of cannabis. In: Fehr KO, Kalant H, editors. Adverse Health and Behavioral Consequences of Cannabis Us. Addiction Research Foundation; 1983. pp. 257–353. [Google Scholar]
- 2.Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature (Lond.) 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- 3.Howlett AC, Johnson MR, Melvin LS, Milne GM. Nonclassical cannabinoid analgetics inhibit adenylate cyclase: development of a cannabinoid receptor model. Mol. Pharmacol. 1988;23:297–302. [PubMed] [Google Scholar]
- 4.Devane WA, Dysarz FA, Johnson MR, Melvin LS, Howlett AC. Determination and characterization of a cannabinoid receptor in rat brain. Mol. Pharmacol. 1988;34:605–613. [PubMed] [Google Scholar]
- 5.Little PJ, Compton DR, Johnson MR, Melvin LS, Martin BR. Pharmacology and stereoselectivity of structurally novel cannabinoids in mice. J. Pharmacol. Exp. Ther. 1988;247:1046–1051. [PubMed] [Google Scholar]
- 6.Herkenham M, Lynn AB, Little MD, Johnson MR, Melvin LS, de Costa BR, Rice KC. Cannabinoid receptor localization in brain. Proc. Natl. Acad. Sci. USA. 1990;87:1932–1936. doi: 10.1073/pnas.87.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerard CM, Mollereau C, Vassart G, Parmentier M. Molecular cloning of a human cannabinoid receptor which is also expressed in testis. Biochem. J. 1991;279:129–134. doi: 10.1042/bj2790129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris LS, Carchman RA, Martin BR. Evidence for the existence of specific cannabinoid binding sites. Life Sci. 1978;22:1131–1138. doi: 10.1016/0024-3205(78)90082-6. [DOI] [PubMed] [Google Scholar]
- 9.Martin BR, Compton DR, Thomas BF, Prescott WR, Little PJ, Razdan RK, Johnson MR, Melvin LS, Mechoulam R, Ward SJ. Behavioral, biochemical, and molecular modeling evaluations of cannabinoid analogs. Pharmacol. Biochem. Behav. 1991;40:471–478. doi: 10.1016/0091-3057(91)90349-7. [DOI] [PubMed] [Google Scholar]
- 10.Howlett AC, Fleming RM. Cannabinoid inhibition of adenylate cyclase: pharmacology of the response in neuroblastoma cell membranes. Mol. Pharmacol. 1984;26:532–538. [PubMed] [Google Scholar]
- 11.Howlett AC. Cannabinoid inhibition of adenylate cyclase: biochemistry of the response in neuroblastoma cell membranes. Mol. Pharmacol. 1985;27:429–436. [PubMed] [Google Scholar]
- 12.Howlett AC, Qualy JM, Khachtrian LL. Involvement of G1 in the inhibition of adenylate cyclase by cannabimimetic drugs. Mol. Pharmacol. 1985;29:307–313. [PubMed] [Google Scholar]
- 13.Howlett AC. Cannabinoid inhibition of adenylate cyclase: relative activity of constituents and metabolites of marihuana. Neuropharmacology. 1987;26:507–512. doi: 10.1016/0028-3908(87)90035-9. [DOI] [PubMed] [Google Scholar]
- 14.Bidaut-Russell M, Howlett AC. Cannabinoid receptor-regulated cyclic AMP accumulation in the rat striatum. J. Neurochem. 1991;57:1769–1773. doi: 10.1111/j.1471-4159.1991.tb06379.x. [DOI] [PubMed] [Google Scholar]
- 15.Gerard CM, Mollereau C, Vassart G, Parmentier M. Nucleotide sequence of a human cannabinoid receptor cDNA. Nucleic Acids Res. 1990;18:7142. doi: 10.1093/nar/18.23.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schatz AR, Kessler FK, Kaminski NE. Inhibition of adenylate cyclase by Δ9-tetrahydrocannabinol in mouse spleen cells: a potential mechanism for cannabinoid-mediated immunosuppression. Life Sci. 1992;51:25–30. doi: 10.1016/0024-3205(92)90414-k. [DOI] [PubMed] [Google Scholar]
- 17.Bartlett MS. Sub-sampling for attributes. J. R. Stat. Soc. Suppl. 1937;4:131. [Google Scholar]
- 18.Dunnett CW. A multiple comparison procedure for comparing several treatments with a control. J. Am. Stat. Assoc. 1955;50:1096–1121. [Google Scholar]
- 19.Gehan-Wilcoxon . In: Survival Distributions: Reliability Applications in the Biomedical Sciences. Gross AJ, Clark A, editors. New York: John Wiley and Sons; 1975. pp. 102–103. [Google Scholar]
- 20.Little PJ, Compton DR, Mechoulam R, Martin BR. Stereochemical effects of 11-OH-Δ8-THC-dimethylheptyl in mice and dogs. Biochem. Behav. 1989;32:661–666. doi: 10.1016/0091-3057(89)90014-2. [DOI] [PubMed] [Google Scholar]
- 21.Harvey DJ, Paton DM. Metabolism of cannabinoids. Rev. Biochem. Toxicol. 1984;6:221–264. [Google Scholar]
- 22.Fuchs BA, Albright JW, Albright JF. Beta-adrenergic receptors on murine lymphocytes: density varies with cell maturity and lymphocyte subtype and is decreased after antigen administration. Cell. Immunol. 1988;114:231–245. doi: 10.1016/0008-8749(88)90318-8. [DOI] [PubMed] [Google Scholar]
- 23.Little P, Martin BR. The effects of Δ9-tetrahydrocannabinol and other cannabinoids on cAMP accumulation in synaptosomes. Life Sci. 1991;48:1133–1141. doi: 10.1016/0024-3205(91)90450-p. [DOI] [PubMed] [Google Scholar]
- 24.Heindel JJ, Keith WB. Specific inhibition of FSH-stimulated cAMP accumulation by Δ9-tetrahythocannabinol in cultures of rat Sertoli cells. Toxicol. Appl. Pharmacol. 1989;101:124–134. doi: 10.1016/0041-008x(89)90218-4. [DOI] [PubMed] [Google Scholar]