Abstract
MicroRNAs (miRNAs) regulate dendritogenesis and plasticity. However, the biological function of miRNAs in axons has not been extensively investigated. Here, using rat primary cortical neurons cultured in a microfluidic chamber, we found that the distal axons of the neurons expressed the miR-17–92 cluster, and proteins that regulate production and activity of mature miRNAs, Dicer and Argonaute 2, respectively, were present in the distal axons. Overexpression of the miR-17–92 cluster in cortical neurons substantially increased axonal outgrowth, whereas distal axonal attenuation of endogenous miR-19a, a key miRNA of the miR-17–92 cluster, with the miRNA hairpin inhibitor suppressed axonal outgrowth. Moreover, overexpression of the miR-17–92 cluster reduced phosphatase and tensin homolog (PTEN) proteins and elevated phosphorylated mammalian target of rapamycin (mTOR) in the distal axons. In contrast, distal axonal attenuation of miR-19a increased PTEN proteins and inactivated mTOR in the axons, but did not affect these protein levels in the cell bodies. Overexpression of PTEN and attenuation of endogenous PTEN prevailed over the enhancement and inhibitory effects of the miR-19a on axonal outgrowth, respectively. Axonal application of LY294002, a phosphoinositide3-kinase inhibitor, or rapamycin, an mTOR inhibitor, abolished axonal outgrowth enhanced by overexpression of the miR-17–92 cluster. Collectively, these findings demonstrate that axonal alteration of miR-17–92 cluster expression regulates axonal outgrowth and that local modulation of PTEN protein levels by miR-19a likely contributes to the axonal outgrowth.
Introduction
MicroRNAs (miRNAs), a family of short noncoding RNA molecules of ∼22 nt, are crucial molecules regulating the magnitude of gene expression (Bartel, 2004; Kim, 2005; Krol et al., 2010). Dicer, a ribonuclease III, cleaves the pre-miRNA into double-stranded RNAs of ∼22 nt (Bartel, 2004; Kim, 2005; Krol et al., 2010). One strand of these double-stranded RNA intermediates is incorporated into RNA-induced silencing complex (RISC) that includes the Argonaute (Ago) protein family (Hammond et al., 2001). Studies in synaptic development indicate that miRNAs control dendritogenesis, synapse formation, and synapse maturation by locally regulating mRNA translation such as small Rho GTPases in synapses and dendrites (Vo et al., 2005; Yu et al., 2008; Schratt, 2009) Emerging data show that neuronal axons contain miRNAs and miRNA biogenesis proteins including Dicer and Ago (Hengst et al., 2006; Aschrafi et al., 2008; Natera-Naranjo et al., 2010), suggesting that axons can locally synthesize miRNAs (Aschrafi et al., 2008; Natera-Naranjo et al., 2010). Using superior cervical ganglia neurons, Aschrafi et al. (2008) reported that brain-specific miR-338 locally regulates mitochondrial activity in axons. Recently, Dajas-Bailador et al. (20120 showed that attenuation of axonal endogenous miR-9 in the embryonic cortical neurons augments axonal outgrowth and that miR-9 targets the microtubule-associated protein 1b. These findings suggest that axonal miRNAs locally mediate axonal outgrowth by regulating their targeted proteins localized to the axon, which is important for the response of the growth cone to guidance cues (Hengst et al., 2006; Aschrafi et al., 2008; Natera-Naranjo et al., 2010). The miR-17–92 cluster, a single polycistronic unit, is present in the distal axons of the superior cervical ganglia neurons (Natera-Naranjo et al., 2010). Phosphatase and tensin homolog (PTEN) is one of the putative target genes of the miR-17–92 cluster (Olive et al., 2009). Deleting the gene that encodes PTEN protein enhances regeneration of axons in adult corticospinal neurons after spinal cord injury (Liu et al., 2010). However, little is known about the biological function of the miR-17–92 cluster and its effect on PTEN proteins in axons of cortical neurons. In the present study, we investigated whether the miR-17–92 cluster locally regulates axonal outgrowth. In addition, we examined axonal PTEN proteins after alteration of miR-17–92 cluster expression. Our data demonstrate that axonal alteration of miR-17–92 cluster expression regulates axonal outgrowth and that local modulation of PTEN protein levels by miR-19a likely contributes to the axonal outgrowth of the embryonic cortical neurons.
Materials and Methods
All experimental procedures were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Henry Ford Hospital.
Cell culture of primary cortical neurons and use of a microfluidic chamber.
Cortical neurons were harvested from embryonic day 18 Wistar rats (either sex; Charles River Laboratories). Cultures were prepared according to a previously described procedure with some modifications (Ueno et al., 2012). Briefly, embryos were removed, and the cerebral cortex dissected, stripped of meninges, and dissociated by a combination of Ca2+- and Mg2+-free HBSS containing 0.125% trypsin digestion for 15 min, then mechanically triturated for ∼20 times. The triturated cells were passed through a 40 μm cell strainer and counted to obtain a concentration of 3 × 107cells/ml.
To separate axons from neuronal soma, a microfluidic chamber (Standard Neuron Device, catalog #SND450, Xona Microfluidics) was used (Taylor et al., 2005, 2009). The small dimension of the microgrooves in the chamber allows axons to sprout from the cell-seeded compartment into the other compartment of the chamber, but prevents the passage of cell bodies (Taylor et al., 2005, 2009). Briefly, cleaned, sterilized, and dried chambers were affixed to poly-d-lysine-coated (Sigma-Aldrich) dishes (35 mm, Corning). The cortical neurons were plated at a density of 6 × 105cells/chamber in DMEM with 5% FBS and, incubated for an initial 24 h. After 24 h, cell culture was initiated with the addition of neurobasal growth medium (Invitrogen), 2% B-27 (Invitrogen), 2 mm GlutaMax, and 1% antibiotic-antimycotic. On day in vitro (DIV) 3, one-half of the medium was replaced with culture medium containing 20 μm 5-fluorodeoxyuridine. The growth media was changed every other day thereafter.
Construction of pCAG-green fluorescent protein miR-17–92 cluster vector and cell transfection.
A primary rat miR-17–92 cluster (786 bp, miRBase) was amplified by PCR. The following primers were used for amplification: forward, 5′-CGGAATTCGTCAGGATAATGTCAAAGTGCTTACA; and reverse, CGGGTACCACCAAACTCAACAGGCCG, with EcoRI and KpnI at 5′ ends. A pCAG-green fluorescent protein (GFP)-miR-17–92 cluster vector was constructed by cloning the primary miR-17–92 cluster fragment into the EcoRI and KpnI sites in the pCAG-GFP vector (Addgene plasmid 11150) (Matsuda and Cepko, 2004). The pCAG-GFP-miR-17–92 vector (2 μg) was introduced into cultured primary neurons (∼10 × 106 neurons) by electroporation using a Nucleofector kit (Mirus). An empty pCAG-GFP vector was used as a negative control.
For overexpression of PTEN, pcDNA3-EGFP-PTEN (2 μg) (Addgene plasmid 13039) was introduced into primary neurons by electroporation. An empty GFP vector was used as a negative control. For attenuation of endogenous PTEN, the neurons were transfected with Silencer Select siRNA-PTEN (s132220, Invitrogen) or a Silencer Select Negative Control (4390843, Invitrogen).
For attenuation of endogenous miRNAs, neurons were transfected with miRNA hairpin inhibitors against rat miR-18a (mature sequence: UAAGGUGCAUCUAGUGCAGAUAG), miR-19a (mature sequence: UGUGCAAAUCUAUGCAAAACUGA), and Caenorhabditis elegans miR-67 (cel-miR-67 mature sequence: UCACAACCUCCUAGAAAGAGUAGA), which lacks homologs in mammals and was used as a negative control (Dharmacon). Briefly, 200 pmol/well miRNA inhibitors were mixed with 100 μl of Nucleofector solution (Mirus), and cell-miRNA mixtures were transferred into a cuvette and electroporated using the program O-03.
For axonal transfection, the N-TER Nanoparticle siRNA Transfection System (Sigma-Aldrich) was used to perform the localized transfection in microfluidic chambers. Briefly, on DIV3, when all of the microgrooves were fully filled with axons, the fluidic isolation environment was provided by the microfluidic chambers, the miR-19a hairpin inhibitor (20 nm) along with nanoparticles packed in N-TER peptide in Neurobasal Medium was applied into the axonal compartment for 72 h. After that, the total RNAs and proteins were extracted from the cell body and axonal compartments. At the same time, cells cultured in additional chambers were used for time-lapse microscopic experiments.
In situ hybridization and immunocytochemistry.
Locked nucleic acid (LNA) probes, specifically against rat miR-18a and miR-19a, and scramble probes (Exiqon) were used for hybridization to detect mature miRNAs according to a published protocol with some modifications (Pena et al., 2009). Briefly, cultured cells were fixed with 4% paraformaldehyde. Digoxigenin-labeled LNA probes at 0.25 μm in hybridization buffer were used. Signals were detected by incubation of the cells in solution containing NBT/BCIP or Fluorophore Amplification Reagent Working Solution (TSA PLUS Fluorescence Kits, PerkinElmer). After in situ hybridization, either single- or double-immunofluorescent staining was performed with a monoclonal antibody against phosphorylated high-molecular-weight neurofilament (pNFH; 1:500; SMI31, Covance), rabbit anti-PTEN (1:200; Cell Signaling Technology), and chicken anti-GFP (1:800; Aves Labs).
Immunofluorescent staining was performed as previously described (Liu et al., 2009). The following primary antibodies were used in the present study: a monoclonal antibody against pNFH (1:500; SMI31, Covance); rabbit anti-PTEN (1:200; Cell Signaling Technology); rabbit anti-Ki67 (1:200; Neo Markers); and chicken anti-GFP (1:800; Aves Lab). After washing with PBS twice, the microfluidic chamber devices were incubated with the primary antibodies listed above for either 1 or 2 nights at 4°C, and with Cy3, Cy5, or FITC-conjugated secondary antibodies for 2 h at room temperature. Nuclei were counterstained with DAPI (1:10,000; Vector Laboratories) for 15 min at room temperature. TUNEL staining was performed, as previously described (Buller et al., 2010).
Time-lapse microscopy.
Primary cortical neurons cultured in microfluidic chambers were incubated on a stage top chamber with 5% CO2 at 37°C (Live-Cell Control Unit), which was placed on the stage of a TE2000-U inverted microscope equipped with a motorized z-stage (Nikon). To track axonal growth, a 40× objective with 1.5× zoom was used for acquiring images in the axonal compartment. Bright-field images were acquired at 30 ms exposure times, whereas fluorescent-field images were obtained at 485 nm excitation at 300 ms exposure times. A stack of images (30 images of 1 μm steps of the z-axis) was acquired only in bright-field view at 5 min intervals for a total of 60 min using a CCD camera (CoolSNAP 5000) and MetaView software (Universal Imaging) (Zhang et al., 2007a, 2009). To minimize phototoxicity on axons, fluorescent-field images were taken only at the beginning and end of the experiment. In preliminary experiments, we performed time-lapse imaging from 0.5 to 4 h and found that a time point longer than 1 h resulted in considerable evaporation of culture medium within both compartments that contained only 500 μl of culture medium. Therefore, the 1 h time point was selected. We selected four to five individual long axons with healthy growth cones for the time-lapse imaging.
To examine the effect of phosphoinositide 3-kinase (PI3K) and mammalian target of rapamycin (mTOR) on axonal outgrowth, pharmacological inhibitors of LY294002 (Calbiochem) and rapamycin (Sigma-Aldrich), respectively, were applied in the axonal compartment. In preliminary experiments, we found that rapamycin at concentrations ≥500 nm suppressed axonal growth, while rapamycin at 200 nm did not inhibit axonal outgrowth under control condition. To examine whether rapamycin specifically blocks miR-17–92 cluster-increased axon growth, we selected the concentration of 200 nm. A dose (20 μm) of LY 294002 was selected based on our published study (Ueno et al., 2012).
Light and fluorescent microcopy.
To further examine the effect of LY 294002 on axonal outgrowth, LY294002 at 20 μm was freshly added into the axonal compartment for consecutive days from DIV1, and axonal outgrowth was recorded at DIV3 to DIV5.
Image acquisition and quantification.
For measurements of axonal elongation outgrowth from DIV3 to DIV5, the distal axons in the axonal compartment were imaged under a 10× objective of an IX71 microscope (Olympus) using a CCD camera (CoolSNAP 5000) and MetaView software (Universal Imaging). Five images per compartment, which encompassed the majority of the compartment, were acquired. The lengths of the 15 longest axons in each compartment were measured by manually tracing individual axon using an MCID system.
For analysis of time-lapse images, 20–25 axonal lengths acquired from six individual chambers of each experimental group were measured with MetaMorph software (Universal Imaging) at different time points. Two of 50 and 4 of 48 axons acquired from the miR-17–92 cluster and empty vector groups, respectively, grew <2 μm during the 1 h experimental period. These axons were excluded from data analysis.
The cell bodies and axons of the cortical neurons were imaged under a 40× objective after the immunocytochemistry staining or immunocytochemistry staining in combination with in situ hybridization using a laser-scanning confocal microscope (LSM 510 NLO, Carl Zeiss) (Zhang et al., 2007b, 2009). Intensity of immunoreactive signals in distal axons was analyzed using the NIH Image analysis program ImageJ, as previously described (Ueno et al., 2012).
Western blot analysis.
Total proteins in the cell body and axonal compartments were extracted. To minimize contamination from the cell body compartment, 100 μl of PBS was added into the cell body compartment, which prevents any buffer in the axonal compartment from flowing back to the cell body compartment. Then, 10 μl of lysis buffer was applied to the axonal compartment for 10 min. During the entire period, the chamber was placed on the ice. To obtain sufficient amount of proteins used for blotting, axonal samples from four to six chambers were pooled for one blot. The protein concentration of the supernatants of the cell body and axonal extracts was determined using a bicinchoninic acid protein assay kit (Pierce Biotechnology). Western blots were performed according to published methods (Ueno et al., 2012). Briefly, equal amounts of total protein for each sample were loaded on 10% SDS-polyacrylamide gels. After electrophoresis, the protein was transferred to nitrocellulose membranes, and the blots were subsequently probed with the following primary antibodies: rabbit polyclonal anti-PTEN (1:1000; Cell Signaling Technology); rabbit polyclonal anti-phosphorylated mTOR (Ser 2448, 1:1000; Cell Signaling Technology); rabbit polyclonal anti-phosphorylated GSK-3β (Ser 21/9, 1:1000; Cell Signaling Technology); rabbit polyclonal anti-PI3K p85 (1:1000; Millipore); and mouse monoclonal anti β-actin (1:10000; Abcam). For detection, horseradish peroxidase-conjugated secondary antibodies were used (1:2000) followed by enhanced chemiluminescence development (Pierce Biotechnology). Protein levels of β-actin and PI3K subunit p85 were used as the internal controls for somata and axons, respectively (Andreassi et al., 2010). Western blots were performed from at least three individual experiments. The optical density of protein signals was quantified using an image processing and analysis program (Scion Image).
Isolation of total RNA and real-time reverse-transcriptase PCR.
To analyze miRNAs, cells were lysed in Qiazol reagent, and total RNA was isolated using the miRNeasy Mini kit (Qiagen). Quantitative RT-PCR was performed on an ABI 7000 and ABI ViiA 7 PCR instrument (Applied Biosystems). miRNAs were reversely transcribed with the miRNA Reverse Transcription kit (Applied Biosystems) and were amplified with the TaqMan miRNA assay (Applied Biosystems), which is specific for mature miRNA sequences. The following specific primers were used: miR-17 (mature sequence: CAAAGUGCUUACAGUGCAGGUAG); miR-18a (mature sequence: UAAGGUGCAUCUAGUGCAGAUAG); miR-19a (mature sequence: UGUGCAAAUCUAUGCAAAACUGA); miR-19b (mature sequence: UGUGCAAAUCCAUGCAAAACUGA); miR-20a (mature sequence: UAAAGUGCUUAUAGUGCAGGUAG); miR-92 (mature sequence: UAUUGCACUUGUCCCGGCCUG); and U6 snRNA (mature sequence: GTGCTCGCTTCGGCAGCACATATACTAAAATTGGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTT). Analysis of gene expression was performed by the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Statistical analysis.
All statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS, version 11.0 SPSS Inc.). One-way ANOVA with post hoc Bonferroni tests was used when comparing more than two groups. Student's t test was used when comparing two groups. Values presented in this study are expressed as the mean ± SE. A p value < 0.05 was considered to be significant.
Results
Distribution of miR-17–92 cluster in axons and somata
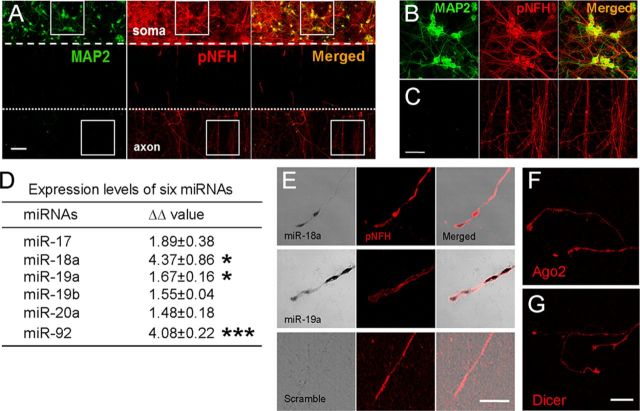
To examine the relative expression of the miR-17–92 cluster in axons and somata of cortical neurons, we cultured cortical neurons in a microfluidic chamber (Standard Neuron Device, Xona Microfluidics), which segregates axons from neuronal cell bodies (Taylor et al., 2005, 2009). Double-immunostaining revealed that in the cell body compartment MAP2+ somata and dendrites and pNFH+ somata and processes were present (Fig. 1A,B), whereas in the axonal compartment there were only pNFH+ processes (Fig. 1A–C). Some pNFH+ somata and processes were MAP2+ in the somal compartment (Fig. 1B). These observations are consistent with the fact that MAP2 is expressed in dendrites and somata but not axons, whereas pNFH is expressed in axons and dendrites (Shi et al., 2004; Mahad et al., 2009). Thus, our immunostaining data confirm that neuronal fibers in the axonal compartment are axons. Using Taqman miRNA-specific primers, we then examined levels of the miR-17–92 cluster in somato-dendrites and -axons of the cortical neurons harvested from the somal and axonal compartments, respectively, by means of real-time quantitative RT-PCR (Liu et al., 2011). Six individual members of the miR-17–92 cluster (miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92) were detected in the distal axons and somato-dendrites with average CT (threshold cycle) values of 24.9 ± 0.6 and 15.4 ± 0.5; 28.3 ± 0.5 and 20.1 ± 0.3; 28.6 ± 0.2 and 19.0 ± 0.1; 23.9 ± 0.1 and 14.3 ± 0.2; 25.3 ± 0.3 and 15.6 ± 0.2; and 25.8 ± 0.1 and 17.6 ± 0.04, respectively. The CT values in the distal axons are much lower than 35, which indicates a low copy number for mature miRNAs (Schmittgen et al., 2008) and are comparable to the CT values detected in axons of sympathetic neurons (Natera-Naranjo et al., 2010). Relative levels of these mature miRNAs in axons and somata were then calculated with the formula −2ΔΔCT after normalizing ΔΔCT values to a reference gene U6 (Natera-Naranjo et al., 2010). Compared with neuronal somata, the distal axons had relatively abundant levels of these six miRNAs (Fig. 1D). However, this comparison may underestimate axonal miRNA levels because not all axons in the cell body compartment entered into the axonal compartment (Fig. 1A,B). Thus, the total RNA isolated from the cell body compartment was not a pure somal fraction and also included an RNA fraction from axons. To further verify the presence of the miR-17–92 cluster in axons, we assessed miR-18a and miR-19a, two key miRNAs of the miR-17–92 cluster, in axons by in situ hybridization using LNA probes specific for miR-18a and miR-19a. Strong signals of miR-18a and miR-19a were detected in axons, while scramble probes did not detect any signals (Fig. 1E). These data indicate the presence of the miR-17–92 cluster in axons of cortical neurons.
Figure 1.
Distribution of the miR-17–92 cluster in axons of cultured primary cortical neurons. A–C, Representative double immunofluorescent images acquired with the confocal microscope show that primary cortical neurons cultured in the compartmentalized microfluidic chamber exhibited MAP2-positive (A, green) and pNFH-positive (A, red) soma and processes in the cell body compartment (A, soma), and pNFH- positive fibers in the axonal compartment (A, axon). B and C are high-magnification images of the boxed areas in A somatic and axonal compartments, respectively. D, Real-time RT-PCR analysis shows relative expression of individual members of miR-17–92 clusters in the axons and cell soma. E, Representative images of in situ hybridization and immunofluorescent staining show the presence of miR-18a (bright field) and miR-19a (bright field) signals in pNFH-positive axons (pNFH and merged), and the absence of miRNA signals with the control probes (bright field) in pNFH-positive axons (pNFH and merged). F, G, Representative immunofluorescent images show Ago2 (F) and Dicer (G) immunoreactive axons. Values in D are given as the mean ± SE from three different experiments with each experiment having three replicates. *p < 0.05 and ***p < 0.001. Scale bars: A, C, 50 μm; E, G, 20 μm.
We then examined axonal localization of Dicer and Argo2 proteins, which are involved in the generation and activity of mature miRNAs, respectively (Schratt, 2009). Immunocytochemistry analysis revealed that axons and growth cones of cortical neurons were Dicer and Ago2 immunoreactive, exhibiting punctate distribution (Fig. 1F,G). The distribution was comparable to axonal miR-18a and miR-19a signals detected by in situ hybridization (Fig. 1E). These results indicate that the distal axons contain miRNA processing machinery, suggesting that axonal Dicer and Ago2 could potentially play a role in local regulation of miRNA syntheses and activity.
Effects of the miR-17–92 cluster on axonal outgrowth
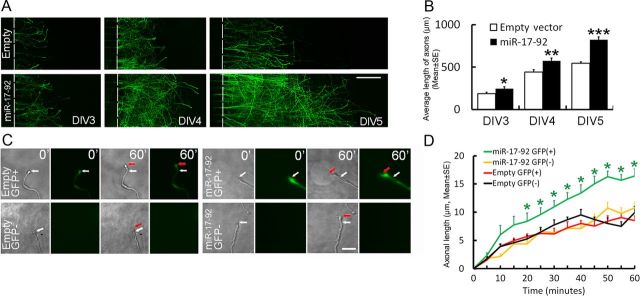
We then examined whether overexpression of the miR-17–92 cluster mediates axonal outgrowth. The cortical neurons were transfected by electroporation using a GFP vector carrying mouse miR-17–92 cluster sequences (GFP-miR-17–92) and a control vector, GFP-empty vector. After transfection, the cortical neurons were cultured in the microfluidic chamber for 5 d and individual members of the miR-17–92 cluster were measured in a pure axonal RNA fraction from the axonal compartment by quantitative real-time RT-PCR using Taqman primers. Compared with neurons transfected with a GFP-empty vector, levels of individual members of the miR-17–92 cluster were substantially elevated in axons of the neurons transfected with the GFP-miR-17–92 cluster (Table 1), indicating that the transfection is effective. We then measured axonal outgrowth daily for 3 d starting on DIV3 after the transfection. During the 3 d period, neurons transfected with the GFP-empty vector exhibited axonal outgrowth (Fig. 2A). However, axonal outgrowth increased by 32 ± 10.9, 30 ± 7.6, and 51 ± 6.4% at DIV3, DIV4, and DIV5, respectively, in neurons transfected with the GFP-miR-17–92 cluster compared with that in the GFP-empty vector group (Fig. 2A,B). To further test the effect of the miR-17–92 cluster on axonal outgrowth, we monitored axonal elongation over a 60 min period on DIV5 using time-lapse microscopy. A substantial increase in axonal outgrowth was detected in GFP-expressing axons of the neurons transfected with the GFP-miR-17–92 cluster compared with GFP-expressing axons of the neurons transfected with the GFP-empty vector (Fig. 2C,D). Furthermore, non-GFP-expressing axons of the neurons transfected with the GFP-miR-17–92 cluster did not exhibit significant axonal elongation compared with GFP-expressing and non-GFP-expressing axons of the neurons transfected with the GFP-empty vector during the same period (Fig. 2C,D). Collectively, these data indicate that forced overexpression of the miR-17–92 cluster is associated with enhancement of axonal outgrowth.
Table 1.
Relative values of individual members of the miR-17–92 cluster in axons and soma after overexpression of the miR-17–92 cluster
| Member | Axon | Soma |
|---|---|---|
| miR-17 | 3.27 ± 0.43** | 1.99 ± 0.10* |
| miR-18a | 2.05 ± 0.56 | 0.93 ± 0.14 |
| miR-19a | 2.06 ± 0.36* | 2.12 ± 1.32 |
| miR-19b | 5.88 ± 0.87** | 0.96 ± 0.07 |
| miR-20a | 37.43 ± 7.34** | 1.63 ± 0.03* |
| miR-92 | 5.82 ± 1.18* | 0.52 ± 0.01 |
RT-PCR data were compared between neurons transfected with the GFP-miR-17–92 cluster and the GFP-empty vector. Data are presented as fold changes compared with the GFP-empty vector as 1. The numbers are mean of four replicates (±SE).
* p < 0.05,
** p < 0.01 miR-17–92 cluster compared with empty vector.
Figure 2.
Effect of miR-17–92 cluster overexpression on axonal outgrowth. A, Representative tiling immunofluorescent images acquired with the fluorescent microscope in the axonal compartment show GFP-positive axons of the cortical neurons transfected with the GFP-empty vector (Empty) and the GFP-miR-17–92 vector (miR-17–92) on DIV3, DIV4, and DIV5 in the culture. B, Quantitative data show the average length of GFP+ axons in the axonal compartment during these 3 d. N = 6/group. *p < 0.05, **p < 0.01, and ***p < 0.001. C, Representative bright-field and fluorescent-field microscopic images acquired with the time-lapse microscope in the axonal compartment show outgrowth of GFP+ and GFP− axons of the cortical neurons transfected with the GFP-empty vector (Empty GFP+ and GFP−) and GFP-miR-17–92 cluster vector (miR-17–92 GFP+ and GFP−) on DIV5. Numbers in each image indicate times at 0 and 60 min. White and red arrows indicate a growth cone at time 0 and 60 min, respectively. D shows quantitative data of axonal outgrowth during 60 min. N = 20 GFP+ and N = 20 GFP− axons for the GFP-miR-17–92 group; N = 21 GFP+ and N = 22 GFP− axons for the GFP-empty vector group. *p < 0.05 versus the GFP-empty vector group and GFP− axons in the GFP-miR-17–92 group. Scale bars: A, 100 μm; C, 20 μm.
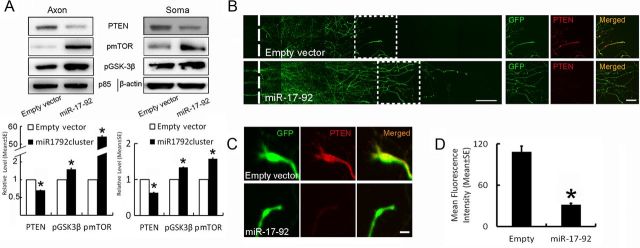
Because the PTEN/mTOR signaling pathway mediates axonal regeneration (Park et al., 2008) and the miR-17–92 cluster targets PTEN (Olive et al., 2009), we explored the possibility that elevation of the miR-17–92 cluster in the distal axons locally modulates PTEN proteins. Extracts from the cell body and axonal compartments on DIV5 were harvested, and protein levels were analyzed by Western blots. Western blotting with an antibody specific against PTEN showed that overexpression of the miR-17–92 cluster reduced PTEN levels by 41% in the distal axons compared with the GFP-empty vector (Fig. 3A). The axonal localization of PTEN was also assessed by immunocytochemistry using the antibody against PTEN. Double-immunofluorescent staining showed that axonal PTEN protein was present in GFP-expressing axons and growth cones of the neurons transfected with the GFP-empty vector (Fig. 3B,C). In contrast, GFP-expressing axons of the neurons transfected with the GFP-miR-17–92 cluster exhibited many fewer axonal PTEN proteins (Fig. 3B–D). These immunocytochemistry results are consistent with our Western blot data (Fig. 3A). Reduction of PTEN activates the PI3K/mTOR signaling pathway (Park et al., 2008). Antibodies specifically against phosphorylated mTOR and phosphorylated GSK-3β have been used for detecting activation of the mTOR pathway (Park et al., 2008; Shi et al., 2011). Western blot analysis revealed that overexpression of the miR-17–92 cluster substantially increased pmTOR and pGSK-3β levels in axons (Fig. 3A), which were inversely related to PTEN levels (Fig. 3A), suggesting that diminishing PTEN activates the mTOR signaling pathway. Overexpression of the miR-17–92 cluster also altered protein levels of PTEN, pmTOR, and pGSK-3β in the cell body fraction (Fig. 3A).
Figure 3.
Effects of miR-17–92 cluster overexpression on axonal PTEN proteins. A, Western blot data show PTEN, pmTOR, and pGSK-3β in the distal axons and cell bodies of neurons transfected with GFP-empty vector (Empty vector) and GFP-miR-17–92 cluster (miR-17–92). Axon and soma in A represent that total proteins were extracted from the axonal and cell body compartments, respectively, while p85 and β-actin were used as internal controls for the distal axons and cell bodies, respectively. N = 3. *p < 0.05 versus the GFP-empty vector. B and C are representative double-immunofluorescent images acquired with the confocal microscope in the axonal compartment showing that GFP+ axons (B, green) and growth cones (C, green) were PTEN immunoreactive (B, C, red). However, PTEN signals were substantially reduced in axons and growth cones of neurons transfected with the GFP-miR-17–92 cluster (B, C, miR-17–92, red) compared with the GFP-empty vector (B, C, empty vector, red). D is quantitative data of PTEN signals in the distal axons. N = 24 axons/empty vector and 17 axons/miR-17–92 cluster vector. Scale bars: B, bottom left, 200 μm; B, bottom right, 20 μm; C, 5 μm. *p < 0.01 versus the GFP-empty vector.
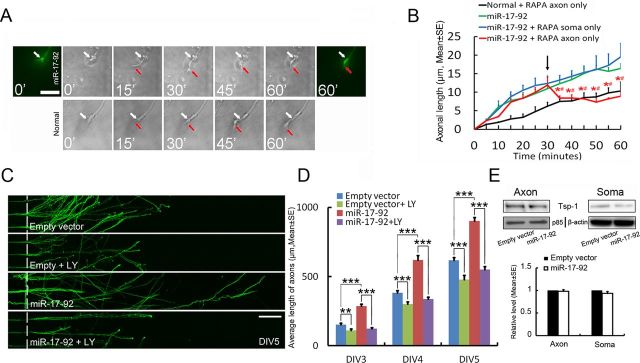
Using axonal application of rapamycin, an mTOR inhibitor (Park et al., 2008; Kobayashi et al., 2012), and time-lapse microscopy, we next assessed whether the miR-17–92 cluster-activated axonal mTOR signaling pathway locally contributes to its axonal outgrowth. Neurons transfected with the GFP-miR-17–92 cluster were cultured in microfluidic chambers that permit the fluidic isolation of axonal microenvironment from the cell body compartment (Taylor et al., 2009). On DIV5, axonal outgrowth was recorded by time-lapse microscopy every 5 min over a 60 min period and rapamycin (200 nm) was applied to the axonal or cell body compartment at 30 min of the recording. An increased axonal outgrowth was observed in GFP-expressing axons of the neurons transfected with the GFP-miR-17–92 cluster compared with axonal outgrowth in nontransfected neurons during the first 30 min period (Fig. 4A,B). Application of rapamycin into the axonal compartment abolished miR-17–92 increased axonal outgrowth during the second 30 min, whereas addition of rapamycin into the cell body compartment did not affect axonal outgrowth increased by the miR-17–92 cluster (Fig. 4B). Treatment of axons of nontransfected neurons with rapamycin did not affect axonal outgrowth (Fig. 4B). Time-lapse data also showed that axonal application of LY 294002 (20 μm), a PI3K inhibitor that inhibits mTOR, completely blocked miR-17–92 cluster-increased axonal growth (4.7 ± 0.2 vs 16.4 ± 1.3 μm in the miR-17–92 cluster group, n = 20 axons/group, p < 0.01). To further examine the effect of LY294002 on miR-17–92 cluster-induced axonal outgrowth, we applied LY294002 into the axonal compartment and measured axonal outgrowth daily for 3 d. Consistent with time-lapse data, axonal application of LY294002 abolished miR-17–92 cluster-induced increases in axonal outgrowth during a 3 d experimental period (Fig. 4C,D). Together, these data demonstrate a function of miR-17–92 cluster in axonal outgrowth in a manner dependent on axonal PI3K/mTOR signals.
Figure 4.
Effects of local application of rapamycin on distal axonal outgrowth. A, Representative bright-field and fluorescent-field images acquired with the time-lapse microscope in the axonal compartment show changes of axonal length in neurons transfected with GFP-miR-17–92 cluster (miR-17–92) and in normal neurons (normal) after axonal application rapamycin at 30 min. Numbers in A represent different times (minutes). White and red arrows indicate a growth cone at time 0 and different time points as indicated, respectively. B, Quantitative data showing axonal outgrowth of neurons transfected with GFP-miR-17–92 cluster and of normal neurons during 60 min when rapamycin (RAPA) was locally applied to the cell body (soma only) and axonal (axon only) compartments at 30 min (arrow). N = 25 axons/normal, N = 20 axons/miR-17–92, N = 23 axons/miR-17–92+RAPA soma only, N = 25 axons/miR-17–92+RAPA axon only. *p < 0.05 the miR-17–92 plus RAPA axon only group versus the miR-17–92 and miR-17–92 RAPA soma only groups; #p < 0.05 the miR-17–92 plus RAPA axon only versus the normal group. C shows representative microscopic images showing the effect of axonal application of LY294002 on the axonal growth of the neurons transfected with the miR-17–92 cluster or empty vector at DIV5. D, Quantitative data of axonal outgrowth in each group during DIV3 to DIV5. N = 5/group. **p < 0.01; ***p < 0.001. E, Western blot data show the effect of overexpression of the miR-17–92 cluster on TSP1 proteins in the cell body and axonal compartments, while p85 and β-actin were used as internal controls for the distal axons and cell bodies, respectively. N = 7/group. Scale bars: A, 20 μm; C, 200 μm.
Among putative targets, the miR-17–92 targets thrombospondin 1 (Tsp1) and connective tissue growth factor (CTGF), and both of them regulate axonal outgrowth (Sandvig et al., 2004; Christopherson et al., 2005; van Almen et al., 2011). However, Western blot analysis did not detect any signals of CTGF in the cell body and axonal fractions (data not shown), while overexpression of the miR-17–92 cluster did not significantly change Tsp1 protein levels (Fig. 4E). These data suggest that the overexpressed-miR-17–92 cluster does not likely target these two proteins in the cortical neurons.
The miR-17–92 cluster has been reported to inhibit apoptosis and to enhance cell viability and proliferation in cancer cells (Hayashita et al., 2005; Mi et al., 2010). To examine whether the miR-17–92 cluster-increased axonal outgrowth is related to the effect of the miR-17–92 cluster on neuronal survival and proliferation, we measured the number of apoptotic and proliferating cells. Overexpression of the miR-17–92 cluster did not significantly affect the number of apoptotic neurons measured by TUNEL-positive cells (3 ± 0.3%, total 1031 cells counted within seven individual chambers) compared with the GFP-empty vector (4 ± 0.5%, total 768 cells counted within seven individual chambers, p > 0.05). Proliferating neurons measured by Ki67-positive cells (3 ± 0.4, total 929 cells counted within seven individual chambers) were also not affected by overexpression of the miR-17–92 cluster compared with the GFP-empty vector group (3 ± 0.4, total 884 cells counted within seven individual chambers, p > 0.05). These data suggest that neuronal proliferation and survival do not contribute to the observed effect of the miR-17–92 cluster on axonal outgrowth.
The effect of miR-19a on axonal outgrowth
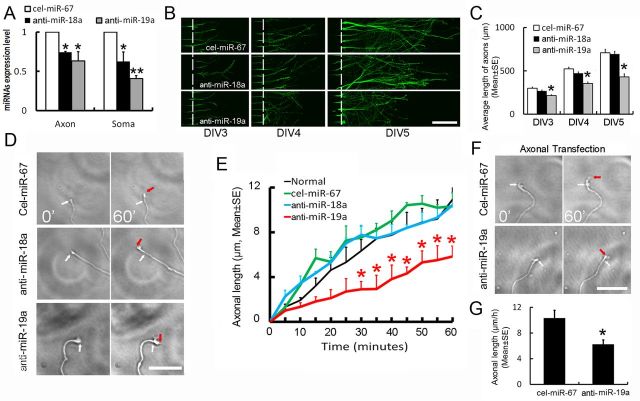
Our in situ hybridization results showed that miR-18a and miR-19a were localized to axons (Fig. 1). These two miRNAs are key members of the miR-17–92 cluster (Olive et al., 2009; Tao et al., 2012). We thus, asked whether endogenous miR-18a and miR-19a contribute to axonal outgrowth. The cortical neurons were transfected by electroporation using miRNA hairpin inhibitors specifically against rat miR-18a and miR-19a, which are complementary to the endogenous mature miRNAs. Neurons transfected with cel-miR-67, which lacks homology in rat, were used as a control. After transfection, the cortical neurons were cultured in the microfluidic chamber for 5 d and levels of miR-18a and miR-19a were measured in a pure axonal RNA fraction from the axonal compartment by quantitative real-time RT-PCR using Taqman primers. Inhibitors of miR-18a and miR-19a substantially reduced axonal miR-18a and miR-19a levels, respectively, compared with cel-miR-67 (Fig. 5A). Attenuation of endogenous miR-19a, but not miR-18a, significantly reduced axonal outgrowth during DIV3 to DIV5 (Fig. 5B,C). Time-lapse microscopy analysis revealed that miR-19a inhibitor suppressed axonal elongation by 50% during 60 min of observation (Fig. 5D,E).
Figure 5.
Effects of attenuation of endogenous miR-18a and miR-19a on axonal outgrowth. A, Quantitative RT-PCR data show relative levels of cel-miR-67, miR-18a, and miR-19a in axonal (Axon) and cell body (Soma) fractions of the neurons transfected with the hairpin inhibitors of miR-18a, miR-19a, and cel-miR-67. N = 3 different experiments and each experiment with triplicates. *p < 0.05 and **p < 0.01 versus cel-miR-67. B, Representative tiling immunofluorescent images acquired with the fluorescent microscope in the axonal compartment show pNFH+ axons of the cortical neurons transfected with the hairpin inhibitors of cel-miR-67 (cel-miR-67), miR-18a (anti-miR-18a), and miR-19a (anti-miR-19a) on DIV3 (A), DIV4 (A), and DIV5 (A) in the culture. C, Quantitative data of axonal outgrowth during 3 d. N = 6/group. *p < 0.01 versus the cel-miR-67 group. D is representative bright-field images acquired with the time-lapse microscope in the axonal compartment showing changes of axonal length in neurons transfected with the hairpin inhibitors at time 0 and 60 min. E, Quantitative data show the miR-19a inhibitor significantly (*p < 0.05) reduced axonal outgrowth compared with the cel-miR-67 group. N = 22 axons/cel-miR-67, N = 25 axons/normal, N = 20 axons/ anti-miR-18a, and N = 20 axons/ anti-miR-19a. F presents representative images acquired with the time-lapse microscope in the axonal compartment showing changes of axonal length in neurons selectively transfected in the distal axons with the hairpin inhibitors of miR-19a (anti-miR-19a) or cel-miR-67 at time 0 and 60 min. G, Quantitative data of axonal outgrowth measured during the 60 min time-lapse experiment. White and red arrows indicate a growth cone at time 0 and 60 min, respectively. *p < 0.05. N = 26 axons/cel-miR-67 and N = 22 axons/ anti-miR-19a inhibitor.
To further examine whether attenuation of axonal miR-19a reduces axonal outgrowth, we used an N-TER Nanoparticle siRNA Transfection System to selectively transfect the distal axons. When the microgrooves within the microfluidic chamber were fully filled by axons on DIV3, miR-19a hairpin inhibitors packed with nanoparticles were applied to the axonal compartment for 72 h. Levels of miR-19a in the cell body and axonal fractions were measured by quantitative RT-PCR using Taqman primers. Axonal transfection resulted in a substantial reduction of miR-19a in the axonal fraction (relative value: 0.42 ± 0.01 vs 1 in normal neurons and vs 1.07 ± 0.04 in neurons transfected with cel-miR-67, n = 3, p < 0.05), but did not alter miR-19a levels in the cell body fraction (1.03 ± 0.13 vs 1 in normal neurons and vs 0.95 ± 0.14 in cel-miR-67, n = 3, p > 0.05), indicating that attenuation of mature miR-19a in the distal axons does not affect miR-19a levels in the cell bodies. Axons with reduced-miR-19a exhibited a substantial decrease of axonal elongation measured by time-lapse microcopy compared with axons transfected with cel-miR-67 during 60 min of recording (Fig. 5F,G). These data indicate that endogenous axonal miR-19a is essential to axonal outgrowth.
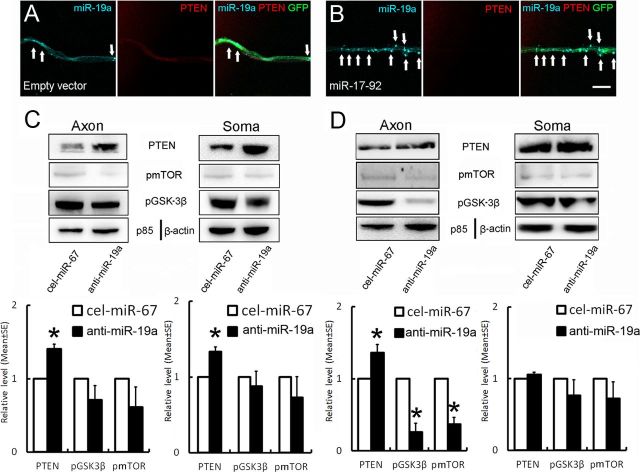
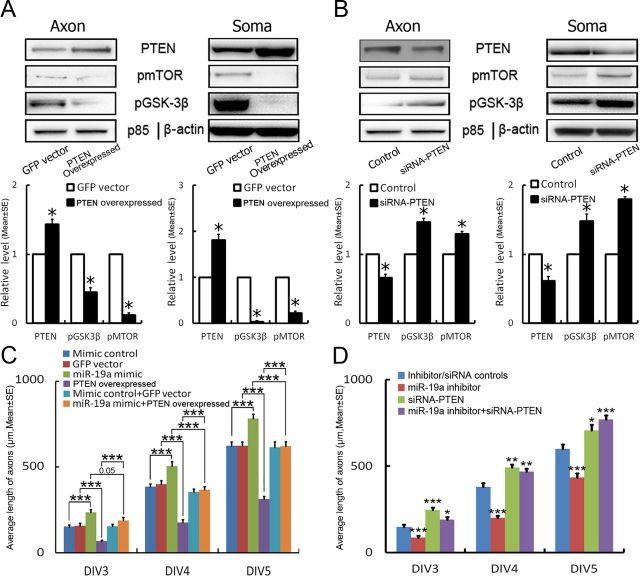
PTEN is a validated target of miR-19a (Olive et al., 2009). Using fluorescence in situ hybridization (FISH) in combination with immunofluorescent staining to simultaneously detect miR-19a and PTEN, we examined colocalization of miR-19a and PTEN in the distal axons. Punctate distribution of miR-19a signals and PTEN immunoreactivity were detected along the length of GFP-expressing axons of the neurons transfected with the GFP-empty vector (Fig. 6A), whereas GFP-expressing axons of the neurons transfected with GFP-miR-17–92 cluster showed increases in miR-19a signals and marked reduction of PTEN immunoreactivity (Fig. 6B). These data suggest that miR-19a signals are inversely associated with PTEN proteins in the distal axons. We therefore investigated whether inhibition of endogenous miR-19a increases PTEN levels. Using Western blot, we assessed PTEN proteins in protein extracts harvested from the cell body and axonal compartments of the neurons transfected by electroporation with the miR-19a hairpin inhibitor. Inhibition of endogenous miR19a significantly increased PTEN proteins in the distal axons and cell bodies (Fig. 6C). However, reduction of pmTOR and pGSK-3β was not significant (Fig. 6C). Because the transfection with electroporation reduced endogenous miR-19a levels in somata and axons, we next examined whether axonal attenuation of miR-19a by the miR-19a inhibitor affects PTEN proteins. Total axonal proteins were extracted from the axonal compartment on DIV5 after axonal transfection with the miR-19a hairpin inhibitor. Western blot analysis showed that attenuation of endogenous axonal miR-19a significantly elevated PTEN levels and reduced levels of pmTOR and pGSK-3β in axons, but did not significantly alter these protein levels in the cell body compartment (Fig. 6D), suggesting that local miR-19a in axons modulates proteins involved in the PTEN/mTOR signaling pathway. Subsequently, we examined whether alteration of PTEN levels modifies the effect of miR-19a on axonal outgrowth by PTEN overexpression in the neurons transfected with a pcDNA3-EGFP-PTEN plasmid or attenuation of endogenous PTEN by siRNA-PTEN. Western blot analysis showed overexpression of PTEN reduced the level of pmTOR and pGSK-3β in somata and axons (Fig. 7A). Conversely, the level of pmTOR and pGSK-3β was decreased in PTEN knock-down neurons (Fig. 7B), indicating that PTEN regulates mTOR and GSK-3β activity. Overexpression of PTEN led to a >50% reduction of axonal outgrowth (Fig. 7C). Cotransfection of PTEN and miR-19a mimics suppressed miR-19a-enhanced axonal outgrowth (Fig. 7C). In contrast, attenuation of endogenous PTEN by siRNA-PTEN robustly increased axonal outgrowth and rescued the inhibitory effect of miR-19a-siRNA on axonal outgrowth (Fig. 7D). Together, these data support that the PTEN is a crucial factor in miR-19a-mediated axonal outgrowth.
Figure 6.
Effects of attenuation of endogenous miR-19a on PTEN proteins. A, B, Representative FISH and immunofluorescent images acquired with the confocal microscope in the axonal compartment show the presence of punctate miR-19a signals (A, B, blue, arrows) and PTEN immunoreactivity (A, B, red) along the length of GFP+ axon (A, B, green) of the cortical neurons transfected with the GFP-empty vector (A, Empty vector) and the GFP-miR-17–92 vector (B, miR-17–92). C, D, Western blot data show levels of PTEN, pmTOR, and pGSK-3β in the distal axons (Axon) and cell bodies (Soma) of neurons transfected by electroporation (C) or axonal transfection (D) with the miR-19a inhibitor, or cel-miR-67 inhibitor, while p85 and β-actin were used as internal controls for the distal axons and cell bodies, respectively. N = 3/group. *p < 0.05. Scale bar, 20 μm.
Figure 7.
The effect of PTEN on axonal outgrowth. A, B, Representative Western blots and quantitative data show protein levels of PTEN, pmTOR, and pGSK-3β in the distal axons (Axon) and cell bodies (Soma) of neurons transfected with the pcDNA3-EGFP-PTEN plasmid (A, PTEN overexpressed), control vector (A, GFP-vector), siRNA-PTEN (B), or control vector (B, Control). N = 3/group. *p < 0.05. C is quantitative data showing the effect of overexpression of PTEN (PTEN overexpressed) on axonal outgrowth of neurons with miR-19 mimics or mimic control during DIV3 to DIV5. D, Quantitative data of the effect of knock-down PTEN (siRNA-PTEN) on axonal outgrowth of neurons transfected with miR-19a inhibitor during DIV3 to DIV5. N = 5/group. *p < 0.05, **p < 0.01, and ***p < 0.001.
Discussion
The present study demonstrates the presence of individual members of the miR-17–92 cluster in the distal axons of embryonic cortical neurons. Overexpression of the miR-17–92 cluster amplified axonal outgrowth, which was associated with reduction of PTEN proteins in axons. In contrast, attenuation of endogenous axonal miR-19a suppressed axonal outgrowth and elevated axonal PTEN levels. These findings provide the first evidence that axonal alteration of miR-17–92 cluster expression regulates axonal outgrowth and PTEN protein levels in developing axons.
The miR-17–92 cluster has been extensively investigated in the tumors (Hayashita et al., 2005; Mi et al., 2010). The miR-17–92 cluster is a typical example of a polycistronic miRNA cluster encoding the miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92, and functions as an oncogene (Hong et al., 2010). By combining microarray expression profiling with miRNA-specific real-time RT-PCR, Natera-Naranjo et al. (2010) recently showed the presence of the miR-17–92 cluster within the axons of superior cervical ganglia neurons. However, biological functions of this cluster in regulating axonal outgrowth have not been studied. Our findings of axonally localized miR-17–92 members in the cortical neurons are consistent with this published study. Furthermore, we found that overexpression of the miR-17–92 cluster in the cortical neurons elevated individual members of the cluster in the distal axons and enhanced axonal outgrowth, whereas local attenuation of endogenous miR-19a by the miRNA hairpin inhibitor in the distal axons markedly reduced axonal outgrowth. These findings suggest that the axonal miR-17–92 cluster mediates axonal outgrowth. Several lines of evidence support that miRNAs can function locally in axons. Following their export from the nucleus, pre-miRNAs are subsequently processed into mature miRNAs in the cytoplasm by Dicer (Schratt, 2009). Mature miRNAs are incorporated into RISC that includes Ago proteins (Schratt, 2009). The presence of miRNA processing machinery, Dicer and RISC, in the distal axons of neurons has been reported, although the mechanism by which miRNAs and pre-miRNAs are shuttled from the cytoplasm into distal axons has not been demonstrated (Hengst et al., 2006; Aschrafi et al., 2008). The present study also shows that Dicer and Ago2 proteins are present in the distal axons of embryonic cortical neurons.
The present study showed that miR-18a and miR-92 were more enriched than other members of the miR-17–92 cluster in axons. Enrichment of miR-92 has also been detected in distal axons of sympathetic neurons (Natera-Naranjo et al., 2010). Neuron navigator 2 is miR-18a putative target and regulates axonal elongation (Muley et al., 2008). During development of human brain, neurons express miR-92 that has putative target genes related to calcium signaling (Somel et al., 2011). Collectively, these data imply a role of miR-18a and miR-92 in regulating genes associated with axonal outgrowth, although the present study did not find that attenuation of endogenous miR-18a affected axonal outgrowth.
Overexpression of the miR-17–92 cluster appears to change profiles of individual members of the cluster in axons, specifically for miR-20a. The presence of Dicer and Ago2 in distal axons could locally regulate miRNA biogenesis. However, we do not know the mechanisms underlying alteration of a particular miRNA profile after overexpression of the miR-17–92 cluster, which warrants future investigation.
Among many miR-17–92 cluster putative target genes encoding proteins, TSP1, CTGF, and PTEN have been indicated in regulating axonal and synaptogenesis (Sandvig et al., 2004; Christopherson et al., 2005; Chadborn et al., 2006; Park et al., 2008). Our data demonstrate the presence of PTEN and TSP1, but not CTGF, proteins in the distal axons, which is consistent with other published findings of the presence of PTEN in axons (Chadborn et al., 2006). Overexpression of the miR-17–92 cluster markedly reduced axonal PTEN proteins and did not significantly alter TSP1 protein levels. Moreover, the reduction of PTEN proteins was associated with increases in pmTOR and pGSK-3β proteins in axons. Reduction of PTEN activates the PI3K/mTOR signaling pathway, and GSK-3β is one of the downstream targets of PTEN (Jiang et al., 2005). Furthermore, we found that transfection of the miR-19a hairpin inhibitor selectively into the distal axons resulted in substantial elevation of axonal PTEN proteins and decreases of pmTOR and pGSK-3β proteins in axons, whereas these protein levels in cell bodies did not change. PTEN has been verified as a target gene of miR-19a (Olive et al., 2009; Hong et al., 2010). Thus, these data suggest that endogenous axonal miR-19a locally regulates PTEN proteins in axons. The PTEN is a primary antagonist of PI3K that is an upstream activator of mTOR (Shi et al., 2011). The PTEN/mTOR signaling pathway regulates axonal regeneration after spinal cord injury (Park et al., 2008; Christie et al., 2010). Phosphorylation of GSK-3β inactivates GSK-3β (Hur and Zhou, 2010). GSK-3β acts as a negative regulator of axon formation (Hur and Zhou, 2010). We previously demonstrated that inhibition of GSK-3β promotes axonal outgrowth in cortical neurons (Ueno et al., 2012). The present study showed that overexpression and knockdown of PTEN led to inactivation and activation of mTOR and GSK-3β, respectively, and overrode the effect of miR-19a on axonal outgrowth. Axonal inhibition of PI3K with LY294002 or mTOR with rapamycin suppressed axonal outgrowth enhanced by overexpression of the miR-17–92 cluster. Collectively, these data suggest that the PTEN/PI3K/mTOR signaling pathway in developing axons plays an important role in miR-17–92 cluster-mediated axonal outgrowth.
The local regulation of gene expression and protein synthesis in axons is required for axonal outgrowth and for the response of growth cones to guidance cues (Martin, 2004; Jung et al., 2012). Molecular mechanisms underlying the regulation are emerging. The present study provides evidence that axonal miR-17–92 cluster modulates PTEN proteins in axons of embryonic cortical neurons, which are likely involved in axonal outgrowth.
Footnotes
This work was supported by National Institutes of Health Grants RO1 AG037506 and RO1 NS75156. Y.U. was a recipient of a research fellowship from the Uehara Memorial Foundation, Tokyo, Japan. We thank Drs. Samie R. Jaffrey and Deglincerti Alessia (Weill Medical College, Cornell University) for providing us protocols and suggestions in the axonal and somal protein preparations; and Dr. Constance L. Cepko (Howard Hughes Medical Institute, Harvard Medical School) for providing us the pCAG-GFP vector.
The authors declare no competing financial interests.
References
- Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Buller B, Liu X, Wang X, Zhang RL, Zhang L, Hozeska-Solgot A, Chopp M, Zhang ZG. MicroRNA-21 protects neurons from ischemic death. FEBS J. 2010;277:4299–4307. doi: 10.1111/j.1742-4658.2010.07818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadborn NH, Ahmed AI, Holt MR, Prinjha R, Dunn GA, Jones GE, Eickholt BJ. PTEN couples Sema3A signalling to growth cone collapse. J Cell Sci. 2006;119:951–957. doi: 10.1242/jcs.02801. [DOI] [PubMed] [Google Scholar]
- Christie KJ, Webber CA, Martinez JA, Singh B, Zochodne DW. PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J Neurosci. 2010;30:9306–9315. doi: 10.1523/JNEUROSCI.6271-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Dajas-Bailador F, Bonev B, Garcez P, Stanley P, Guillemot F, Papalopulu N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat Neurosci. 2012 doi: 10.1038/nn.3082. doi: 10.1038/nn.3082. Advance online publication. Retrieved March 20, 2013. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, Xiao C, Hu WY, Han J, Sun P. The miR-17–92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70:8547–8557. doi: 10.1158/0008-5472.CAN-10-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur EM, Zhou FQ. GSK3 signalling in neural development. Nat Rev Neurosci. 2010;11:539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Guo W, Liang X, Rao Y. Both the establishment and the maintenance of neuronal polarity require active mechanisms: critical roles of GSK-3beta and its upstream regulators. Cell. 2005;120:123–135. doi: 10.1016/j.cell.2004.12.033. [DOI] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–324. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Wilson AC, Chao MV, Mohr I. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translation repressor. Genes Dev. 2012;26:1527–1532. doi: 10.1101/gad.190157.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Liu K, Lu Y, Lee JK, Samara R, Willenberg R, Sears-Kraxberger I, Tedeschi A, Park KK, Jin D, Cai B, Xu B, Connolly L, Steward O, Zheng B, He Z. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat Neurosci. 2010;13:1075–1081. doi: 10.1038/nn.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Chopp M, Zhang RL, Hozeska-Solgot A, Gregg SC, Buller B, Lu M, Zhang ZG. Angiopoietin 2 mediates the differentiation and migration of neural progenitor cells in the subventricular zone after stroke. J Biol Chem. 2009;284:22680–22689. doi: 10.1074/jbc.M109.006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Chopp M, Zhang RL, Tao T, Wang XL, Kassis H, Hozeska-Solgot A, Zhang L, Chen C, Zhang ZG. MicroRNA profiling in subventricular zone after stroke: MiR-124a regulates proliferation of neural progenitor cells through notch signaling pathway. PLoS One. 2011;6:e23461. doi: 10.1371/journal.pone.0023461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta]CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mahad DJ, Ziabreva I, Campbell G, Lax N, White K, Hanson PS, Lassmann H, Turnbull DM. Mitochondrial changes within axons in multiple sclerosis. Brain. 2009;132:1161–1174. doi: 10.1093/brain/awp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC. Local protein synthesis during axon guidance and synaptic plasticity. Curr Opin Neurobiol. 2004;14:305–310. doi: 10.1016/j.conb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Li Z, Chen P, He C, Cao D, Elkahloun A, Lu J, Pelloso LA, Wunderlich M, Huang H, Luo RT, Sun M, He M, Neilly MB, Zeleznik-Le NJ, Thirman MJ, Mulloy JC, Liu PP, Rowley JD, Chen J. Aberrant overexpression and function of the miR-17–92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci U S A. 2010;107:3710–3715. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muley PD, McNeill EM, Marzinke MA, Knobel KM, Barr MM, Clagett-Dame M. The atRA-responsive gene neuron navigator 2 functions in neurite outgrowth and axonal elongation. Dev Neurobiol. 2008;68:1441–1453. doi: 10.1002/dneu.20670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. Rna. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17–92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, He Z. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig A, Berry M, Barrett LB, Butt A, Logan A. Myelin-, reactive glia-, and scar-derived CNS axon growth inhibitors: expression, receptor signaling, and correlation with axon regeneration. Glia. 2004;46:225–251. doi: 10.1002/glia.10315. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- Shi GD, OuYang YP, Shi JG, Liu Y, Yuan W, Jia LS. PTEN deletion prevents ischemic brain injury by activating the mTOR signaling pathway. Biochem Biophys Res Commun. 2011;404:941–945. doi: 10.1016/j.bbrc.2010.12.085. [DOI] [PubMed] [Google Scholar]
- Shi SH, Cox DN, Wang D, Jan LY, Jan YN. Control of dendrite arborization by an Ig family member, dendrite arborization and synapse maturation 1 (Dasm1) Proc Natl Acad Sci U S A. 2004;101:13341–13345. doi: 10.1073/pnas.0405370101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G, Li N, Hu Y, Chen W, Pääbo S, Khaitovich P. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Wu D, Li P, Xu B, Lu Q, Zhang W. microRNA-18a, a member of the oncogenic miR-17–92 cluster, targets Dicer and suppresses cell proliferation in bladder cancer T24 cells. Mol Med Rep. 2012;5:167–172. doi: 10.3892/mmr.2011.591. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y, Chopp M, Zhang L, Buller B, Liu Z, Lehman NL, Liu XS, Zhang Y, Roberts C, Zhang ZG. Axonal outgrowth and dendritic plasticity in the cortical peri-infarct area after experimental stroke. Stroke. 2012;43:2221–2228. doi: 10.1161/STROKEAHA.111.646224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Almen GC, Verhesen W, van Leeuwen RE, van de Vrie M, Eurlings C, Schellings MW, Swinnen M, Cleutjens JP, van Zandvoort MA, Heymans S, Schroen B. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell. 2011;10:769–779. doi: 10.1111/j.1474-9726.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314:2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RL, LeTourneau Y, Gregg SR, Wang Y, Toh Y, Robin AM, Zhang ZG, Chopp M. Neuroblast division during migration toward the ischemic striatum: a study of dynamic migratory and proliferative characteristics of neuroblasts from the subventricular zone. J Neurosci. 2007a;27:3157–3162. doi: 10.1523/JNEUROSCI.4969-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RL, Zhang ZG, Wang Y, LeTourneau Y, Liu XS, Zhang X, Gregg SR, Wang L, Chopp M. Stroke induces ependymal cell transformation into radial glia in the subventricular zone of the adult rodent brain. J Cereb Blood Flow Metab. 2007b;27:1201–1212. doi: 10.1038/sj.jcbfm.9600430. [DOI] [PubMed] [Google Scholar]
- Zhang RL, Chopp M, Gregg SR, Toh Y, Roberts C, Letourneau Y, Buller B, Jia L, P Nejad Davarani S, Zhang ZG. Patterns and dynamics of subventricular zone neuroblast migration in the ischemic striatum of the adult mouse. J Cereb Blood Flow Metab. 2009;29:1240–1250. doi: 10.1038/jcbfm.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]