Abstract
Childhood maltreatment is likely to influence fundamental biological processes and engrave long-lasting epigenetic marks, leading to adverse health outcomes in adulthood. We aimed to elucidate the impact of different early environment on disease-related genome-wide gene expression and DNA methylation in peripheral blood cells in patients with posttraumatic stress disorder (PTSD). Compared with the same trauma-exposed controls (n = 108), gene-expression profiles of PTSD patients with similar clinical symptoms and matched adult trauma exposure but different childhood adverse events (n = 32 and 29) were almost completely nonoverlapping (98%). These differences on the level of individual transcripts were paralleled by the enrichment of several distinct biological networks between the groups. Moreover, these gene-expression changes were accompanied and likely mediated by changes in DNA methylation in the same loci to a much larger proportion in the childhood abuse (69%) vs. the non-child abuse-only group (34%). This study is unique in providing genome-wide evidence of distinct biological modifications in PTSD in the presence or absence of exposure to childhood abuse. The findings that nonoverlapping biological pathways seem to be affected in the two PTSD groups and that changes in DNA methylation appear to have a much greater impact in the childhood-abuse group might reflect differences in the pathophysiology of PTSD, in dependence of exposure to childhood maltreatment. These results contribute to a better understanding of the extent of influence of differences in trauma exposure on pathophysiological processes in stress-related psychiatric disorders and may have implications for personalized medicine.
Keywords: epigenome, biomarkers, psychiatry, development
Childhood maltreatment is a complex problem that exerts an enormous impact on individuals, families, and society, and is of great significance for public health. Maltreatment during childhood likely influences fundamental biological processes and engraves long-lasting epigenetic marks, leading to adverse health outcomes in adulthood (1). Exposure to adverse life events in childhood has not only been linked to an increased susceptibility for a number of psychiatric disorders, but also to cardiovascular disease, diabetes, and chronic lung disease, possibly via long-term influences on the immune system (2–9). This finding suggests that early adverse experiences not only alter neurobiological systems leading to an increased risk for psychiatric disorders, but may have a long-lasting effect on a number of organ systems. Experience of childhood abuse might influence these biological systems via epigenetic modifications conferring lifelong susceptibility to disease. In fact, in humans, distinct epigenetic and gene-expression changes have been observed in postmortem brains of suicide victims with a history of childhood abuse compared with suicide victims without a history of childhood abuse or control subjects (10).
Early life trauma is a strong risk factor for stress-related psychiatric disorders, which themselves have been shown to be associated with distinct changes in gene-expression and epigenetic profiles, both in the brain as well as in peripheral tissues (11–13). Posttraumatic stress disorder (PTSD) is one example of a common and disabling disorder that occurs after exposure to potentially life-threatening traumatic events during childhood and adulthood. Individuals who experience early life stress have been shown to develop PTSD in adulthood more often than individuals with no history of early life stress (14–16), and are also more likely to be exposed to traumatic events in adulthood (17, 18). PTSD and other stress-related disorders can thus occur both in the presence and absence of early life trauma.
It has been suggested that the impact of trauma can depend on the type and timing of the adverse events, and this in turn determines downstream consequences, such as perturbation of biological pathways (19–21). Although individuals with PTSD with or without a history of exposure to childhood maltreatment meet diagnostic criteria for the same disorder, it is not clear whether disease would be associated with similar biological modifications.
The central aim of this study was to search for evidence at the genome-wide level of such distinct disease-related biological modifications associated with the presence or absence of a history of childhood maltreatment. This goal was addressed by interrogating the influence of early life trauma on gene-expression and epigenetic signatures in immune cells occurring in patients with PTSD as an example of a stress-related psychiatric disorder.
Results
In the present study we selected 169 individuals, all of whom had experienced at least two types trauma other than childhood abuse. Of these individuals, 108 had a trauma history but did not have a life-time history of PTSD or current PTSD symptoms, and these were thus considered trauma-exposed controls. Sixty-one individuals met criteria for current PTSD, of which 32 reported a history of childhood maltreatment and 29 did not report childhood abuse. Clinical and epidemiological demographics of the participants across the three investigated groups are depicted in Table 1. To control for the observed significant group differences in age, sex, ethnicity, adult trauma severity, and substance abuse, these variables were used as covariates in the association analyses.
Table 1.
Distribution and comparisons of demographic and clinical variables of the samples included in the study
| Sample | Demographics | Group comparisons | ||||
| Controls (n = 108) | PTSD + child abuse (n = 32) | PTSD no child abuse (n = 29) | P value | P value | P value | |
| Controls ∼ PTSD + child abuse | Controls ∼ PTSD no child abuse | PTSD + child abuse ∼ PTSD no child abuse | ||||
| Age – mean (SD) | 44.23 (1.2) | 39.56 (1.8) | 43.69 (2.0) | 0.059 | 0.044 | 0.134 |
| Sex | ||||||
| Men | 30 (27.8%) | 5 (15.6%) | 13 (44.8%) | 0.244 | 0.113 | 0.023 |
| Women | 78 (72.25%) | 27 (84.4%) | 16 (55.2%) | |||
| Ethnicity | ||||||
| African American | 101 (93.5%) | 24 (75%) | 25 (86.2%) | 0.006 | 0.245 | 0.343 |
| Other | 7 (6.5%) | 8 (25%) | 4 (13.8%) | |||
| PSS – mean (SD) | 3.49 (0.32) | 35.06 (1.2) | 29.83 (1.7) | 1.89 × 10−72 | 1.14 × 10−52 | 0.011 |
| CAPS current – mean (SD) | 26.19 (3.3) | 112.73 (9.8) | 79.71 (10.2) | 1.68 × 10−17 | 4.46 × 10−9 | 0.054 |
| PTSD diagnosis | ||||||
| No | 108 (100%) | 0 | 0 | 2.59 × 10−32 | 1.4 × 10−26 | 1 |
| Yes | 0 | 32 (100%) | 29 (100%) | |||
| BDI mean (SD) | 8.08 (0.7) | 30.13 (2.3) | 25.27 (2.1) | 2.36 × 10−24 | 8.47 × 10−18 | 0.128 |
| Child abuse (CTQ) – mean(SD) | 34.35 (1.0) | 76.23 (3.5) | 37.24 (2.0) | 4.69 × 10−32 | 0.206 | 5.15 × 10−9 |
| No of types of child abuse (moderate to severe) | ||||||
| None | 75 (69.4%) | 0 | 29 (100%) | 1.19 × 10−21 | 0.317 | 2.54 × 10−13 |
| One | 21 (19.4%) | 0 | 0 | |||
| Two | 12 (11.2%) | 32 (100%) | 0 | |||
| Total types adult trauma (TEI) – mean (SD) | 4.63 (0.23) | 7.91 (0.6) | 6.49 (0.6) | 4.69 × 10−9 | 0.005 | 0.154 |
| Substance abuse current | ||||||
| No | 103 (95.4%) | 28 (87.5%) | 25 (86.2%) | 0.111 | 0.077 | 0.881 |
| Yes | 5 (4.6%) | 4 (12.5%) | 4 (13.8%) | |||
| PTSD treatment lifetime | ||||||
| No | 108 (100%) | 24 (72%) | 24 (82.8%) | 8.9 × 10−5 | 0.03 | 0.352 |
| Yes | 0 | 8 (28%) | 4 (17.2%) | |||
Effects of Childhood Maltreatment on PTSD-Associated Transcriptional Profiles.
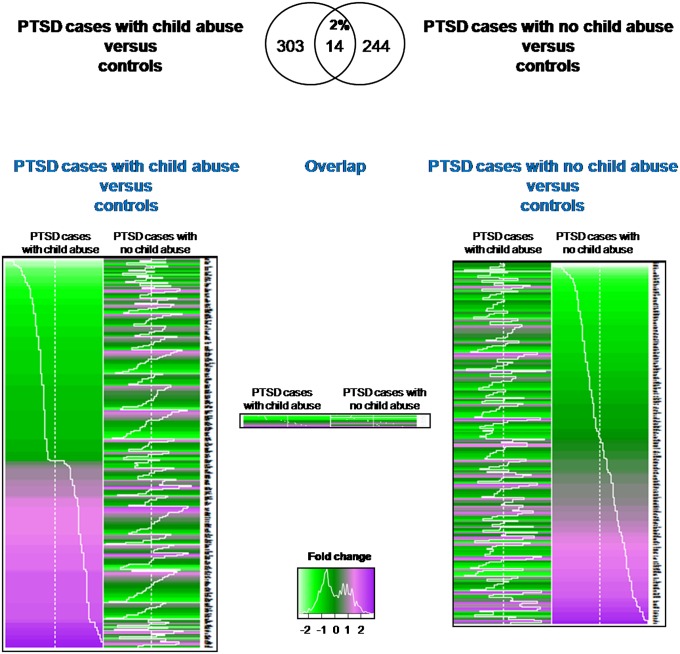
To test for gene-expression differences between individuals with PTSD and trauma-exposed controls, linear regression models were built to assess the influence of the case status on expression profiles. A total of 303 transcripts were differentially expressed between PTSD individuals with child abuse vs. controls, and 244 transcripts were differentially expressed between PTSD individuals without child abuse vs. controls after corrections for multiple testing (adjusted P < 0.05) (Fig. 1 and Dataset S1). An overlap of only 14 transcripts was observed (2%), indicating distinct biomarker profiles in PTSD in the presence or absence of child abuse. This overlap is no more than expected by random chance (Fisher’s exact P value = 0.095). Post hoc analysis identified that only 4% of the differentially expressed transcripts might be confounded by lifetime PTSD treatment and only 1.5% of the transcripts might be confounded by different proportions of immune cells types, suggesting that these factors are not major confounds in the current analysis (Dataset S1). Among the differentially expressed transcripts, we successfully validated the gene-expression changes in the corresponding groups by using quantitative PCR (qPCR) in five of five tested transcripts (Dataset S2).
Fig. 1.
Influence of child abuse on gene expression profiles in peripheral blood in PTSD. (A) Venn diagram representing the overlap of transcripts significantly differentially regulated between individuals with PTSD and child abuse vs. trauma-exposed controls and individuals with PTSD without child abuse vs. trauma-exposed controls. (B) Heatmap of differentially expressed transcripts. Gene-expression fold-changes in comparison with the controls is depicted. Up-regulation is depicted in magenta and down-regulation is depicted in bright green.
Correlation of Gene-Expression Differences with DNA Methylation Changes.
Childhood maltreatment has an influence on biological processes via epigenetic modifications (22). To identify if the above gene-expression changes were correlated with DNA methylation differences in loci encoding these transcripts, we investigated DNA methylation profiles of the transcripts differentially regulated in either of the PTSD case groups versus controls (n = 547) using the Illumina 450 k HumanMethylation array. Transcripts with at least one CpG within their respective gene locus on the Illumina HumanMethylation 450 k array (n = 304) were assessed for methylation differences in the same subjects tested for gene-expression differences. Although at least one significantly differentially methylated CpG site was identified in 69.3% of transcripts unique to PTSD cases with child abuse, only 33.6% of transcripts unique to PTSD cases without child abuse also showed differences on the DNA methylation level for at least one CpG after correction for multiple testing. This difference was even more accentuated in transcripts for which the locus showed significant differences in DNA methylation in five or more CpGs; here, 11.7% of the transcripts showed such matching epigenetic differences in the PTSD child abuse group, but this was only the case for 0.8% of the transcripts from the PTSD non-child abuse-only group (Fig. 2 A and B, and Dataset S3). DNA methylation results were validated for seven CpG sites in two distinct loci using an independent method, the Sequenom EpiTYPER (Dataset S4).
Fig. 2.
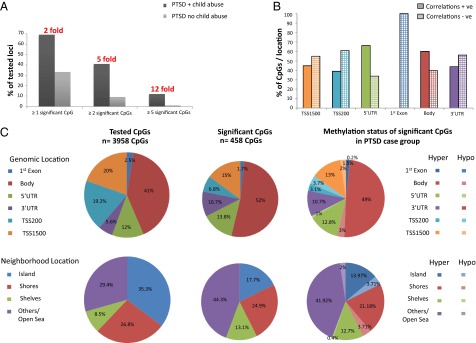
Influence of child abuse on DNA methylation in peripheral blood in PTSD. (A) DNA methylation differences are more frequent in PTSD with child abuse. The bar graph shows the percentage of transcripts differentially regulated in the two respective group comparisons having one or more, two or more, or five or more differentially methylated CpGs in the locus. DNA methylation changes underlying the observed gene expression differences were 2- to 12-fold higher in the PTSD with child abuse group compared with the PTSD with no child abuse group. (B) Correlations of DNA methylation with gene expression. The bar graph shows the percent of CpGs with positive vs. negative correlations with gene expression of the closest transcript in the whole sample stratified by location with respect to the regulated gene. Inverse correlations close to the transcription start sites and an increasing proportion of positive correlations in proximal and distal regulatory sites were observed. (C) Distribution of the tested (left panels) and significant (middle panels) CpGs and direction of methylation across all differentially methylated CpGs in both groups. The right panels denote the relative distributions of all tested and significant CpG sites with respect to the closest gene and the CpG islands and the directions of methylation differences of the significant CpGs. Of the significant CpGs, 90% were hypermethylated and 10% were hypomethylated in the PTSD case group with respect to the controls.
The relation between DNA methylation and gene expression is complex. In line with other studies (23, 24), we observe negative correlations of CpGs with gene expression close to the transcription start sites and an increasing proportion of positive correlations in proximal and distal regulatory sites (Fig. 2B and Dataset S5).
Compared with all tested CpGs within the differentially expressed transcripts, the CpGs that also show significantly different methylation levels are more often located close to the 3′ UTR and in the body of the gene as well as in the shelves of the CpG islands and the open sea. In addition, the vast majority of these CpGs were hypermethylated in the case group (Fig. 2C). In addition, even though the absolute differences in methylation levels between the groups are small (for most less than 5%), the direction of the differences in DNA methylation between the case and the control groups agrees to a large extent (73% for individual CpGs and 94% at the locus level) with the observed group differences in gene expression (see Fig. 3 and Dataset S5 for more detail).
Fig. 3.
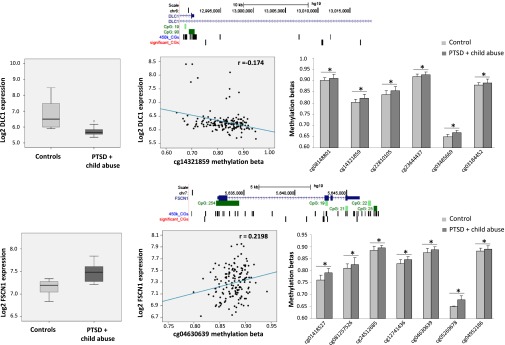
Two examples of directional associations between DNA methylation and gene-expression changes. This figure shows gene-expression and DNA methylation data for two genes, DLC1 (deleted in liver cancer 1 gene) and FSCN1 (fascin homolog 1, actin-bundling protein). The position of the significant CpGs in relation to the gene, CpG islands and other CpGs on the 450 k methylation array are on top of the series of diagrams for each gene (with DLC1 on top and FSCN1 on the bottom). Below, we depict the difference in gene expression for the respective group comparison (Left), the correlation of methylation levels of a representative CpG in that locus with the expression level of the gene in the whole sample (Center), and the group differences in DNA methylation of the significant CpGs in the respective locus (Right). These data illustrate that the direction of the differences in DNA methylation and gene expression between the groups are as expected from the direction of correlation between gene expression and DNA methylation in the overall sample. * indicates adjusted P value of less than or equal to 0.05.
These results indicate that changes in gene expression are more often associated with changes in DNA methylation in PTSD occurring after child abuse than PTSD occurring in the absence of child abuse, and that these changes in DNA methylation may indeed mediate the observed gene-expression changes.
Enrichment of Biological Processes Among Differentially Expressed Transcripts.
As we observed differences in gene-expression profiles and epigenetic marks in PTSD occurring in the presence or absence of childhood maltreatment, we next sought to better comprehend how the transcripts interacted with each other. We built subnetworks from the transcript relationships using a modified gene-set enrichment analysis to identify overrepresented networks in Ariadne Pathway Studio 8.0.
Although functional annotation revealed overlapping pathways between the groups, such as cell survival, cell development, cell migration, cell adhesion, T-cell activation, and immunity networks, several nonoverlapping networks were observed between the groups. The most notable differences included the enrichment of the central nervous system development and tolerance induction pathways in the PTSD group with child abuse and the apoptosis and growth rate networks in the PTSD group without child abuse (Fig. S1 and Dataset S6).
Interestingly, despite the completely divergent transcripts differentially expressed between the two groups, several overlapping networks were identified, suggesting a possible common downstream effect of disease in blood cells.
Discussion
The objective of this study was to interrogate biomarker profiles at the level of gene expression and DNA methylation in individuals with PTSD with or without childhood abuse. We observed that although robust gene-expression differences could be identified for both PTSD groups compared with trauma-exposed controls, the significant changes hardly overlapped at the transcript-specific level (2%) and only modestly (35%) at the biological pathway level. In fact, the differentially expressed transcripts in each group showed highly variable regulation and often the opposite pattern in the respective other group (Fig. 1). Moreover, the extent of epigenetic modifications (i.e., DNA methylation changes) concurrent with these gene-expression changes were up to 12-fold higher in the childhood trauma-exposed PTSD group. Our results imply that biological perturbations in individuals with PTSD who have a history of childhood maltreatment may systematically and meaningfully differ from those individuals with PTSD who do not have a history of childhood maltreatment. These findings suggest that taking into account exposure to childhood maltreatment is of great importance in the search for and application of gene-expression and DNA methylation signatures as biomarkers for PTSD and other trauma-related disorders.
Despite the fact that the differentially expressed transcripts in each PTSD group were distinct, similar cellular processes enriched across both transcript groups, suggesting that these initially different changes could be associated with perturbation of common downstream biological pathways. On the other hand, several specific cellular processes were significantly overrepresented only in one of the PTSD groups, indicating that different biological pathways maybe altered in PTSD with or without child abuse.
Long-term effects of child abuse may be mediated by epigenetic modifications, especially DNA methylation changes (10, 25, 26). The fact that we see up to 12 times as many differentially expressed transcripts also showing differences in DNA methylation in the early abuse group seems consistent with this hypothesis and may even suggest that the biological mechanisms leading to the observed differences in gene expression are distinct depending on the type of trauma exposure. Although the measured changes in DNA methylation were small (98% were less than 5%), the fact that we observe the expected directional association of DNA methylation with gene expression for 94% of the loci (Fig. 3 and Dataset S5), suggests that even such modest methylation differences could be of functional relevance. Comparison of DNA methylation profiles between brain and blood has demonstrated that interindividual DNA methylation differences were the highest in peripheral blood and, even though several DNA methylation signatures were tissue-specific, interindividual variability patterns were consistent across the tissues, suggesting that peripheral blood might reflect at least some of the DNA methylation changes in the brain (27). Furthermore, a recent study in rhesus macaques demonstrated that DNA methylation changes occurring in response to early-life adversity were persistent into adulthood and reflected in both prefrontal cortex and T cells (28). Our finding that the top enriched biological process in the transcripts from peripheral blood cells in PTSD with childhood trauma in central nervous system development may be an indication of such potential blood/brain overlap.
The analyses were performed using a common set of trauma-exposed controls, not matched by exposure to childhood maltreatment. This approach allowed identifying distinct biomarker profiles between the two case groups. The use of two distinct but trauma-matched controls groups would make it more difficult to determine if the differences originate in the case or the control groups. We acknowledge that this approach has limitations and therefore performed a post hoc analysis using matched controls with or without child abuse and did not see any differences in the percentage of overlap in differentially expressed genes (Dataset S7) between the two PTSD case group comparisons, indicating that these results are not likely confounded by the study design. Furthermore, comparisons of the transcripts defining the signature of child abuse [by (i) direct comparison within PTSD cases and (ii) comparison of individuals with and without child abuse using the whole sample] with transcripts significantly different between any of the two PTSD case groups and controls showed a minimal overlap (0.7%), suggesting that the gene-expression signature of PTSD with and without child abuse is distinct from the signature of child abuse within PTSD cases (Dataset S7).
Another potential confound of these results is the possibility that the observed gene-expression and DNA methylation profiles are only a reflection of differences in peripheral blood cell types. Comparison with transcripts correlated with the relative amount of the main immune cells subtypes (CD15–CD16, CD14, CD4, and CD8) revealed that only up to 1.5% of the transcripts might be confounded by differential cell counts. The possibility of the impact of other specific immune-cell subtypes, not measured in our analysis can, however, not be excluded. Furthermore, because the gene-expression levels and methylation patterns were only measured at one time-point, it is difficult conclude whether the expression and methylation changes are indeed the direct consequence of exposure to child abuse per se or a reflection of other factors changed by the response to child abuse or related to the disease itself.
To the best of our knowledge, this genome-wide study assessing the influence of exposure to childhood maltreatment on gene expression and DNA methylation profiles in peripheral blood in PTSD is unique. Our findings are in line with a recent study demonstrating that epigenetic alterations in the hippocampus among suicide completers with or without childhood trauma were distinct (29), although this study did not explore related changes in gene expression. These data provide further evidence that there are substantial differences in disease-related biological perturbations in the presence or absence of exposure to maltreatment in childhood. The fact that several nonoverlapping biological pathways seem to be affected in the two PTSD groups and that changes in DNA methylation appear to have a much greater impact in the childhood maltreatment group may indicate differences in the pathophysiology of PTSD, in dependence of exposure to type of trauma. This finding could not only have implications for biomarker research for stress-related disorders but may help to elucidate pathophysiological differences in dependence of trauma exposure in immune disturbances often accompanying these disorders (3, 4, 30, 31). If these distinct changes are not limited to peripheral blood cells but extend to other organ systems, as indicated by some studies (27, 28, 32), they might also aid in the search for disease mechanisms and therapeutic intervention.
Materials and Methods
Samples.
Participants in this study belonged to a larger study investigating the contribution of genetic and environmental factors in PTSD (33). All study procedures were approved by the institutional review boards of Emory University School of Medicine and Grady Memorial Hospital and all subjects gave written informed consent to the study.
The 396 participants were selected from a larger study, which has been described previously (34). Microarray gene-expression results had been reported in a previous report (34) for a subset of these patients (n = 211). For this study, we selected patients from a now expanded set of microarray gene-expression results (adding 185 new samples with gene-expression arrays to the previously reported ones) and we have now also added DNA methylation array data for all 396 individuals.
Psychological Assessments.
PTSD symptomatic scale and clinician administered PTSD scale.
The modified PTSD symptomatic scale (mPSS) was used as a measure of PTSD symptoms (35). PSS frequency items were summed to obtain a continuous measure, with values of above 20 considered as clinically significant PTSD symptoms. In addition, a categorical PTSD classification was made by using the scores of the B, C, and D clusters of the PSS. Individuals classified as having PTSD needed to have clinically significant symptoms in all three clusters, representing intrusive, avoidance/numbing, and hyperarousal (36). The clinician-administered PTSD scale (CAPS) was available for a subset of 349 of the 396 individuals (37). There was a significant strong positive correlation between PSS and current CAPS in this dataset (Pearson r = 0.69, P = 3.15 × 10−29). The CAPS was also used to assess the presence or absence of lifetime PTSD.
Beck depression inventory.
The Beck depression inventory (BDI) was administered to measure current depression symptoms (38). Current depression and PTSD symptoms were highly correlated (r = 0.653, P = 1.56 × 10−42). Diagnostic tests for variance inflation factor and tolerance revealed a variance inflation factor of 1.72 and tolerance of 0.58, indicating no evidence for multicollinearity between depressive and PTSD symptoms.
Substance abuse.
As described previously (34), two self-report items were used to assess for current alcohol and substance use-related problems.
Trauma-Exposure Measurements.
Childhood trauma questionnaire.
The childhood trauma questionnaire (CTQ) was used as a measure of child abuse. The CTQ is a consistent and stable self-report inventory assessing five types of childhood trauma: sexual, physical, and emotional abuse, and emotional and physical neglect (39). In accordance with prior research, we restricted our analyses to the three abuse subscales only (40). As described previously, participants were dichotomized into two groups for each of the three categories of abuse (presence or absence of moderate to severe physical, sexual, or emotional abuse). Finally, we created a composite variable across the three abuse types, grouping participants into (i) those with no exposure to any childhood abuse (scores less than cut-off for all three abuses), and (ii) those with moderate to severe exposure to at least two types of childhood abuse (scores above cut-off for at least two of the abuses).
Trauma events inventory.
The trauma events inventory (TEI) is the primary measure of non-child abuse trauma in this study (41). Total numbers of different types of trauma among the participants was summed up for each category into a continuous variable. Although the TEI instrument also includes exposure to non-child abuse traumatic events in childhood, the mean age of exposure was 23.81 (0.70) y; hence, in the current study this refers to mostly adult trauma.
Sample Selection.
Of the sample of 396 described above, 169 individuals who had experienced at least two types of moderate-to-severe childhood abuses or at least two other types of traumas during their lifetime were included in the analysis for this study. Of these, individuals with PSS ≤10 and moderate-to-severe child abuse or two other types of traumas were selected as trauma-exposed individuals with neither current nor a history of lifetime PTSD symptoms (trauma controls) (n = 108). From a total of 61 individuals with current PTSD symptoms (PSS > 20 and significant symptoms in all PTSD B, C, and D clusters), we selected individuals (i) who had experienced at least two types of moderate-to-severe childhood abuses (n = 32) and (ii) who had experienced at least two other types of traumas but no child abuse (n = 29).
Statistical Analysis.
Gene-expression data.
Raw microarray scan files from Illumina HT-12 v3.0 arrays (Illumina) were exported using the Illumina Beadstudio program and loaded into R for downstream analysis (www.R-project.org). Evaluation of the different microarray steps was done using the Illumina internal controls. Samples which were >5% SD were excluded. The data were transformed and normalized using the variance stabilizing normalization (42). A total of 15,877 probes passing the filter criteria of Illumina probe detection P value of <0.01 in 5% of the individuals were used for subsequent analysis. To correct for confounding as a result of batch effects, the data were normalized using an empirical Bayes method for batch correction (43). Reproducibility of the gene-expression data were assessed using six pairs of technical replicates, yielding average Pearson correlations of 0.996. General linear models were constructed by regressing the gene-expression profiles against the PTSD group status and adjusting for sex, age, ethnicity, substance abuse, and treatment. The significance of association was estimated by two-tailed P values using the ANOVA F test. Results were corrected for multiple testing by 10,000 permutations using the permutation of regressor residuals test (http://cran.r-project.org/web/packages/glmperm/index.html). Briefly, the general linear models for each transcript were built as described above and the residuals of the regressions were permuted 10,000 times for each transcript using the shuffle-Z method to obtain the empirical P values corrected for multiple testing as described previously (34).
DNA methylation data.
Raw methylation Beta values from the HumanMethylation 450k BeadChip (Illumina) were determined via the Illumina Beadstudio program and loaded into R. Internal Illumina controls were used to assess the quality of staining, extension, hybridization, bisulfite conversion, and specificity. Samples with probe detection call rates <90% and those with an average intensity value of either <50% of the experiment-wide sample mean or <2,000 arbitrary units (AU) were excluded from further analysis, allowing 163 samples for subsequent analysis. Unsupervised hierarchical clustering was performed to identify extreme outliers and global trends in methylation. One sample of male DNA was included on each BeadChip as a technical control throughout the experiment and assessed for reproducibility, with average Pearson correlation coefficient of 0.993 across all replicates. Signals from methylated (M) and unmethylated (U) bead types were used to calculate a beta value as β = M/(U + M). Hybridization and chip batch effects were accounted for using an empirical Bayes method (43). The samples were quantile normalized and peak-corrected using the IMA package functions in R (44, 45). Hybridization and chip batch effects were accounted for using an empirical Bayes method (43). Methylation differences were calculated using generalized linear models in R by regressing the β-values against the PTSD group and adjusting for age, sex, ethnicity, and substance abuse. Results were corrected for multiple testing by 10,000 permutations using the permutation of regressor residuals test (http://cran.r-project.org/web/packages/glmperm/index.html).
Supplementary Material
Acknowledgments
The authors thank Maik Koedel and members of the Grady Trauma Project at Emory—particularly Allen Graham, Kimberly Kerley, Jennifer Davis, Angelo Brown, Alicia Nelson, Jennifer Winkler, Will Holland, Sarah Spann, Kimberly Dement, Dana Goodenough, and Emily Reiser—for excellent technical assistance. This work was supported by the Max-Planck Society; the Behrens-Weise Foundation (E.B.B.); National Institute of Mental Health Grants MH071537 (to K.J.R.) and MH085806 (to A.K.S.); and the Burroughs Wellcome Fund.
Footnotes
Conflict of interest statement: K.J.R. has an unrelated role as cofounder of Extinction Pharmaceuticals for development of N-methyl-d-aspartate-based therapeutics.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217750110/-/DCSupplemental.
References
- 1.Szyf M. The early life social environment and DNA methylation: DNA methylation mediating the long-term impact of social environments early in life. Epigenetics. 2011;6(8):971–978. doi: 10.4161/epi.6.8.16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danese A, et al. Biological embedding of stress through inflammation processes in childhood. Mol Psychiatry. 2011;16(3):244–246. doi: 10.1038/mp.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danese A, et al. Adverse childhood experiences and adult risk factors for age-related disease: Depression, inflammation, and clustering of metabolic risk markers. Arch Pediatr Adolesc Med. 2009;163(12):1135–1143. doi: 10.1001/archpediatrics.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danese A, et al. Elevated inflammation levels in depressed adults with a history of childhood maltreatment. Arch Gen Psychiatry. 2008;65(4):409–415. doi: 10.1001/archpsyc.65.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danese A, Pariante CM, Caspi A, Taylor A, Poulton R. Childhood maltreatment predicts adult inflammation in a life-course study. Proc Natl Acad Sci USA. 2007;104(4):1319–1324. doi: 10.1073/pnas.0610362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pradhan AD, Manson JE, Rifai N, Buring JE, Ridker PM. C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus. JAMA. 2001;286(3):327–334. doi: 10.1001/jama.286.3.327. [DOI] [PubMed] [Google Scholar]
- 7.Ridker PM, Cushman M, Stampfer MJ, Tracy RP, Hennekens CH. Inflammation, aspirin, and the risk of cardiovascular disease in apparently healthy men. N Engl J Med. 1997;336(14):973–979. doi: 10.1056/NEJM199704033361401. [DOI] [PubMed] [Google Scholar]
- 8.Sin DD, Man SF. Why are patients with chronic obstructive pulmonary disease at increased risk of cardiovascular diseases? The potential role of systemic inflammation in chronic obstructive pulmonary disease. Circulation. 2003;107(11):1514–1519. doi: 10.1161/01.cir.0000056767.69054.b3. [DOI] [PubMed] [Google Scholar]
- 9.Widom CS, DuMont K, Czaja SJ. A prospective investigation of major depressive disorder and comorbidity in abused and neglected children grown up. Arch Gen Psychiatry. 2007;64(1):49–56. doi: 10.1001/archpsyc.64.1.49. [DOI] [PubMed] [Google Scholar]
- 10.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drury SS, et al. Telomere length and early severe social deprivation: Linking early adversity and cellular aging. Mol Psychiatry. 2012;17(7):719–727. doi: 10.1038/mp.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberlander TF, et al. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 13.Ressler KJ, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470(7335):492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bremner JD, Southwick SM, Johnson DR, Yehuda R, Charney DS. Childhood physical abuse and combat-related posttraumatic stress disorder in Vietnam veterans. Am J Psychiatry. 1993;150(2):235–239. doi: 10.1176/ajp.150.2.235. [DOI] [PubMed] [Google Scholar]
- 15.Breslau N, Chilcoat HD, Kessler RC, Davis GC. Previous exposure to trauma and PTSD effects of subsequent trauma: Results from the Detroit Area Survey of Trauma. Am J Psychiatry. 1999;156(6):902–907. doi: 10.1176/ajp.156.6.902. [DOI] [PubMed] [Google Scholar]
- 16.Cougle JR, Timpano KR, Sachs-Ericsson N, Keough ME, Riccardi CJ. Examining the unique relationships between anxiety disorders and childhood physical and sexual abuse in the National Comorbidity Survey-Replication. Psychiatry Res. 2010;177(1-2):150–155. doi: 10.1016/j.psychres.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Brewin CR, Andrews B, Valentine JD. Meta-analysis of risk factors for posttraumatic stress disorder in trauma-exposed adults. J Consult Clin Psychol. 2000;68(5):748–766. doi: 10.1037//0022-006x.68.5.748. [DOI] [PubMed] [Google Scholar]
- 18.Widom CS. Posttraumatic stress disorder in abused and neglected children grown up. Am J Psychiatry. 1999;156(8):1223–1229. doi: 10.1176/ajp.156.8.1223. [DOI] [PubMed] [Google Scholar]
- 19.Andersen SL, Teicher MH. Stress, sensitive periods and maturational events in adolescent depression. Trends Neurosci. 2008;31(4):183–191. doi: 10.1016/j.tins.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Heim C, Newport DJ, Mletzko T, Miller AH, Nemeroff CB. The link between childhood trauma and depression: Insights from HPA axis studies in humans. Psychoneuroendocrinology. 2008;33(6):693–710. doi: 10.1016/j.psyneuen.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Lupien SJ, McEwen BS, Gunnar MR, Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci. 2009;10(6):434–445. doi: 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- 22.Klengel T, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16(1):33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenet F, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE. 2011;6(1):e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuang TJ, Chen FC, Chen YZ. Position-dependent correlations between DNA methylation and the evolutionary rates of mammalian coding exons. Proc Natl Acad Sci USA. 2012;109(39):15841–15846. doi: 10.1073/pnas.1208214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies MN, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13(6):R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunnar M, Quevedo K. The neurobiology of stress and development. Annu Rev Psychol. 2007;58:145–173. doi: 10.1146/annurev.psych.58.110405.085605. [DOI] [PubMed] [Google Scholar]
- 27.Sommershof A, et al. Substantial reduction of naïve and regulatory T cells following traumatic stress. Brain Behav Immun. 2009;23(8):1117–1124. doi: 10.1016/j.bbi.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Provençal N, et al. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci. 2012;32(44):15626–15642. doi: 10.1523/JNEUROSCI.1470-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labonté B, et al. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. 2012;69(7):722–731. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith AK, et al. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(6):700–708. doi: 10.1002/ajmg.b.31212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uddin M, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci USA. 2010;107(20):9470–9475. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee RS, et al. A measure of glucocorticoid load provided by DNA methylation of Fkbp5 in mice. Psychopharmacology (Berl) 2011;218(1):303–312. doi: 10.1007/s00213-011-2307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillespie CF, et al. Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry. 2009;31(6):505–514. doi: 10.1016/j.genhosppsych.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehta D, et al. Using polymorphisms in FKBP5 to define biologically distinct subtypes of posttraumatic stress disorder: Evidence from endocrine and gene expression studies. Arch Gen Psychiatry. 2011;68(9):901–910. doi: 10.1001/archgenpsychiatry.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foa EB, Tolin DF. Comparison of the PTSD symptom scale-interview version and the clinician-administered PTSD scale. J Trauma Stress. 2000;13(2):181–191. doi: 10.1023/A:1007781909213. [DOI] [PubMed] [Google Scholar]
- 36.Jovanovic T, et al. Impaired fear inhibition is a biomarker of PTSD but not depression. Depress Anxiety. 2010;27(3):244–251. doi: 10.1002/da.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blake DD, et al. A clinician rating scale for assessing current and lifetime PTSD: The CAPS-1. Behav Therapist. 1990;13:187–188. [Google Scholar]
- 38.Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Arch Gen Psychiatry. 1961;4:561–571. doi: 10.1001/archpsyc.1961.01710120031004. [DOI] [PubMed] [Google Scholar]
- 39.Bernstein DP, et al. Development and validation of a brief screening version of the Childhood Trauma Questionnaire. Child Abuse Negl. 2003;27(2):169–190. doi: 10.1016/s0145-2134(02)00541-0. [DOI] [PubMed] [Google Scholar]
- 40.Bradley RG, et al. Influence of child abuse on adult depression: Moderation by the corticotropin-releasing hormone receptor gene. Arch Gen Psychiatry. 2008;65(2):190–200. doi: 10.1001/archgenpsychiatry.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hovens JE, Bramsen I, van der Ploeg HM, Reuling IE. Test-retest reliability of the trauma and life events self-report inventory. Psychol Rep. 2000;87(3 Pt 1):750–752. doi: 10.2466/pr0.2000.87.3.750. [DOI] [PubMed] [Google Scholar]
- 42.Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 43.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 44.Dedeurwaerder S, et al. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3(6):771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- 45.Wang D, et al. IMA: An R package for high-throughput analysis of Illumina’s 450K Infinium methylation data. Bioinformatics. 2012;28(5):729–730. doi: 10.1093/bioinformatics/bts013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.