Abstract
Demand for nonnutritive sweeteners continues to increase due to their ability to provide desirable sweetness with minimal calories. Acesulfame potassium and saccharin are well-studied nonnutritive sweeteners commonly found in food products. Some individuals report aversive sensations from these sweeteners, such as bitter and metallic side tastes. Recent advances in molecular genetics have provided insight into the cause of perceptual differences across people. For example, common alleles for the genes TAS2R9 and TAS2R38 explain variable response to the bitter drugs ofloxacin in vitro and propylthiouracil in vivo. Here, we wanted to determine whether differences in the bitterness of acesulfame potassium could be predicted by common polymorphisms (genetic variants) in bitter taste receptor genes (TAS2Rs). We genotyped participants (n = 108) for putatively functional single nucleotide polymorphisms in 5 TAS2Rs and asked them to rate the bitterness of 25 mM acesulfame potassium on a general labeled magnitude scale. Consistent with prior reports, we found 2 single nucleotide polymorphisms in TAS2R31 were associated with acesulfame potassium bitterness. However, TAS2R9 alleles also predicted additional variation in acesulfame potassium bitterness. Conversely, single nucleotide polymorphisms in TAS2R4, TAS2R38, and near TAS2R16 were not significant predictors. Using 1 single nucleotide polymorphism each from TAS2R9 and TAS2R31, we modeled the simultaneous influence of these single nucleotide polymorphisms on acesulfame potassium bitterness; together, these 2 single nucleotide polymorphisms explained 13.4% of the variance in perceived bitterness. These data suggest multiple polymorphisms within TAS2Rs contribute to the ability to perceive the bitterness from acesulfame potassium.
Key words: bitterness, genetics, noncaloric sweetener, Project GIANT-CS, saccharin, taste phenotype
Introduction
Taste is the number one driver of food choices (Glanz et al. 1998; IFIC 2011), and sweetness is innately liked by humans (reviewed by Steiner et al. 2001), even prior to birth (de Snoo 1937). Accordingly, many highly liked foods contain high endogenous amounts of natural sugars or have sugars or other sweeteners added during processing. However, although consumers continue to desire sweetened products, there is also demand for reduced added sugar in foods due to associated health risks such as cardiovascular disease, diabetes, and obesity (Hill and Prentice 1995; Howard and Wylie-Rosett 2002). To retain desired levels of sweetness while reducing calories, bulk carbohydrates are often replaced with nonnutritive sweeteners. Replacing sugar with nonnutritive sweeteners may help with managing energy intake (Tordoff and Alleva 1990; Duffy and Anderson 1998; Raben et al. 2002; De la Hunty et al. 2006), although not all evidence supports this view (Stellman and Garfinkel 1986; Anderson et al. 2012). These nonnutritive sweeteners can be natural (e.g., rebaudioside A) or synthetic (e.g., saccharin), each with varying level of potency (DuBois et al. 1991; Hayes 2008).
Acesulfame potassium (AceK) was approved by the US Food and Drug Administration for use in dry foods in 1994, and its approval as a general-purpose sweetener followed in 2002. According to Mintel market research data, AceK was the most used nonnutritive sweetener in new product launches between 2004 and 2010. However, in addition to eliciting sweet sensations, many nonnutritive sweeteners also have objectionable side tastes, such as bitterness, that are experienced by some individuals but not others (Kamerud and Delwiche 2007). This bitterness is concentration dependent (Schiffman et al. 1979; Horne et al. 2002), so one solution to reduce aversive bitterness is to use mixtures of these high potency sweeteners, either with each other or with bulk carbohydrate sweeteners (Hanger et al. 1996). This method has been used with some degree of success commercially (e.g., Coke Zero), although anecdotal reports suggest some individuals still find the taste of these blends objectionable. Thus, better understanding of the mechanisms underlying this variability may facilitate improved product formulation, with the potential to substantially impact health and wellness.
There are 25 bitter taste receptor genes (TAS2Rs) in humans (Adler et al. 2000; Chandrashekar et al. 2000; Meyerhof et al. 2010), and these genes are highly polymorphic compared with the rest of the genome (Kim et al. 2005). Beginning with the deorphanization of hT2R4 and hT2R38 a decade ago (Chandrashekar et al. 2000; Kim et al. 2003), substantial progress has been made in identifying ligands for the majority of these receptors (Meyerhof et al. 2010). With regard to nonnutritive sweeteners, Kuhn et al. (2004) demonstrated receptors encoded by TAS2R31 (formerly called TAS2R44) and TAS2R43 are activated by saccharin and AceK in vitro. Moreover, perceived bitterness of these 2 sulfonyl amide sweeteners is greatly reduced by cross-adaptation to aristolochic acid, a purely bitter hT2R31/hT2R43 agonist (Kuhn et al. 2004). These data are largely consistent with earlier psychophysical data showing that the bitterness of AceK and saccharin covaries with each other but not with propylthiouracil (Horne et al. 2002). Like propylthiouracil (e.g., Hayes et al. 2008) and grapefruit juice (Hayes et al. 2011), the bitterness of AceK and saccharin varies across individuals, and this variation has a genetic basis (Pronin et al. 2007; Roudnitzky et al. 2011). Roudnitzky et al. (2011) generated long-range haplotypes across 5 highly related TAS2Rs on chromosome 12 and identified a single common haplotype (out of 7 common haplotypes) that was associated with AceK and saccharin bitterness. By mutating single residues in artificial chimeric receptors, they were able to demonstrate in vitro that the Arg35Trp (R35W) polymorphism in TAS2R31 was causal. However, they also found that even in the presence of the high functioning Trp35 allele, other TAS2R31 mutations abolished the ability of hT2R31 to respond to AceK and saccharin.
As outlined above, numerous studies have demonstrated that the perception of bitter taste in humans is moderated by genetic variation in TAS2R genes (Duffy, Davidson, et al. 2004; Pronin et al. 2007; Reed et al. 2010; Hayes et al. 2011; Roudnitzky et al. 2011). However, many human behavioral studies have focused on single genetic variants, which neglects the fact that both bitter tastants and bitter receptors can be highly promiscuous (Meyerhof et al. 2010). This approach is problematic because the examination of any single variant may be obscured by noise in other variants. That is, effects of one isolated allele can be overshadowed by an aggregate effect of several other alleles that are high or low functioning (see Lotsch et al. 2009 for a nontaste example). Here, we explore the influence of putatively functional polymorphisms in multiple TAS2Rs on AceK bitterness in individual single nucleotide polymorphism (SNP) analyses before modeling these effects simultaneously.
Materials and methods
Overview
Data presented here are part of a larger, ongoing study of the genetics of oral sensation (Project GIANT-CS). This study involves 4 laboratory sessions across days; here, we only describe the first day of testing. On Day 1, the study was explained to participants and consent was obtained. Participants then completed a food-liking questionnaire. Next, anthropometric data and salivary DNA samples were collected, followed by digital microscopy of the anterior tongue. Participants were oriented to the psychophysical scale, and sampled 6 perceptually complex tastants and irritants, rating them for multiple qualities. Finally, participants completed a standard propylthiouracil (PROP) phenotyping protocol with PROP, salt, and tones. After leaving the laboratory, participants completed several personality questionnaires via web form. Total time in the laboratory for session 1 was ~1 h; all data were collected one-on-one by project staff.
Participants
Prospective participants were prescreened to ensure they qualified. Eligibility criteria included the following: between 18 and 45 years old; not pregnant or breastfeeding; nonsmoker (had not smoked in the last 30 days); no known defects of smell or taste; no lip, cheek, or tongue piercings; no history of any condition involving chronic pain; not currently taking any prescription pain medication; no reported history of choking or difficulty swallowing; and no history of thyroid disease. Participants also needed to be willing to provide a DNA sample via saliva. Written informed consent was obtained from all participants. All procedures were approved by the Pennsylvania State University Institutional Review Board (protocol number #33176).
DNA samples were available from 147 participants. Race and ethnicity was self-reported using categories provided by the 1997 OMB Directive 15. To minimize potential population stratification, which can potentially cause false negatives and false positives in gene association studies (Hamer and Sirota 2000), individuals with Asian (n = 18), African (n = 5), or unknown (n = 15) ancestry were excluded from the present analyses. Thus, we report data from 108 participants (34 men) of European ancestry, with a mean age of 27.4 (±8.1 SD) years. Results were not substantively different in the mixed ancestry sample, but we report only the results for the European–American participants to facilitate interpretation of the linkage disequilibrium (LD) plots.
Psychophysical scaling
A general labeled magnitude scale (gLMS) was used to collect perceived intensity of suprathreshold stimuli (Snyder, Fast et al. 2004). This scale ranges from 0 (“no sensation”) to 100 (“the strongest imaginable sensation of any kind”), with intermediate descriptors at 1.4 (“barely detectable”), 6 (“weak”), 17 (“moderate”), 35 (“strong”), and 51 (“very strong”). All participants participated in an orientation to the scale, making ratings for a list of 15 imagined or remembered sensations that included both oral and nonoral items (Hayes et al. 2013). The orientation procedure and scale instructions were intended to promote use of the scale in a generalized context not limited to oral sensations. All psychophysical data were collected using Compusense five, version 5.2 (Guelph).
Sampling and rating of perceptually complex stimuli
Six food-grade stimuli were presented in 10-mL aliquots: 0.56M potassium chloride (Spectrum) (salty/bitter), 0.41mM quinine HCl (Sigma–Aldrich) (bitter), 25mM Acesulfame K (Spectrum) (sweet/bitter), 100mM monosodium glutamate (MSG) (Ajinomoto) + 50 mM inosine monophosphate (IMP) (Ajinomoto) (umami/savory), 0.5M sucrose (Domino) (sweet), and 25 μM capsaicin (Sigma–Aldrich) (burning/stinging). Only AceK and PROP (see next section) data are reported here; data for the other stimuli will be reported elsewhere. A pilot study and prior experience were used to determine appropriate concentrations. The concentrations were selected to produce a sensation near “moderate” on a gLMS for the main perceptual quality of the stimulus.
After being told, “You may receive stimuli causing more than one quality. Please attend to all sensations on all trials,” participants swished the 10-mL sample for 3 s and then expectorated prior to rating. Separate intensity ratings were obtained for sweetness, bitterness, sourness, burning/stinging, savory/umami, and saltiness on the gLMS. Presentation order was a counterbalanced Williams’ design. Participants rinsed with room temperature reverse osmosis (RO) water prior to the first sample and between each sample. A minimum interstimulus interval of 30 s was enforced between samples.
Measuring PROP phenotype
PROP phenotype was determined using a standard concentration series with PROP, sodium chloride (salt), and sound, as described elsewhere (e.g., Duffy, Peterson, et al. 2004; Dinehart et al. 2006; Hayes et al. 2010). Briefly, participants rated the intensity of 10 PROP solutions, 10 salt solutions, and twenty-five 1-kHz tones. Stimuli were blocked so participants received 5 tones, 5 salt solutions, 5 tones, 5 salt solutions, 5 tones, 5 PROP solutions, 5 tones, 5 PROP solutions, and 5 tones. Block order was fixed; stimulus order within a block was counterbalanced. Half log steps were used for PROP solutions (3.2, 1, 0.32, 0.1, and 0.032mM) and the salt solutions (0.01, 0.032, 0.1, 0.32, and 1 M). The 1-kHz tones were generated with a Maico MA39 audiometer calibrated to deliver the specified sound pressure level binaurally; stimuli ranged from 50 to 90 dB in 10 dB steps. The tastants were prepared with USP grade 6-n-propylthiouracil (Sigma) and kosher salt in RO water. Participants rinsed with room temperature RO water between each sample, waiting a minimum of 30 s before next sample. Overall intensity ratings for each stimulus were obtained with a gLMS. Mean intensity of the top PROP concentration (3.2mM) was used as a continuous variable.
Genetic analysis
DNA was collected from saliva using Oragene collection kits according to manufacturer instructions (Genotek Inc.). SNPs in TAS2R4, TAS2R16, and TAS2R38 on chromosome 7 and TAS2R9 and TAS2R31 on chromosome 12 were determined using Sequenom MassARRAY technology (Sequenom). All primers were purchased from Integrated DNA Technologies. Genotypes were assigned automatically via MassARRAY software (Sequenom) and independently inspected by 2 technicians. As a standard procedure, 15% of samples are rerun to ensure reliability.
For the Arg35Trp (rs10845295) SNP in TAS2R31, attempts were made to obtain custom assays using 2 different technologies (Sequenom MassARRAY and custom made to order TaqMan assays); neither approach was successful despite repeated efforts. Thus, a tag SNP approach is used here for TAS2R31, as the only published method for determining the Arg35Trp SNP, direct sequencing, is beyond the scope of the current project.
Statistical analysis
Data were analyzed using SAS 9.2. Prior to analyses, the smallest possible rating on the gLMS (0.5) was added to all psychophysical ratings to eliminate zeros, and data were log-transformed. For analysis of individual SNPs, analysis of variance (ANOVA) was performed via proc mixed in SAS. Post hoc comparisons were made via the Tukey–Kramer method. For the significant SNPs, we then tested for deviations from a simple additive model using the 2-step approach recommended by Carey (2007). Finally, we used multiple regression to assess the independent influence of multiple putatively functional SNPs on the phenotype simultaneously. Because we did not observe evidence of nonadditivity (see Results), a simple count method was used to code each SNP; a participant was given an allele score of 0, 1, or 2 corresponding to the number of putatively high function alleles—based on the results of the ANOVAs for each SNP—the individual possessed (e.g., a simple additive model for each SNP). The recoded SNP variables were then used to predict the perceived bitterness of AceK via proc reg in SAS with a separate term for each SNP.
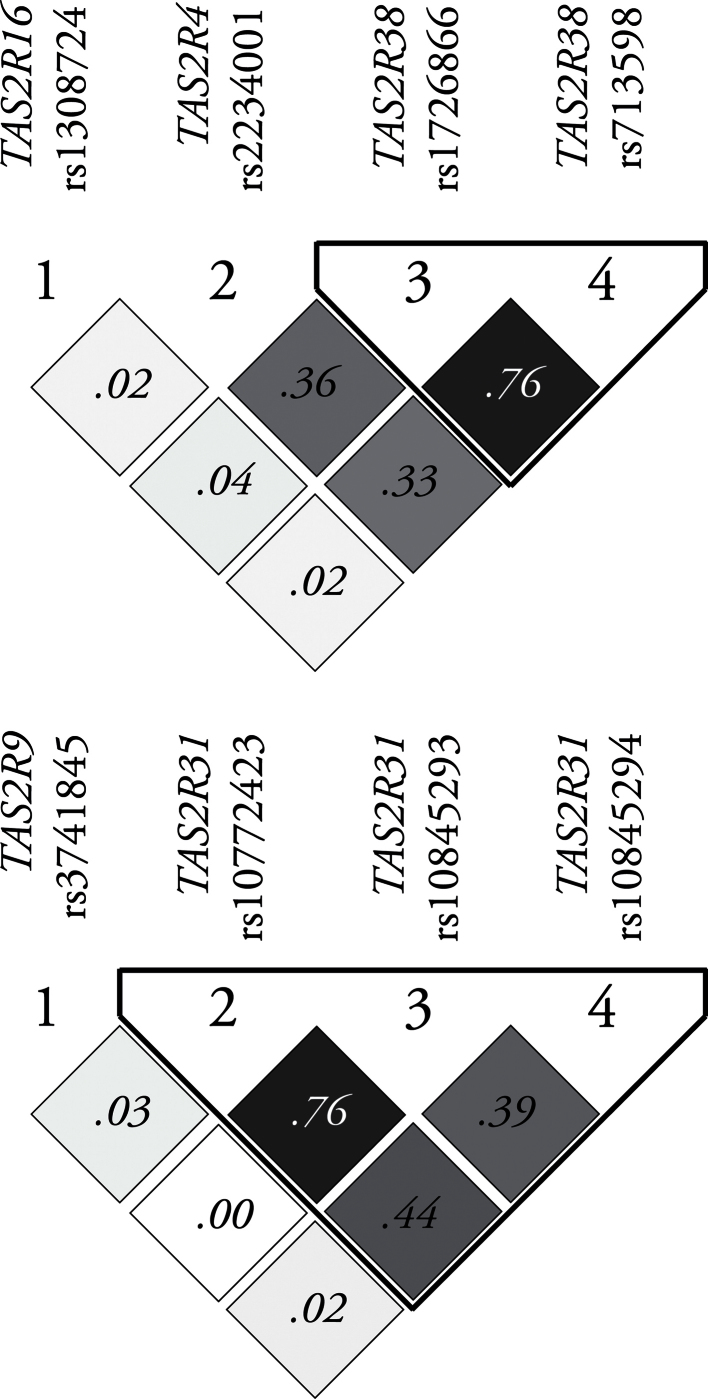
For TAS2R38, polymorphisms at amino acid residues 49 (Ala49Pro) and 262 (Val262Ala) are known to form 2 common haplotypes: the Proline–Alanine (PA_) haplotype is ancestral, bestowing the ability to sense thiourea (N–C=S) compounds, whereas the Alanine–Valine (AV_) variant is less functional (Wooding et al. 2004). The program Haploview (Barrett et al. 2005) was used to examine the extent of LD between each pair of markers and to determine haplotype block structure. Haplotype blocks were defined according to solid spine of LD criteria (Barrett et al. 2005). Haplotype pairs were assigned to each participant of European ancestry using PHASE (Stephens et al. 2001; Stephens and Donnelly 2003). PHASE estimates the probabilities of all likely pairs of haplotypes (diplotypes) assigned to each individual from genotype data. Of these, diplotypes assigned with a probability of ≥0.80 were selected for further analysis. LD plots (Figure 1) show rounded R 2 values in individual squares, and shading is used to represent the exact R 2 value, with darker shades of gray indicating larger R 2 values. All genotyping, construction, and assignment of haplotypes were done blind to outcome variables.
Figure 1.
LD plot for TAS2R SNPs on chromosomes 7 (top) and 12 (bottom). Numbers indicate rounded R 2 values and shading indicates exact R 2 values generated via Haploview.
Results
TAS2R9 alleles predict AceK bitterness
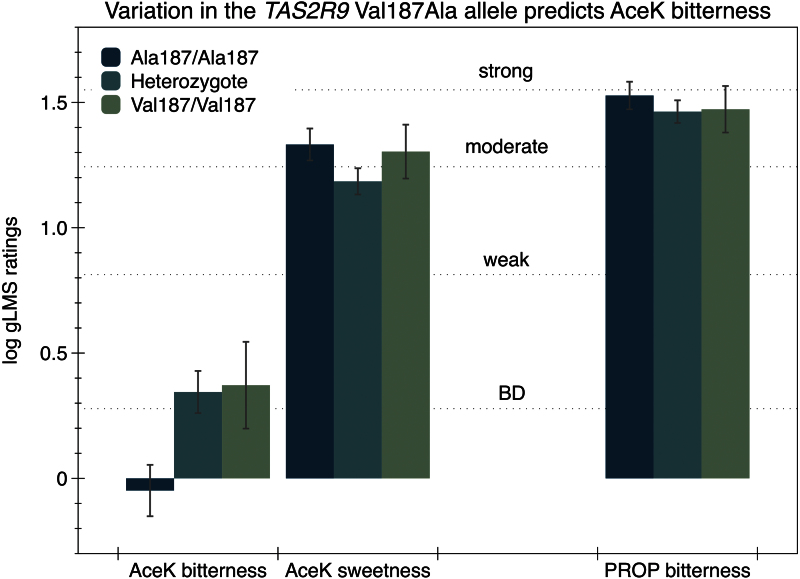
Dotson et al. (2008) reported hT2R9 responds to the bitter drugs ofloxacin, procainamide, and pirenzepine, and this response varies with a missense polymorphism in the TAS2R9 gene (Val187Ala; rs3741845; chr 12). Here, we find evidence that this allele is also functional for AceK bitterness, as ANOVA revealed this SNP was significantly associated with the bitterness of AceK [F(2,102) = 4.89; P = 0.009]. Group means of logged ratings are shown in Figure 2; the Ala187 homozygotes (n = 37) reported less bitterness than heterozygotes (n = 55) (Tukey–Kramer P = 0.011) and the Val187 homozygotes (n = 13) (Tukey–Kramer P = 0.097). The heterozygotes and Val187 homozygotes did not differ (Tukey–Kramer P = 0.987). In the additive regression model, this SNP explained 7.0% of the variance in logged AceK bitterness (P = 0.006); there was no evidence of dominance (P = 0.16).
Figure 2.
Effect of the TAS2R9 Val187Ala polymorphism on the bitterness and sweetness of AceK and bitterness of PROP. The bitterness of AceK was significantly different across genotype; no effect was observed for AceK sweetness or PROP bitterness (P values provided in text). Adjectives refer to semantic labels on a gLMS (see text). BD refers to “barely detectable.”
In contrast, there was no evidence that the Val187Ala allele predicted AceK sweetness [F(2,102) = 1.73; P = 0.18]. Also, as would be expected, the Val187Ala allele did not predict the bitterness of PROP [F(2,102) = 0.42; P = 0.66]. This SNP is not in LD with any other SNPs on chromosome 12 (Figure 1; bottom).
TAS2R31 alleles predict variation in AceK bitterness
We explored the role of 3 TAS2R31 SNPs previously implicated in AceK bitterness by Roudnitzky et al. (2011). Below, we describe individual analyses for each SNP; however, these 3 SNPs are in LD, as shown in Figure 1 (bottom). Prior evidence (Roudnitzky et al. 2011) suggests that these SNPs are not causal, but are in strong LD with the causal Trp35 allele. We were unable to directly measure the Arg35Trp polymorphism, as attempts to obtain custom primers for either Sequenom or TaqMan methods were unsuccessful, necessitating a tag SNP approach here.
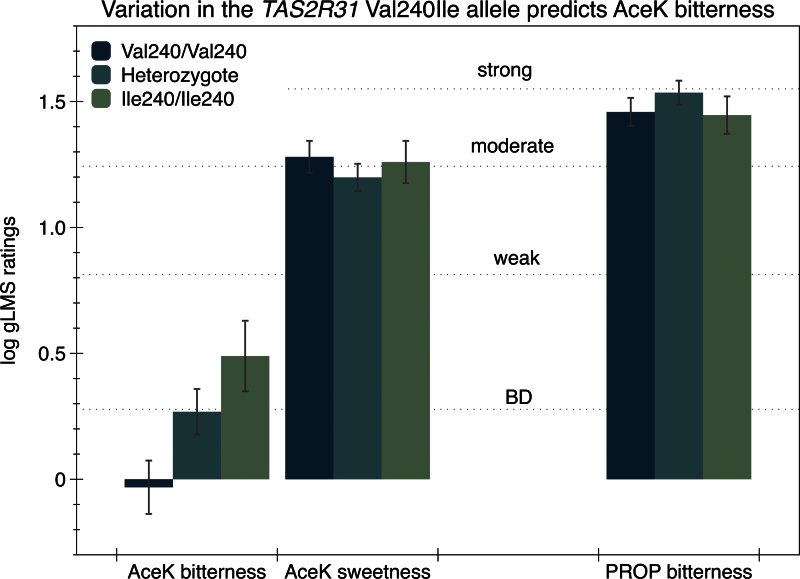
In ANOVA, the Val240Ile SNP (rs10772423; chr 12) was significantly associated with the bitterness of AceK [F(2,100) = 4,80; P = 0.010). As shown in Figure 3, the Val240 homozygotes (n = 35) reported less bitterness from AceK than the Ile240 homozygotes (n = 20) (Tukey–Kramer P = 0.010). The Val/Ile heterozygotes (n = 48) fell in the middle, as the heterozygote group mean tended to be higher than the Val240 homozygotes (Tukey–Kramer P = 0.085), but did not differ from the Ile240 homozygotes (Tukey–Kramer P = 0.38). In the additive regression model, this SNP explained 8.7% of the variance in logged AceK bitterness (P = 0.003); there was no evidence of dominance (P = 0.75). Again, we failed to find evidence this SNP predicted AceK sweetness [F(2,100) = 0.1.43; P = 0.24] or PROP bitterness [F(2,100) = 0.77; P = 0.47].
Figure 3.
Effect of the TAS2R31 Val240Ile polymorphism on the bitterness and sweetness of AceK and bitterness of PROP. The bitterness of AceK was significantly different across genotype; no effect was observed for AceK bitterness or PROP bitterness (P values provided in text).
We also found a similar effect pattern for the Ala227Val SNP (rs10845293; chr 12) [F(2,98)= 3.55; P = 0.032]. The group means of the logged bitterness ratings for the Val227 homozygotes (n = 33), Ala/Val heterozygotes (n = 49), and Ala227 homozygotes (n = 19) are shown in Supplementary Figure 1. Because this SNP was in LD with the Val240Ile SNP, we did not test for additivity or dominance. We found no evidence that the Ala227Val SNP was associated with AceK sweetness [F(2,98) = 0.15; P = 0.86] or the perceived bitterness ratings of PROP [F(2,98) = 1.27; P = 0.29].
Finally, we also explored the influence of the Gln217Glu SNP (rs10845294; chr 12) on AceK bitterness. We did not have a sufficiently large cohort to detect a significant effect [F(2,102) = 1.28; P = 0.28] given the low number of Glu217 homozygotes (n = 6) compared with heterozygotes (n = 39) and Gln217 homozygotes (n = 60). Previously, Roudnitzky et al. (2011) showed that having either the Glu217 or Gln217 residue did not change the activation of hT2R31 in vitro when the Trp35 variant was present. The sweetness of AceK did not significantly differ across Gln217Glu [F(2,102) = 1.41; P = 0.25] nor was this SNP associated with the reported bitterness ratings of PROP [F(2,102) = 0.08; P = 0.93].
Collectively, these results confirm that TAS2R31 contains a functional polymorphism that predicts at least some of the variation in the suprathreshold bitterness of AceK. Present data also suggest that hT2R31 mediates the bitterness of AceK but not PROP. Additionally, the TAS2R31 alleles do not appear to influence sweetness, at least at the concentration tested here.
TAS2R38 diplotypes predict variation in the bitterness of PROP but not AceK
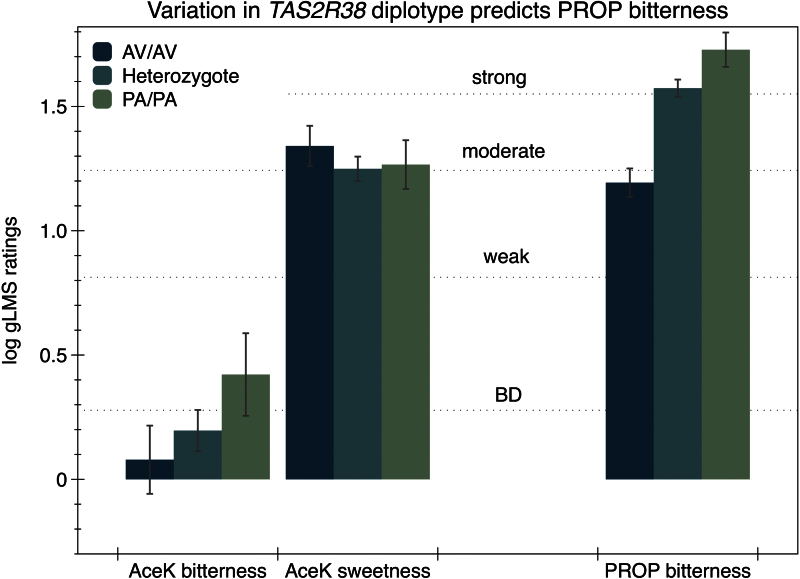
Two well-characterized SNPs in TAS2R38 on chromosome 7 were measured here: Ala49Pro (rs713598) and Val262Ala (rs1726866). These SNPs are well known to exhibit strong LD and there is evidence of 2 common haplotypes (Wooding et al. 2004). This was verified in our data via Haploview. Using PHASE, the following common diplotypes (probability ≥ 0.80) were assigned in our data: AV_ homozygotes, AV_/PA_ heterozygotes, and PA_ homozygotes, and tested whether they predicted AceK bitterness. Individuals with rare diplotypes (n = 11) were excluded a priori from the analysis as these rare haplotypes are known to have intermediate phenotypes that differ both from each other and the common haplotypes (Bufe et al. 2005; Hayes et al. 2008; Mennella et al. 2011), so binning them together in 1 group is not justified. Here, these included AA/PA (4), AA/AV (3), PV/AV (2), PV/PA (1), and PV/PV (1) individuals. We did not observe any evidence of a relationship between common TAS2R38 diplotypes and the bitterness of AceK [F(2,94) = 1.28; P = 0.28] or sweetness of AceK [F(2,94) = 0.49; P = 0.62] (Figure 4).
Figure 4.
Effect of the AV/PA TAS2R38 polymorphism on the bitterness and sweetness of AceK and bitterness of PROP. PROP bitterness differed by diplotype; differences in AceK bitterness or sweetness across diplotype were not significant (P values provided in text).
As expected, the bitterness of PROP differed with TAS2R38 diplotype [F(2,94)= 22.02; P < 0.0001]. The AV_ homozygotes (n = 22) reported significantly less bitterness than the heterozygotes (n = 60; Tukey–Kramer P < 0.001) or PA_ homozygotes (n = 15) (Tukey–Kramer P < 0.001); mean bitterness for the heterozygous individuals was similar to the PA_homozygotes (Tukey–Kramer P = 0.12), consistent with numerous other reports (Duffy, Davidson, et al. 2004; Bufe et al. 2005; Hayes et al. 2008; Mennella et al. 2010; Calo et al. 2011).
Relationship between PROP bitterness and AceK bitterness and sweetness
Prior reports conflict as to whether the bitterness of sulfonyl amide sweeteners is related to the bitterness of PROP (Bartoshuk 1979; Horne et al. 2002). Here, we found minimal evidence that PROP bitterness was predictive of AceK bitterness (R 2 = 2.9%; P =0.077). Conversely, the bitterness of PROP was positively associated with AceK sweetness, predicting 10.2% of the variation (P < 0.001).
AceK bitterness was not predicted by putatively functional SNPs in TAS2R4 or near TAS2R16
We also tested a putatively functional SNP in TAS2R4 and a putatively functional SNP 9.4 kb downstream from TAS2R16 (Hayes et al. 2011). These 2 SNPs did not exhibit LD with each other or with the TAS2R38 haplotype (Figure 1, top). There was little to no evidence that the bitterness of AceK varied with rs2234001 in TAS2R4 [F(2,97)= 0.69; P = 0.50]. There was no evidence that the rs1308724 SNP near TAS2R16 was a significant predictor of AceK bitterness [F(2,98)= 0.16; P = 0.85].
Effect of multiple loci on AceK bitterness
Although the 25 TAS2Rs are highly polymorphic, empirical evidence that these polymorphisms are functional has only been shown for 5 receptor genes (TAS2R4, TAS2R16, TAS2R19, TAS2R31, and TAS2R38). Here, we tested one or more candidate SNPs in 4 of these genes in regard to the bitterness of AceK. Based on our findings for the individual genes above, we used a simple regression model to assess the simultaneous influence of a single polymorphism in each gene—TAS2R9 (Val187Ala) and TAS2R31 (Val240Ile)—on AceK bitterness. Because the effects of these 2 SNPs did not appear to deviate from additivity, a score of 1 was assigned for each putatively functional allele for each of the SNPs, resulting in 2 variables coded 0, 1, and 2. AceK bitterness was regressed against these 2 recoded variables. This model explained 13.4% of the variance in bitterness (P < 0.001), and the recoded variables for Val187Ala (P = 0.021) in TAS2R9 and Val240Ile (P = 0.008) in TAS2R31 were both significant.
Discussion
Current data support the idea that not all humans perceive bitterness from AceK, which is congruent with earlier findings. Using polymorphisms found in TAS2Rs, which were previously shown to be functional for AceK and other nonnutritive sweeteners (i.e., saccharin) in vitro (Kuhn et al. 2004; Roudnitzky et al. 2011) and in vivo (Pronin et al. 2007), our results suggest multiple bitter taste receptors on 2 different chromosomes (7 and 12) contribute to the perceived bitterness of AceK in humans. The use of multiple loci simultaneously increased our ability to explain variance in the quantitative trait measured here (AceK bitterness).
The TAS2R38 diplotype has repeatedly been shown to associate with PROP bitterness (Duffy, Davidson, et al. 2004; Bufe et al. 2005; Duffy et al. 2010), which we confirm here. The first SNP in this haplotype, Ala49Pro, is known to be in strong LD with the other 2 TAS2R38 SNPs, Val262Ala and Val296Ile (not measured here), such that more than 95% of individuals carry either the PAV or AVI variant (Kim et al. 2003; Hayes et al. 2008). In in vitro heterologous expression systems, site-directed mutation indicates that the amino acid at site 49 is the primary determinant of PROP response, with an additional influence arising from the residue at site 262; the residue at site 296 does not seem to matter, at least in cultured cells (Bufe et al. 2005). Large-scale molecular psychophysics in vivo (Mennella et al. 2011) partially confirm these data. Specifically, by comparing humans with rare haplotypes behaviorally, Mennella and colleagues were able to parse apart the independent contributions of each site. Among those with similar diplotypes at sites 49 and 262, the 296 position also contributed to perceived bitterness, as Val296 carriers were more sensitive to PROP than Ile296. This suggests all 3 sites Pro49Ala, Ala262Val, and Val296Ile contribute to PROP response in vivo. These data reinforce the need to confirm data from in vitro heterologous expression systems with behavior in the whole organism (i.e., via animal or human psychophysics).
Here, a 2-site haplotype approach (PA_ vs. AV_) did not predict AceK bitterness, consistent with data showing that AceK does not activate hT2R38 in heterologous expression systems (Meyerhof et al. 2010). However, psychophysical data in humans do not always agree with in vitro expression study cells (compare Bufe et al. 2005; Mennella et al. 2011). Previously, we reported that the bitterness of grapefruit juice varies as a function of a polymorphism in TAS2R19 (Hayes et al. 2011), whereas neither limonin nor naringin activates hT2R19 in functional expression systems (Meyerhof et al. 2010). Thus, it remains important to confirm negative in vitro findings psychophysically in humans.
Here, we found that PROP bitterness was positively associated with the sweetness of AceK, but not the bitterness. PROP bitterness is a marker for overall heightened taste response across qualities (Hayes and Keast 2011). For sweetness, this is consistent with prior data showing PROP bitterness is positively associated with the sweetness of sucrose (Hayes et al. 2008), aspartame (Duffy et al. 2006), AceK (Horne et al. 2002), and sweet foods (Lanier et al. 2005). In regard to bitterness from sulfonyl amide sweeteners, prior reports conflict. When dichotomizing individuals using threshold methods (i.e., tasters vs. nontasters), PROP nontasters report less suprathreshold bitterness from saccharin (Bartoshuk 1979). Conversely, Lawless and colleagues (Horne et al. 2002) failed to observe a relationship between saccharin and AceK bitterness and the bitterness of PROP in 2 experiments with relatively small sample sizes (n = 30 and 38). Here, we confirm that PROP bitterness predicts AceK sweetness but not bitterness in a larger sample, suggesting the failure to find an association between the bitterness of AceK and PROP is not simply a matter of power.
The relationship between PROP bitterness and AceK sweetness, but not AceK bitterness, speaks directly to the dual nature of PROP bitterness as a marker of taste function. That is, PROP bitterness confounds 2 separate but distinct sources of variation: TAS2R38 polymorphisms and overall taste response (aka hypergeusia) (Hayes and Keast 2011). Previously, we showed PROP bitterness predicts variation in the intensity of nonbitter tastants, even after controlling for TAS2R38 genotype (Hayes et al. 2008), supporting its traditional use as a marker of overall orosensory response. However, molecular and behavioral data also indicate TAS2R38 variation clearly predicts differential response to compounds that contain the thiourea (N–C=S) moiety (Bufe et al. 2005; Meyerhof et al. 2010). Given this duality, it is not unreasonable that PROP bitterness should predict AceK sweetness but not bitterness in studies with small numbers of participants. That is, for AceK sweetness, PROP bitterness is capturing overall heightened taste response (Hayes and Keast 2011), as does for other sweet stimuli (Lanier et al. 2005). Conversely, for AceK bitterness, the perceived intensity varies not only as a function of overall taste response but also as a function of variation in TAS2R31 and possibly other TAS2R genes. Thus, estimated effect sizes for PROP should be lower or absent for bitterness than sweetness, especially if random, unmeasured, variation in TAS2R31 or TAS2R9 obfuscates weak effects in small- to medium-sized studies.
Present data support prior evidence that the Val187Ala SNP in TAS2R9 is functional (Dotson et al. 2008). Of 64 bitter stimuli tested in vitro by Dotson et al. (2008), only 3 synthetic pharmaceuticals—ofloxacin, pirenzapine, and procanimide—activated hT2R9. They reported that when Ala was replaced with Val at position 187, the receptor was no longer activated over a wide range of concentrations. They also note the rs3741845 SNP results in amino acid change in a region thought to alter the binding pocket of hT2R receptors (Dotson et al. 2008). Here, we provide the first evidence that this polymorphism may contribute to the perceived bitterness of AceK, although this finding needs to be confirmed. Previously, Dotson and colleagues reported that saccharin did not activate hT2R9 in vitro; however, the top concentration used in their study was ~100 times lower than the amount required to elicit hT2R31 response in vitro (Pronin et al. 2007). This suggests our data do not directly contradict those of Doston et al. with regard to hT2R9 and sulfonyl amide sweeteners, as their null finding may simply be a matter of dose. Notably, in their systematic screening efforts of 104 compounds in vitro, Meyerhof et al. (2010) were unable to identify any potential hT2R9 ligands. Their screening battery did not include the pharmaceutical agents identified by Dotson et al., and even if they had, Meyerhof et al. used the Val187 variant in their heterologous expression system, which would not be expected to detect the ligands tested by Dotson et al. However, based on our in vivo data, we would have expected Meyerhof’s team to identify AceK as a potential hT2R9 ligand in vitro. However, their team also observed poor expression of hT2R9 receptors on the surface of their cells, and none of 104 compounds in their test battery activated hT2R9. Thus, it is currently unknown whether AceK is able to activate the Val187 hT2R9 variant in vitro at biologically relevant doses.
Paradoxically, the gain of function allele here (Val187) is the loss of function allele in the Dotson et al. (2008) report. Previously, it has been hypothesized that mutations in TAS2Rs may drive a gain of function for an alternative ligand (e.g., Wooding et al. 2004). Although there is no definitive example of this to date, it has long been known that phenylthiocarbamide (PTC) tasters are nonresponsive to the bitterness of Antidesma bunius berries, whereas PTC nontasters report bitterness from these berries (Henkin and Gillis 1977). Recently, this finding was confirmed for the TAS2R38 genotype and A. bunius berries, although the specific ligand itself was not isolated (Reed D, personal communication). Present results need to be confirmed in vitro, but if replicated, this would represent the first demonstrated case of a SNP that broadens the molecular receptive range of a hT2R for heterozygotes by enabling the detection of an additional class of alternative ligands.
Here, we confirm polymorphisms in the TAS2R31 gene explain variation in AceK perception in humans. Previous work indicated recognition thresholds differ across individuals (Pronin et al. 2007; Roudnitzky et al. 2011); here, we extend these findings to include suprathreshold intensity. The distinction between threshold and suprathreshold psychophysics is critical, as classical thresholds frequently fail to predict affective response and ingestive behavior (Duffy, Peterson, et al. 2004; Lucas et al. 2011; Harwood et al. 2012). Our attempts to directly genotype individuals for the Arg35Trp (R35W) (rs10845295) were unsuccessful for technical reasons. Instead, we measured several other SNPs in TAS2R31. On the basis of single point mutation studies in vitro, Roudnitzky et al. (2011) found that when Ile240 was changed to Val240, this substitution had no effect on the activation of hT2R31 by AceK. However, in the haplotypes actually observed in the population, the Ile240 allele was in strong LD with Arg35. Likewise, Val227 was in disequilibrium with Arg35. Thus, the SNPs measured here—Val240Ile and Ala227Val—can reasonably be used as tag SNPs for the causal SNP at residue 35.
The present study also extends prior work by looking at TAS2Rs located on separate chromosomes. This suggests that the putatively functional SNPs identified here make an independent contribution to the bitterness of AceK. That is, because they occur on distinct chromosomes, present results cannot be the result of a single haplotype across highly conserved genes. Thus, multilocus approaches like the one used here may have substantial utility when linking TAS2Rs to ingestive behavior, given the likelihood for functional recovery that may otherwise overwhelm the effects on individual SNP analyses.
Here, we show our multilocus model predicts 13.4% of the variation observed in perceived bitterness of AceK. With the inclusion of 2 SNPs from different receptor genes, it would seem that other receptors or polymorphisms not tested here may contribute to the inability to perceive bitterness from AceK in some individuals. Approximately half of our participants rated AceK bitterness at zero. It is possible that offering participants other response options in addition to bitterness, such as “metallic” or “other,” may have captured additional off-tastes typically associated with AceK. However, we find this interpretation unsatisfying, as we would also expect untrained participants to dump any aversive, unpleasant sensations into the bitter response option (Clark and Lawless 1994). Previous reports support that some proportion of individuals report little or no bitterness from AceK, but they do not report the exact proportion (Horne et al. 2002). There is still a large amount of unexplained variation in our data. This could be due to other factors like fungiform papillae density (Zuniga et al. 1993), central gain (Green and George 2004), or other unmeasured polymorphisms in TAS2R genes. In particular, it seems possible that rare TAS2R31 variants may reduce response in vivo. Indeed, in vitro evidence indicates that even when the Trp35 allele is present, mutations at amino acid residues 45, 237, 276, and 281 all cause a loss of function (Roudnitzky et al. 2011). In the future, whole gene sequencing of TAS2Rs may be required to better explain phenotypic variation. Also, as with any candidate SNP study, we should note that unmeasured third variables can obscure SNP findings (e.g., population stratification) and that the associations reported here may not be causal, arising instead from LD with other unmeasured polymorphisms.
Conclusions
Using suprathreshold psychophysics in humans, we explained variation in the perceived intensity of AceK bitterness using a candidate SNP approach across multiple TAS2R genes. These data suggest more than 1 hT2R receptor is responsible for the perception of AceK bitterness. Tag SNPs believed to be in complete LD with the putatively causal SNP in TAS2R31 predicted variation in AceK bitterness. In addition, a polymorphism Ala187Val in TAS2R9 not been previously reported as being functional for AceK was shown to predict bitterness in vivo. Conversely, putatively functional SNPs in TAS2R4, TAS2R38, and near TAS2R16 did not predict bitterness. Polymorphisms in 2 bitter receptor genes on different chromosomes both appeared to contribute to the suprathreshold bitterness of AceK, and a simple multilocus model was able to predict 13% of the variance in perceived bitterness. However, present data also suggest additional polymorphisms may contribute to the inability to perceive AceK bitterness. More research is needed to determine if other receptors confer additional response, and whether rare polymorphisms in the genes studied here may attenuate responses to AceK in vivo.
Supplementary material
Supplementary material can be found at http://www.chemse.oxfordjournals.org/
Funding
This work was partially supported by a National Institutes of Health grant from the National Institute National of Deafness and Communication Disorders [DC010904] to the corresponding author, United States Department of Agriculture Hatch Project PEN04332 funds, funds from the Pennsylvania State University and a VA shared equipment grant to JEM.
Conflict of interest disclosure
The corresponding author has been paid for developing and delivering educational presentations on taste perception and taste biology for the food industry; his laboratory also conducts routine consumer acceptability tests for the food industry to facilitate practical training for students. None of these companies have provided any direct or indirect support for the work described here. The authors declare no other relationships or activities that may have influenced the work described here.
Supplementary Material
Acknowledgments
This manuscript was completed in partial fulfillment of the requirements for a Master of Science degree at the Pennsylvania State University by the first author. The first author developed the data collection protocol in collaboration with the corresponding author, collected a large portion of the psychophysical data, ran all statistical analyses with input from the corresponding author, and wrote the majority of the manuscript. The authors would also like to thank Dr Emma Feeney, Nadia K. Byrnes, and Meghan Kane for collecting additional psychophysical data, Samantha M. Bennett for assistance with protocol development, Kayla Beaucage for genotyping our DNA samples, and Dr Rohan Palmer for his assistance in imputing haplotypes. We also thank our study participants for their time and participation.
References
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJP, Zuker CS. 2000. A novel family of mammalian taste receptors. Cell. 100(6):693–702 [DOI] [PubMed] [Google Scholar]
- Anderson GH, Foreyt J, Sigman-Grant M, Allison DB. 2012. The use of low-calorie sweeteners by adults: impact on weight management. J Nutr. 142(6):1163S–1169S [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM. 1979. Bitter taste of saccharin related to the genetic ability to taste the bitter substance 6-n-propylthiouracil. Science. 205(4409):934–935 [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PAS, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. 2005. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 15(4):322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 21(2):263–5 [DOI] [PubMed] [Google Scholar]
- Carey G. 2007. The association design and a continous phenotype [Internet]. Institute for Behavioral Genetics, University of Colorado. Updated 4 April 2007. Available from: http://psych.colorado.edu/~carey/Courses/PSYC5102/handouts/Association-Quantitative.pdf
- Calo C, Padiglia A, Zonza A, Corrias L, Contu P, Tepper BJ, Barbarossa IT. 2011. Polymorphisms in TAS2R38 and the taste bud trophic factor, gustin gene co-operate in modulating PROP taste phenotype. Physiol Behav. 104(5):1065–1071 [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJP. 2000. T2Rs function as bitter taste receptors. Cell. 100(6):703–711 [DOI] [PubMed] [Google Scholar]
- Clark CC, Lawless HT. 1994. Limiting response alternatives in time-intensity scaling: an examination of the halo-dumping effect. Chem Senses. 19(6):583–594 [DOI] [PubMed] [Google Scholar]
- De la Hunty A, Gibson S, Ashwell M. 2006. A review of the effectiveness of aspartame in helping with weight control. Nutr Bull. 31(2):115–128 [Google Scholar]
- de Snoo K. 1937. Das trikende Kind im Uterus. Monatsschrift fur Geburtshilfe und Gynakologie. 105 88–97 [Google Scholar]
- Dinehart M, Hayes J, Bartoshuk L, Lanier S, Duffy V. 2006. Bitter taste markers explain variability in vegetable sweetness, bitterness, and intake. Physiol Behav. 87(2):304–313 [DOI] [PubMed] [Google Scholar]
- Dotson CD, Zhang L, Xu H, Shin YK, Vigues S, Ott SH, Elson AET, Choi HJ, Shaw H, Egan JM. 2008. Bitter taste receptors influence glucose homeostasis. PLoS ONE. 3(12):e3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois GE, Walters DE, Schiffman SS, Warwick ZS, Booth BJ, Pecore SD, Gibes K, Carr BT, Brands LM. 1991. Concentration—response relationships of sweeteners: a systematic study. In Walters D.E., Orthoefer F.T., Dubois G.E. (eds), ACS Symposium Series 450. Sweeteners: Discovery, Molecular Design and Chemoreception. Boston, MA, American Chemical Society, pp. 261–276 [Google Scholar]
- Duffy VB, Anderson G. 1998. Position of the American Dietetic Association: use of nutritive and nonnutritive sweeteners. J Am Diet Assoc. 98(5):580–587 [DOI] [PubMed] [Google Scholar]
- Duffy VB, Davidson AC, Kidd JR, Kidd KK, Speed WC, Pakstis AJ, Reed DR, Snyder DJ, Bartoshuk LM. 2004. Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcohol Clin Exp Res. 28(11):1629–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy VB, Hayes JE, Davidson AC, Kidd JR, Kidd KK, Bartoshuk LM. 2010. Vegetable intake in college-aged adults is explained by oral sensory phenotypes and TAS2R38 genotype. Chemosens Percept. 3(3):137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy VB, Hayes JE, Dinehart ME. 2006. Genetic differences in sweet taste perception. In: Spillane WJ, editor. Optimising the sweet taste in foods. Cambridge (UK): Woodhead Publishing, pp. 30–53 [Google Scholar]
- Duffy VB, Peterson JM, Bartoshuk LM. 2004. Associations between taste genetics, oral sensation and alcohol intake. Physiol Behav. 82(2–3):435–445 [DOI] [PubMed] [Google Scholar]
- Glanz K, Basil M, Maibach E, Goldberg J, Snyder D. 1998. Why Americans eat what they do: taste, nutrition, cost, convenience, and weight control concerns as influences on food consumption. J Am Diet Assoc. 98(10):1118–1126 [DOI] [PubMed] [Google Scholar]
- Green BG, George P. 2004. ‘Thermal taste’ predicts higher responsiveness to chemical taste and flavor. Chem Senses. 29(7):617–628 [DOI] [PubMed] [Google Scholar]
- Hamer D, Sirota L. 2000. Beware the chopsticks gene. Mol Psychiatry. 5(1):11–13 [DOI] [PubMed] [Google Scholar]
- Hanger L, Lotz A, Lepeniotis S. 1996. Descriptive profiles of selected high intensity sweeteners (HIS), HIS blends, and sucrose. J Food Sci. 61(2):456–459 [Google Scholar]
- Harwood ML, Ziegler GR, Hayes JE. 2012. Rejection thresholds in chocolate milk: evidence for segmentation. Food Qual Prefer. 26(1):128–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE. 2008. Transdisciplinary perspectives on sweetness. Chemosens Percept. 1(1):48–57 [Google Scholar]
- Hayes JE, Allen AL, Bennett SM. 2013. Direct comparison of the generalized Visual Analog Scale (gVAS) and general Labeled Magnitude Scale (gLMS). Food Qual Prefer. 28(1):36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Bartoshuk LM, Kidd JR, Duffy VB. 2008. Supertasting and PROP bitterness depends on more than the TAS2R38 gene. Chem Senses. 33(3):255–265 [DOI] [PubMed] [Google Scholar]
- Hayes JE, Keast RSJ. 2011. Two decades of supertasting: where do we stand? Physiol Behav. 104(5):1072–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Sullivan BS, Duffy VB. 2010. Explaining variability in sodium intake through oral sensory phenotype, salt sensation and liking. Physiol Behav. 100(4):369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Wallace MR, Knopik VS, Herbstman DM, Bartoshuk LM, Duffy VB. 2011. Allelic variation in TAS2R bitter receptor genes associates with variation in sensations from and ingestive behaviors toward common bitter beverages in adults. Chem Senses. 36(3):311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin RI, Gillis WT. 1977. Divergent taste responsiveness to fruit of the tree Antidesma bunius . Nature. 265(5594):536–537 [DOI] [PubMed] [Google Scholar]
- Hill JO, Prentice AM. 1995. Sugar and body weight regulation. Am J Clin Nutr. 62(1):264S–273S [DOI] [PubMed] [Google Scholar]
- Horne J, Lawless HT, Speirs W, Sposato D. 2002. Bitter taste of saccharin and acesulfame-K. Chem Senses. 27(1):31–38 [DOI] [PubMed] [Google Scholar]
- Howard BV, Wylie-Rosett J. 2002. Sugar and cardiovascular disease. Circulation. 106(4):523–527 [DOI] [PubMed] [Google Scholar]
- IFIC 2011. Consumer attitudes toward food safety, nutrition & health. International Food Information Council Foundation; Washington DC: [Google Scholar]
- Kamerud JK, Delwiche JF. 2007. Individual differences in perceived bitterness predict liking of sweeteners. Chem Senses. 32(9):803–810 [DOI] [PubMed] [Google Scholar]
- Kim U, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. 2003. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 299(5610):1221–1225 [DOI] [PubMed] [Google Scholar]
- Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. 2005. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum Mutat. 26(3):199–204 [DOI] [PubMed] [Google Scholar]
- Kuhn C, Bufe B, Winnig M, Hofmann T, Frank O, Behrens M, Lewtschenko T, Slack JP, Ward CD, Meyerhof W. 2004. Bitter taste receptors for saccharin and acesulfame K. J Neurosci. 24(45):10260–10265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier SA, Hayes JE, Duffy VB. 2005. Sweet and bitter tastes of alcoholic beverages mediate alcohol intake in of-age undergraduates. Physiol Behav. 83(5):821–831 [DOI] [PubMed] [Google Scholar]
- Lotsch J, Geisslinger G, Tegeder I. 2009. Genetic modulation of the pharmacological treatment of pain. Pharmacol Ther. 124(2):168–184 [DOI] [PubMed] [Google Scholar]
- Lucas L, Riddell L, Liem G, Whitelock S, Keast R. 2011. The influence of sodium on liking and consumption of salty food. J Food Sci. 76(1):S72–S76 [DOI] [PubMed] [Google Scholar]
- Mennella JA, Pepino MY, Duke FF, Reed DR. 2010. Age modifies the genotype-phenotype relationship for the bitter receptor TAS2R38. BMC Genet. 11(1):60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella JA, Pepino MY, Duke FF, Reed DR. 2011. Psychophysical dissection of genotype effects on human bitter perception. Chem Senses. 36(2):161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. 2010. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 35(2):157–170 [DOI] [PubMed] [Google Scholar]
- Pronin AN, Xu H, Tang H, Zhang L, Li Q, Li X. 2007. Specific alleles of bitter receptor genes influence human sensitivity to the bitterness of aloin and saccharin. Curr Biol. 17(16):1403–1408 [DOI] [PubMed] [Google Scholar]
- Raben A, Vasilaras TH, Møller AC, Astrup A. 2002. Sucrose compared with artificial sweeteners: different effects on ad libitum food intake and body weight after 10wk of supplementation in overweight subjects. Am J Clin Nutr. 76(4):721–729 [DOI] [PubMed] [Google Scholar]
- Reed DR, Zhu G, Breslin PAS, Duke FF, Henders AK, Campbell MJ, Montgomery GW, Medland SE, Martin NG, Wright MJ. 2010. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum Mol Genet. 19(21):4278–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudnitzky N, Bufe B, Thalmann S, Kuhn C, Gunn HC, Xing C, Crider BP, Behrens M, Meyerhof W, Wooding SP. 2011. Genomic, genetic and functional dissection of bitter taste responses to artificial sweeteners. Hum Mol Genet. 20(17):3437–3449 [DOI] [PubMed] [Google Scholar]
- Schiffman SS, Reilly DA, Clark TB. 1979. Qualitative differences among sweeteners. Physiol Behav. 23(1):1–9 [DOI] [PubMed] [Google Scholar]
- Snyder D, Fast K., Bartoshuk LM.2004. Valid comparisons of suprathreshold sensations. J Conscious Stud. 11(7–8):96–112 [Google Scholar]
- Steiner JE, Glaser D, Hawilo ME, Berridge KC. 2001. Comparative expression of hedonic impact: affective reactions to taste by human infants and other primates. Neurosci Biobehav Rev. 25(1):53–74 [DOI] [PubMed] [Google Scholar]
- Stellman SD, Garfinkel L. 1986. Artificial sweetener use and one-year weight change among women. Prev Med. 15(2):195–202 [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. 2003. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 73(5):1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 68(4):978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordoff MG, Alleva AM. 1990. Effect of drinking soda sweetened with aspartame or high-fructose corn syrup on food intake and body weight. Am J Clin Nutr. 51(6):963–969 [DOI] [PubMed] [Google Scholar]
- Wooding S, Kim U, Bamshad MJ, Larsen J, Jorde LB, Drayna D. 2004. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 74(4):637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga JR, Davis SH, Englehardt RA, Miller IJ, Schiffman SS, Phillips C. 1993. Taste performance on the anterior human tongue varies with fungiform taste bud density. Chem Senses. 18(5):449–460 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.