Abstract
Nanotechnology has allowed the construction of various nanostructures for applications, including biomedicine. However, a simple target-specific, economical, and biocompatible drug delivery platform with high maximum tolerated doses is still in demand. Here, we report aptamer-tethered DNA nanotrains (aptNTrs) as carriers for targeted drug transport in cancer therapy. Long aptNTrs were self-assembled from only two short DNA upon initiation by modified aptamers, which worked like locomotives guiding nanotrains toward target cancer cells. Meanwhile, tandem “boxcars” served as carriers with high payload capacity of drugs that were transported to target cells and induced selective cytotoxicity. aptNTrs enhanced maximum tolerated dose in nontarget cells. Potent antitumor efficacy and reduced side effects of drugs delivered by biocompatible aptNTrs were demonstrated in a mouse xenograft tumor model. Moreover, fluorophores on nanotrains and drug fluorescence dequenching upon release allowed intracellular signaling of nanotrains and drugs. These results make aptNTrs a promising targeted drug transport platform for cancer theranostics.
Keywords: self-assembly, DNA nanomedicines, targeted anticancer drug delivery, in vivo
Although chemotherapeutic drugs are widely used in cancer therapy, they lack specificity and can induce cytotoxicity in both cancerous and healthy cells, causing side effects (1), limited maximum tolerated dose (MTD), and reduced therapeutic efficacy (2, 3). A theranostic (4) platform with targeted and efficient drug transport would solve these problems, and, by its programmability, DNA nanotechnology has been used for the rational assembly of one-, two-, and three-dimensional nanostructures (5–8), which have been further studied for biomedical applications, including the passive targeted transport of theranostic agents (9–17). In addition, aptamers, as specific recognition elements, have been studied for active targeted transport of conventional chemotherapeutic drugs (11, 12, 18–21). Nucleic acid aptamers are single-stranded oligonucleotides with unique intramolecular conformations and specific recognition abilities to cognate targets, including mammalian cancer cells (22–26). Recent biotechnological advancements have led to a variety of targeted drug transport (TDT) strategies based on aptamer–drug conjugates or aptamer–nanomaterial assemblies (11, 12, 18–21, 27). However, these strategies have unique limitations that could hamper the transition to clinical application, including (i) complicated design, laborious and uneconomical bulky preparation of myriad ssDNA as building blocks to construct sophisticated nucleic acid-based nanomaterials, or laborious and inefficient preparation of aptamer–drug conjugates (9, 11, 14, 15, 17, 18); (ii) limited drug payload capacity and the attendant high cost, hampering production scale-up (9, 11, 14, 15, 17, 18, 20, 27); (iii) poor biodegradability, causing chronic accumulation of nanomaterials in vivo (28, 29); and (iv) limited universality by the requirement of specific aptamer for drug loading (20).
However, we have designed and engineered a DNA nanostructure able to circumvent these limitations. Specifically, we report an aptamer-tethered DNA nanotrain (aptNTr), which is a long linear DNA nanostructure self-assembled simply from two relatively short DNA building blocks upon initiation of aptamer-tethered trigger probes, through a hybridization chain reaction (8) (Fig. 1A). The programmable, periodic, and biodegradable nature of these nanostructures provides unprecedented opportunities for biomedical applications (30–32). The conditional formation of aptNTrs upon initiation from engineered aptamer-trigger probes ensures that each resultant nanotrain is tethered with an aptamer moiety on one end of the nanoconstruct. These aptamer moieties, capable of selective recognition of cognate target cancer cells, operate like locomotives guiding a series of tandem dsDNA “boxcars” toward target cells. Importantly, the periodically aligned boxcar segments provide a large number of spatially addressable sites, allowing high-capacity loading of therapeutics or bioimaging agents. These features are expected to reduce the cost for DNA preparation, increase MTD, reduce side effects, and improve therapeutic efficacy in cancer therapy. The intrinsic biodegradability of DNA is expected to avoid the otherwise in vivo chronic accumulation of nanomaterials. Moreover, aptamers in these nanostructures are interchangeable, and this system should be able to apply to RNA-based systems and some other drugs, ensuring the wide applicability of this platform.
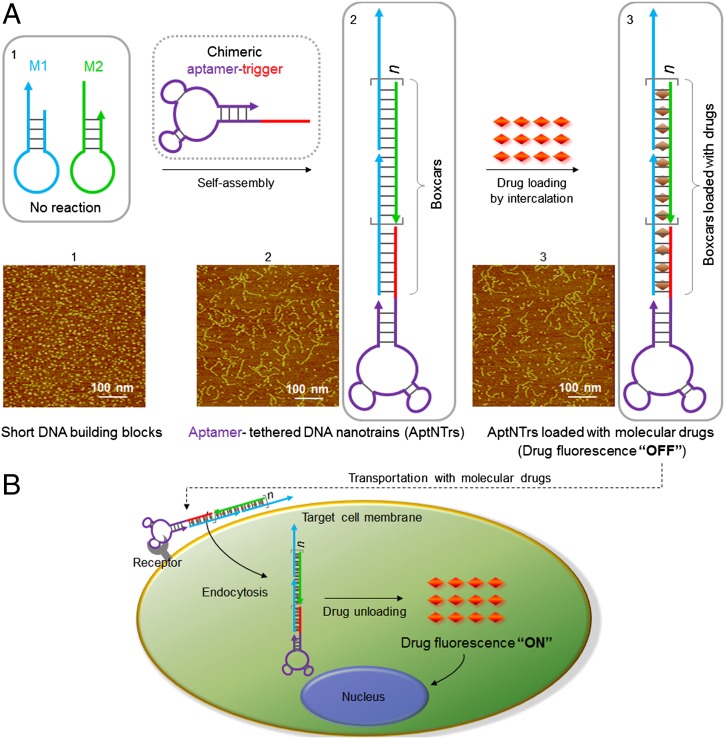
Fig. 1.
Schematics of the self-assembly of aptamer-tethered DNA nanotrains (aptNTrs) for transport of molecular drugs in theranostic applications. (A) Self-assembly of aptNTrs from short DNA building blocks (1) upon initiation from a chimeric aptamer-tethered trigger probe. The resultant long nanotrains (2) were tethered with aptamers working as locomotives on one end, with multiple repetitive “boxcars” to be loaded with molecular drugs (3). AFM images (1–3) showed the morphologies of the corresponding nanostructures [1, M1+M2; 2, sgc8–NTrs; 3, sgc8–NTrs loaded with molecular drugs (Dox)]. (B) The drugs were specifically transported to target cancer cells via aptNTrs, unloaded, and induced cytotoxicity to target cells. The fuorescence of drugs loaded onto nanotrains was quenched (“OFF”), but was recovered upon drug unloading (“ON”), enabling this platform to signal target recognition and drug unloading.
Our results show that these long aptNTrs could be easily prepared through self-assembly. Specific aptamer–target interaction permitted selective recognition and internalization into target cancer cells. The tandem “boxcars,” as described above, allowed for high drug payloads, and the drug-loaded aptNTr complexes showed high stability. aptNTrs can be constructed using aptamers targeting a variety of cancer cells and can be loaded with many types of therapeutic or bioimaging agents, indicating the wide applicability. In addition to good biocompatibility shown under our experimental conditions, these nanotrains selectively transported anticancer drug payload to target cancer cells and offloaded drugs to induce potent cytotoxicity, while dramatically reducing drug toxicity in nontarget cells, indicating selective cytotoxicity and the enhancement of MTD. In vivo evaluation of this TDT system using a mouse xenograft tumor model demonstrated potent antitumor efficacy and reduced side effects of drugs delivered via aptNTrs. Moreover, an array of fluorophores modified on building blocks allowed imaging of intracellular behaviors of nanotrains in target cancer cells. Upon drug offloading, drug fluorescence dequenching also served as a real-time imaging and signaling mechanism of drug release at target cells. Overall, these results make aptNTrs a promising TDT platform for targeted cancer theranostics.
Results and Discussion
Construction and Characterization of aptNTrs.
As a proof of concept, two hairpin monomers (M1, M2, sequences in Table S1) were designed such that the stored energy in the loops is protected by the corresponding stems, preventing their polymerization in the absence of an initiation probe. To construct aptNTrs, aptamer sgc8, which can bind to target human protein tyrosine kinase 7 (PTK7), overexpressed on target CEM (human T-cell acute lymphocytic leukemia) cell membrane but not on nontarget Ramos cells (24, 33), was chosen as a model. To initiate NTr self-assembly, a DNA trigger probe was modified on the 5′-end of sgc8. The selective recognition ability of this chimeric aptamer–trigger probe (sgc8–trigger) was verified (Fig. S1). Introduction of sgc8–trigger to a mixture of M1 and M2 initiates the autonomous polymerization of these building blocks through mutual hybridization, resulting in the self-assembly of sgc8-tethered DNA nanotrains (sgc8–NTrs) (Fig. 1A). The molar ratio of sgc8–trigger to monomers in the initial reaction mixture was optimized (Fig. S2), and a 1:10 ratio was used in subsequent studies. The aptamer-initiated nanotrain formation was demonstrated using atomic force microscopy (AFM). In contrast to unpolymerized monomers (Fig. 1A, 1), nanotrains of up to hundreds of nanometers were observed (Fig. 1A, 2). Agarose gel electrophoresis followed by fluorescence imaging of FITC labeled on sgc8–trigger and ethidium bromide for all DNA species further verified the assembly of long nanotrains in the presence of sgc8–trigger (Fig. 2A) and the incorporation of sgc8 to these nanotrains (Fig. S3). The long nanochains provided correspondingly high numbers of “boxcar” compartments for subsequent drug loading.
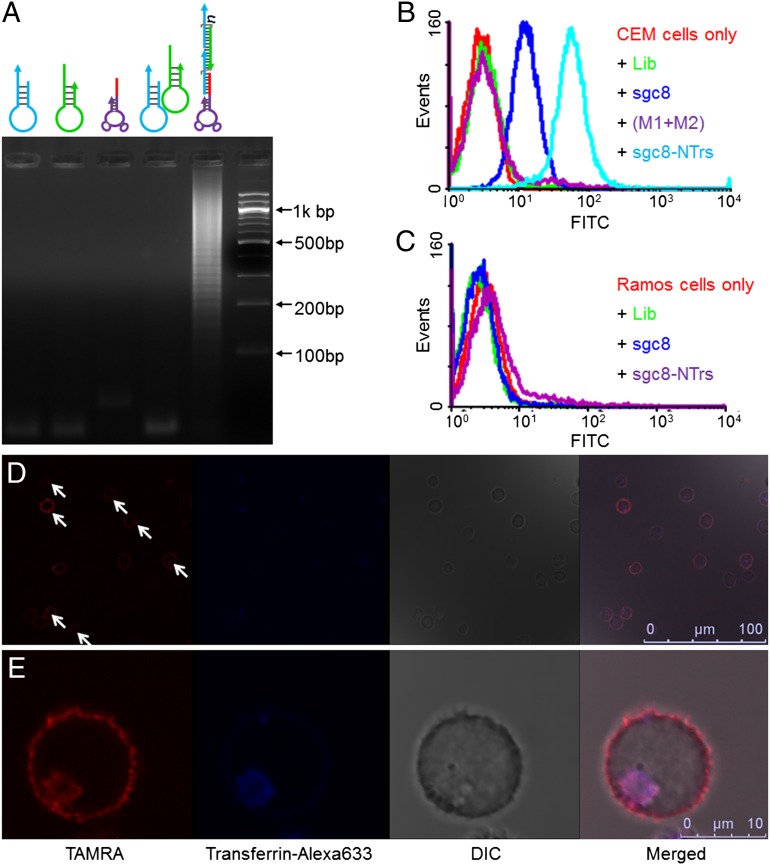
Fig. 2.
Characterization of the formation, selective cancer cell recognition, and internalization of sgc8–NTrs. (A) Agarose gel electrophoresis showing the self-assembly of sgc8–NTrs initiated by sgc8–trigger. (B and C) Flow-cytometric results showing the selective recognition abilities of sgc8–NTrs to CEM cells (B), but not to Ramos cells (C). The presence of multiple monomers on one nanotrain resulted in signal amplification of sgc8–NTr-bound CEM cells [lib, sgc8, M1, M2: labeled with FITC; (M1+M2): unpolymerized M1 and M2]. (D and E) Confocal laser-scanning microscopy images displaying the internalization of sgc8–NTrs into target CEM cells. Cells were incubated with sgc8–NTrs (100 nM sgc8–trigger equivalents) at 37 °C for 2 h, followed by transferrin–Alexa 633 staining. The intracellular TAMRA fluorescence signal (denoted by arrows in D) colocalized with Alexa 633 signal indicates the internalization of sgc8–NTrs through endocytosis (M2: labeled with TAMRA). (Scale bars: D, 100 µm; E, 10 µm.)
Selective Recognition Ability of aptNTrs.
The selective binding ability of sgc8–NTrs to target CEM cells, but not to nontarget Ramos cells, was verified through flow-cytometric analysis (Fig. 2 B and C), providing a basis for the locomotive action of aptamer moiety guiding nanotrains toward target sites. The amplified fluorescence signal intensities of CEM cells bound by sgc8–NTrs further suggested the presence of multiple building blocks in one nanotrain. To study the universality of aptamers for aptNTr construction, another aptamer, AS1411, which can specifically recognize many types of cancer cells (34), was used to construct AS1411-tethered nanotrains, which showed specific recognition of target Huh7 cells (human hepatoma cells) (Fig. S4). However, in this study, sgc8–NTrs were used for subsequent studies. For efficient drug transport, it is essential that macromolecular drug transporters be internalized by diseased cells. Previous work showed that sgc8 was specifically internalized by target CEM cells via endocytosis (35). Compared with the results of a study using sgc8–NTrs at 4 °C (Fig. S5), tetramethylrhodamine (TAMRA)-labeled sgc8–NTrs were selectively internalized into target CEM cells at physiological temperature (37 °C; Fig. 2 D and E). Thus, the large sizes, selective recognition, and internalization capability of aptNTrs all point to their potential as TDT drug transporters with high payload capacity (Fig. 1B).
High Drug Payload Capacity of aptNTrs.
The sgc8–NTrs were then evaluated as carriers for cargo loading. It was previously noted that the DNA building blocks in these nanotrains provide many spatially addressable sites for functionalization, allowing cargos to be loaded by either chemical modification or physical association. In this study, two types of cargos were loaded: 13-nm gold nanoparticles (AuNPs) and chemotherapeutic drugs. The AuNPs were attached to the thiol groups modified on the 5′-ends of M1 and M2. The AuNPs-loaded nanotrains were then confirmed by transmission electron microscopy (Fig. S6). This allows loading nanotrains with cargos that can be conjugated with AuNPs. Chemotherapeutic drugs were also loaded on the nanotrains. Chemotherapy is still one of the primary cancer therapies, but the lack of specificity and potential side effects, as well as limited MTD and therapeutic efficacy, of these drugs (1, 2) make targeted and high-capacity drug transport particularly important. Several widely-used anthracycline anticancer drugs, including doxorubicin (Dox), daunorubicin (DNR), and epirubicin (EPR), were used as drug cargo models. Because it is well known that these drugs can preferentially intercalate into double-stranded 5′-GC-3′ or 5′-CG-3′, resulting in the quenching of drug fluorescence (fluorescence “OFF”) (20, 27, 36), M1 and M2 were designed such that all their sequences would form drug intercalation sites (ACG/CGT) in nanotrains, with each pair of M1 and M2 contributing an average of 16 sites. Consequently, each individual nanotrain needs only one aptamer locomotive for targeting, whereas all of the remaining dsDNA “boxcars,” as characterized above, are used to carry a high payload of drugs, thus reducing the amount of DNA otherwise required to transport a specific amount of drugs. Drug loading on nanotrains by intercalation, as verified by fluorescence spectrometry, showed the gradual quenching of fluorescence with increasing equivalents of sgc8–NTrs (Fig. 3A for Dox, and Fig. S7 for DNR and EPR). Because the molar ratio of sgc8–trigger to M1/M2 in the initial reaction mixture was 1:10, one nanotrain should theoretically average about 160 drug loading sites. Under these conditions, drug fluorescence with a sgc8–NTr/drug molar ratio of 1/50 was dramatically quenched (Fig. 3A). This verified the high drug payload capacity, and 1/50 of this ratio was used in subsequent studies. It is worth noting that high drug payload capacity and the use of short DNA building blocks should substantially reduce the overall cost of reproducing this type of DNA-based TDT nanostructures.
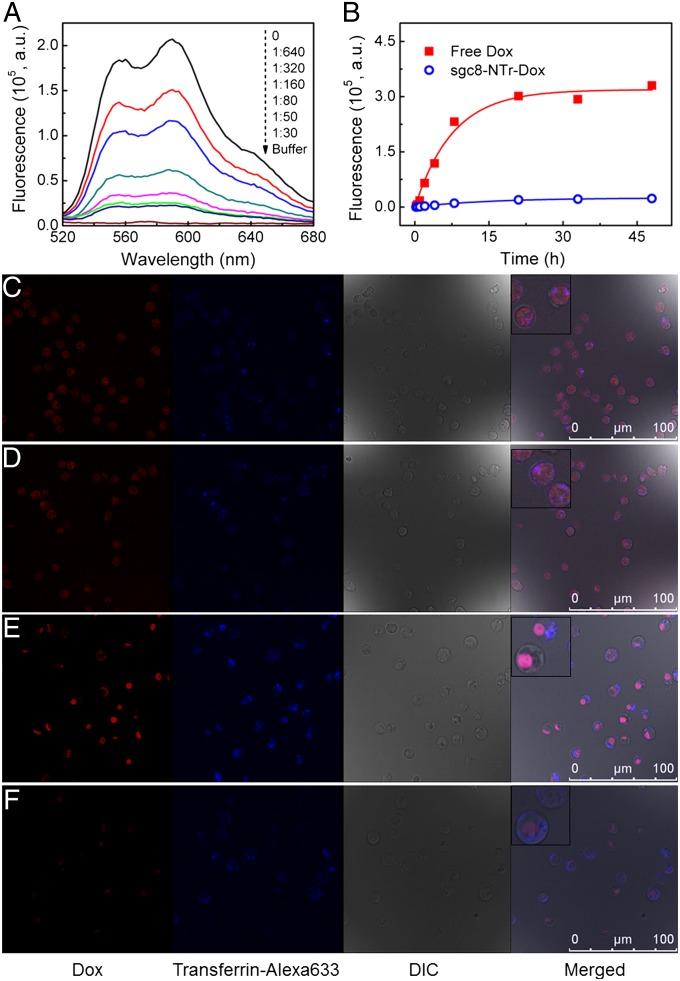
Fig. 3.
Targeted drug transport using aptNTrs with high payload capacity and stability. (A) Fluorescence spectra of Dox (2 µM) with increasing equivalents of sgc8–NTrs (shown by values from top to bottom). The fluorescence quenching indicates drug loading into sgc8–NTrs. The apparent Dox fluorescence quenching with as low as 1/50 NTr equivalents reflects high drug payload capacity. (B) Scattered data points showing the fluorescence intensities of Dox diffused from free Dox or sgc8–NTr–Dox (Dox: 30 µM) during dialysis to outside PBS buffer at different time points. Data were fit to a drug release model by nonlinear regression (fit lines shown in solid). The negligible drug diffusion from nanotrains, in contrast to fast diffusion from free Dox, indicates the high stability of sgc8–NTr–Dox complexes. (C–F) Confocal laser-scanning microscopy images displaying the intracellular signaling of drug unloading and selective drug transport to target cells by sgc8–NTrs. Target CEM cells (C and D) and nontarget Ramos cells (E and F) were treated with free Dox (C and E; 2 µM) and sgc8–NTr–Dox (D and F; 2 µM Dox equivalents), followed by transferrin–Alexa 633 staining. The comparable intracellular Dox fluorescence intensities in C and D, but significantly lower intensities in F than E, indicate the selectivity of drug transport via sgc8–NTrs (Insets: enlarged cells). (Scale bar: 100 µm.)
Dox was used as a model drug in our further study, and we next evaluated the stability of sgc8–NTr–Dox complexes through a drug diffusion experiment using MINI Dialysis Units, and results showed negligible drug diffusion from sgc8–NTrs in contrast to fast diffusion from a free drug solution, indicating the high stability of sgc8–NTr–Dox complexes (Fig. 3B). Furthermore, the stability and integrity of these complexes were also demonstrated by AFM images displaying comparable morphologies and length frequency distributions of unloaded sgc8–NTrs and sgc8–NTr–Dox complexes (Fig. 1A, 2 and 3, and Fig. S8).
Selective Drug Transport via aptNTrs and Real-Time Monitoring of Intracellular Drug Unloading.
To evaluate the selectivity of sgc8–NTrs for transport of molecular drugs, drug uptake was studied with CEM and Ramos cells (Fig. 3 C–F). Cells were treated with free Dox, as a control, or Dox transported by sgc8–NTrs, respectively, followed by microscopic examination. In addition, transferrin–Alexa 633 was used to locate the endosomes (35). The recovery of Dox fluorescence (fluorescence “ON”) after release from nanotrains enabled the real-time signaling of intracellular drug unloading. Strong Dox fluorescence signals were observed in both CEM and Ramos cells treated with free Dox (Fig. 3 C and E). When cells were treated with sgc8–NTr–Dox (Fig. 3 D and F), Dox fluorescence intensity comparable to that of the corresponding cells treated with free Dox was observed only in CEM cells, but not in Ramos cells. This indicates the selectivity of Dox transport mediated by aptNTrs. Presumably, Dox unloading from internalized nanotrains is through simple diffusion and facilitated by intracellular factors such as pH, ionic environment, and nuclease degradation (9, 10, 20). Moreover, in CEM cells treated with sgc8–NTr–Dox, colocalization of Dox and Alexa 633 signals was observed, indicating that some Dox was unloaded after sgc8–NTr–Dox was internalized through endocytosis. The cytosolic Dox signal outside the endosomes might have resulted from the escape of Dox from the endosomes, or from Dox uptaken by cells following release from nanotrains at the cell membrane. To further examine the intracellular behaviors of drugs and nanotrains in target cells, a fluorophore cyanine 5 (Cy5), was used as a model and chemically modified on M1 and M2 to construct sgc8–NTrs. CEM cells were then treated with free Dox and sgc8–NTr–Dox for different time lengths, followed by confocal microscopy observation of Dox and Cy5 fluorescence. In cells treated with free Dox (Fig. S9A), drug was diffused into cells within a short period. Whereas in cells treated with sgc8–NTr–Dox, Dox fluorescence intensity was gradually enhanced (Fig. S9B). Moreover, compared with free Dox, Dox transported by aptNTrs was initially colocalized with nanotrains and then gradually unloaded and distributed in other intracellular areas in a time-dependent manner. Overall, sgc8–NTrs selectively delivered Dox into target cells, and fluorophores modified on nanotrains as well as drug fluorescence dequenching enabled both intracellular signaling of drug unloading and intracellular monitoring of drug and nanotrain behaviors at target cancer cells, providing the basis for future theranostic applications.
In Vitro Selective Cytotoxicity of Anticancer Drugs Transported by aptNTrs.
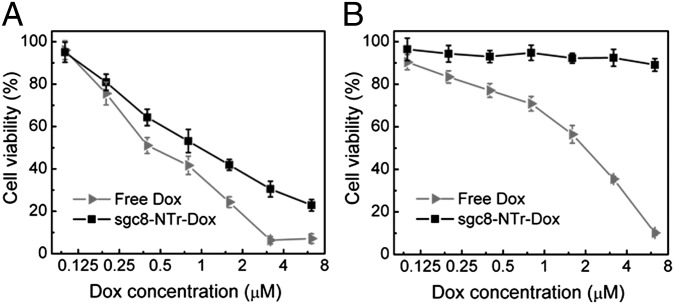
Having established that Dox can be selectively transported into target cells by nanotrains, the resultant cytotoxicity was evaluated by a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay. In this assay, both target CEM cells and nontarget Ramos cells were treated with free Dox and sgc8–NTr–Dox complexes, respectively. Free drug showed dose-dependent cytotoxicity in both CEM cells and Ramos cells. In contrast, only in target CEM cells (Fig. 4) did Dox transported by sgc8–NTrs induce dose-dependent cytotoxicity comparable to that of free Dox. This demonstrated the robust cytotoxic efficacy of sgc8–NTr–Dox in target cells and the excellent selective cytotoxicity of this molecular drug transported by aptNTrs. In contrast, the lack of cytotoxicity of sgc8–NTrs in either CEM or Ramos cells indicates the biocompatibility of these transporters under our experimental conditions (Fig. S10A). We next studied whether aptNTrs maintained selectivity in cancer cell recognition as well as drug delivery under a physiological environment, where nucleases could cleave DNA-based drug carriers before reaching target cells, resulting in loss in selectivity. In this study using cell culture medium containing FBS at 37 °C, the selective recognition ability of sgc8–NTrs was verified using target CEM cells and nontarget Ramos cells (Fig. S10 B–D), and potent cytotoxicity was confirmed for Dox delivered via sgc8–NTrs in CEM cells, but much less cytotoxicity in nontarget Ramos cells (Fig. S10 E and F). This indicates the selectivity of cancer cell recognition and drug delivery under a simulated physiological environment, providing the basis for subsequent in vivo evaluation of this TDT platform. In addition, the selective cytotoxicity of DNR and EPR transported by sgc8–NTrs was demonstrated with CEM cells and Ramos cells using an MTS assay (Fig. S11). In nontarget Ramos cells that simulate normal tissue cells in vivo, the dramatic reduction of cytotoxicities induced by drugs delivered by aptNTrs compared with free Dox suggests the increase of MTD in nontarget cells in vitro, and is expected to increase MTD of drugs transported by aptNTrs in vivo. The selective and potent in vitro therapeutic efficacy indicated that aptNTr is a promising TDT platform.
Fig. 4.
Selective cytotoxicity of molecular drugs (Dox) transported by aptNTrs. (A and B) MTS assay results showing that Dox transported by sgc8–NTrs (sgc8–NTr–Dox) selectively induced potent cytotoxicity and inhibited cell proliferation in target CEM cells (A), but not in nontarget Ramos cells (B), in contrast to nonselective cytotoxicity induced by free Dox in both target and nontarget cells.
In Vivo Efficacy of Anticancer Drugs Transported via aptNTrs.
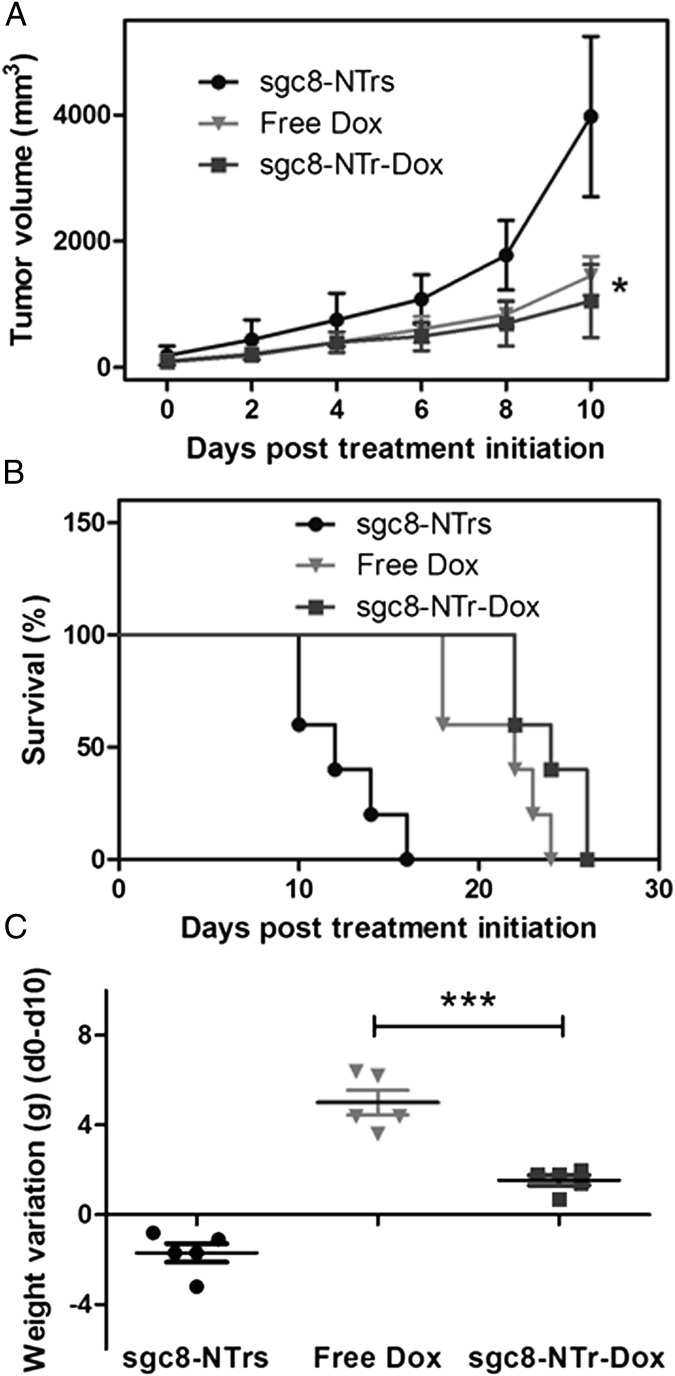
We next evaluated the in vivo therapeutic efficacy (i.e., anticancer therapeutic potency and side effects) of anticancer drugs (Dox) delivered by this TDT platform using a CEM s.c. mouse xenograft tumor model, which was developed by s.c. injection of CEM cells in the back of NOD.Cg-Prkdc (scid) IL2 mice. After dorsal tumor nodules grew to ∼100 mm3, mice were divided into three groups for comparative efficacy studies, in which the following regimens were administered by i.v. injections every other day: (i) sgc8–NTrs, (ii) free Dox, and (iii) sgc8–NTr–Dox. The Dox dosage was kept the same in groups i and ii at 2 mg/kg, which has been reported for use in this mouse strain (27), and the aptNTr dosage in group i was accordingly maintained the same to that in group iii. Tumor size and body weight were monitored every other day. Results (Fig. 5A) showed that, compared with blank drug carriers (sgc8–NTrs), both sgc8–NTr–Dox and free Dox caused significant inhibition of tumor growth, with stronger potency of sgc8–NTr–Dox formulation than free Dox. The stronger potency of sgc8–NTr–Dox than free Dox was attributed to such features of aptNTrs as specific targeting ability and larger molecular weight, which lead to relatively long drug clearance time from blood, relatively high concentration of accumulated drugs, and long drug retention time in tumor. Consistently, both sgc8–NTr–Dox and free Dox led to longer mouse survival time than sgc8–NTrs, with longer survival time in sgc8–NTr–Dox-treated group than free Dox-treated group (Fig. 5B). The inhibition of tumor growth and the elongation of mouse survival time demonstrated the potent anticancer efficacy of drugs delivered via aptNTrs. One primary aim of developing TDT platforms is to reduce the potential nonspecific toxicity in normal tissues and the resultant side effects of drugs. To study whether aptNTrs reduced the side effects of Dox, mouse body weight variations before and after drug administration were examined. Results in Fig. 5C indicate that mice treated with free Dox lost significantly more weight than those treated with sgc8–NTr–Dox, whereas those treated with sgc8–NTrs showed slight body weight increase. These results clearly demonstrated the reduction of drug side effects using aptNTrs, as well as the biocompatibility of aptNTrs. As shown before, increased MTD of sgc8–NTr–Dox compared with free Dox was demonstrated using an in vitro nontarget Ramos cells simulating normal tissue cells in vivo (Fig. 4B and Fig. S11 B and D). We thus speculate that, with the use of MTD of sgc8–NTr–Dox (higher than that of free Dox), stronger therapeutic potency would be achieved than using MTD of free Dox in this study. Detailed study of this would be our interest in future. Overall, these data demonstrated the potent antitumor efficacy and the reduced side effects of drugs delivered via aptNTrs.
Fig. 5.
Potent antitumor efficacy and reduced side effects of drugs transported via aptNTrs. CEM xenograft mouse tumor model was developed by s.c. injection of CEM cells in the back of NOD.Cg-Prkdc (scid) IL2 mice. Mice were divided into three groups that are, respectively, treated by i.v. injections of (i) sgc8–NTrs, (ii) free Dox, and (iii) sgc8–NTr–Dox, with 2 mg/kg Dox or Dox equivalent dosages in ii and iii and accordingly 23 mg/kg sgc8–NTrs in i. (A) Tumor volume up to day 10 after treatment initiation (mean ± SD; n = 5). Asterisk on day 10 represents significant differences between tumor volumes of free Dox- and sgc8–NTr–Dox-treated mice (*P < 0.05, n = 5; Student t test). (B) Survival percentage of mice after treatment initiation. (C) Mouse body weight loss at day 10 compared with day 0, after treatment initiation (mean ± SD; n = 5). Asterisk represents significant differences between weight loss of free Dox- and sgc8–NTr–Dox-treated mice (***P < 0.001, n = 5; one-way ANOVA with Newman–Keuls post hoc test).
Conclusion
In summary, we have developed a TDT platform using self-assembled aptNTrs. This platform presents some remarkable features: (i) easy design and preparation: automated DNA synthesis, only three DNA building blocks needed and the consequent simple DNA sequence design and preparation, and simple DNA nanotrain self-assembly and aptNTr–drug complex formation; (ii) high payload capacity; the unique configuration of aptNTrs allowed all of the duplex “boxcar” DNA, which possessed many addressable sites in one single nanotrain, to maximally contribute to cargo loading, resulting in high payloads of drugs or bioimaging agents; (iii) reduction of the cost of DNA preparation in reproducing this type of drug carriers, due to (a) the use of short DNAs in aptNTrs leading to a relatively high DNA synthesis yield compared with the use of long ones, and (b) the maximal contribution of DNA in aptNTrs to cargo loading resulting in the use of a relatively low amount of DNA to deliver a specific amount of cargo; (iv) aptamer-tethered nanotrains allow for specific targeting in cancer therapy, as demonstrated by selective cancer cell recognition and anticancer drug delivery, selective in vitro cytotoxicity, enhanced MTD, and reduced side effects with potent antitumor efficacy in vivo; (v) bioimaging agents coupled on nanotrains and drug fluorescence dequenching upon release allow for real-time signaling of behaviors of nanotrains and drugs at target cells; and (vi) by simple aptamer or drug substitution, our design can be applied to a variety of target cell types and drugs. This platform should also be applicable to RNA-based systems. These make this TDT platform widely applicable. Furthermore, the degradability of DNA would prevent a chronic accumulation of nanomaterials with MWs above the renal filtration cutoff, and the long linear nanostructure of this drug transporter is expected to increase circulation time in vivo, as shown in studies using filomicelles (37). Collectively, these features are poised to make aptNTrs uniquely attractive for the development of novel TDT platforms in cancer theranostics.
Materials and Methods
Preparation of sgc8–NTrs and Drug Loading into Nanotrains.
Probes M1, M2, and sgc8–trigger were individually snap cooled (heated at 95 °C for 3 min, incubated on ice for 3 min), and then left at room temperature for 2 h. The mixture of sgc8–trigger, M1 (5 µM), and M2 (5 µM) was left at room temperature for 24 h. sgc8–NTr–Dox was prepared by mixing Dox (Fisher Scientific) and prepared sgc8–NTrs in Dulbecco’s PBS (Sigma-Aldrich) supplemented with MgCl2 (5 mM).
Binding Assay Using Flow Cytometry.
The binding abilities of DNA probes or sgc8–NTrs were determined by incubating dye-labeled aptamers or as-prepared aptamer-tethered nanotrains (200 nM aptamer equivalents) with cells (2 × 105) in binding buffer or cell culture medium (200 µL) containing FBS [10% (vol/vol)] on ice or at 37 °C for 30 min, followed by washing with washing buffer and suspending in binding buffer (200 µL). Random sequences (lib) were used as controls. The fluorescence intensities of cells were determined with a FACScan cytometer (Becton Dickinson Immunocytometry Systems). Data were analyzed with the WinMDI or the FlowJo software.
In Vitro Cytotoxicity Assay.
In vitro cytotoxicity was determined using CellTiter 96 Cell Proliferation Assay (Promega). Briefly, cells (5 × 104 cells per well) were treated with sgc8–NTrs, free drug, or drug-loaded on sgc8–NTrs (sgc8–NTr-drug) in medium (without FBS, unless denoted otherwise; 37 °C; 5% CO2) for 2 h; and then cells were precipitated by centrifugation. The 80% supernatant medium was removed, and fresh medium (10% FBS, 200 μL) was added for further cell growth (48 h). After removing cell medium, CellTiter reagent (20 μL) diluted in fresh medium (100 μL) was added to each well and incubated for 1–2 h. The absorbance (490 nm) was recorded using a plate reader (Tecan Safire microplate reader). Cell viability was calculated as described by the manufacturer.
In Vivo Anticancer Efficacy Evaluation.
NOD.Cg-Prkdc (scid) IL2 mice were purchased from The Jackson Laboratory and maintained under pathogen-free conditions. The animal use protocol was approved by the University of Florida Institutional Animal Care and Use Committee on animal care. The mouse xenograft tumor model was developed by s.c. injecting 8 × 106 in vitro-propagated CEM cells (in 100 µL of PBS buffer) into Cg-Prkdc (scid) IL2 mice on the back. Dorsal tumor nodules were allowed to grow to a volume of ∼100 mm3 before treatment initiation. Tumor-bearing mice were randomly assigned to three groups, with five mice in each group: (i) treated with sgc8–NTrs; (ii) treated with free Dox; and (iii) treated with sgc8–NTrs–Dox complexes. The Dox dosage was kept the same in groups ii and iii at 2 mg/kg, and the aptNTr dosage in group i was accordingly maintained the same to that in group iii. Drugs were injected through tail veins every other day, and tumor length and width for each mouse were measured with calipers every other day. Tumor volume was calculated using the following equation:
The body weight of each mouse was also measured every other day to monitor the potential drug toxicity. Mice were killed when tumor volume exceeded 2,000 mm3 or developed ulceration.
Supplementary Material
Acknowledgments
We thank Dr. Kathryn R. Williams for manuscript review, Dr. Mingxu You, Prof. Zhuo Chen, Prof. Mao Ye, and Prof. Xiaobing Zhang for insightful discussions, Mr. Yuliang Zhang for assistance with AFM imaging, Xiaokui Zhang for assistance with mouse work, and Mr. Craig Moneypenny for assistance with confocal microscopy study. This work is supported by National Key Scientific Program of China Grant 2011CB911000, National Natural Science Foundation of China Grant 21221003, China National Instrumentation Program Grant 2011YQ03012412, and by US National Institutes of Health Grants GM079359 and CA133086.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220817110/-/DCSupplemental.
References
- 1.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: Molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. 2004;56(2):185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 2.Cardinale D, et al. Anthracycline-induced cardiomyopathy: Clinical relevance and response to pharmacologic therapy. J Am Coll Cardiol. 2010;55(3):213–220. doi: 10.1016/j.jacc.2009.03.095. [DOI] [PubMed] [Google Scholar]
- 3.MacKay JA, et al. Self-assembling chimeric polypeptide-doxorubicin conjugate nanoparticles that abolish tumours after a single injection. Nat Mater. 2009;8(12):993–999. doi: 10.1038/nmat2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lammers T, Aime S, Hennink WE, Storm G, Kiessling F. Theranostic nanomedicine. Acc Chem Res. 2011;44(10):1029–1038. doi: 10.1021/ar200019c. [DOI] [PubMed] [Google Scholar]
- 5.Seeman NC. Nanomaterials based on DNA. Annu Rev Biochem. 2010;79(1):65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bath J, Turberfield AJ. DNA nanomachines. Nat Nanotechnol. 2007;2(5):275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 7.Pinheiro AV, Han D, Shih WM, Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat Nanotechnol. 2011;6(12):763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci USA. 2004;101(43):15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Q, et al. DNA origami as a carrier for circumvention of drug resistance. J Am Chem Soc. 2012;134(32):13396–13403. doi: 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- 10.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18(1):33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 11.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335(6070):831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 12.Tan W, et al. Molecular aptamers for drug delivery. Trends Biotechnol. 2011;29(12):634–640. doi: 10.1016/j.tibtech.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7(6):389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schüller VJ, et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano. 2011;5(12):9696–9702. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 15.Li J, et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5(11):8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 16.Tan SJ, Kiatwuthinon P, Roh YH, Kahn JS, Luo D. Engineering nanocarriers for siRNA delivery. Small. 2011;7(7):841–856. doi: 10.1002/smll.201001389. [DOI] [PubMed] [Google Scholar]
- 17.Chang M, Yang C-S, Huang D-M. Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy. ACS Nano. 2011;5(8):6156–6163. doi: 10.1021/nn200693a. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y-F, et al. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. ChemBioChem. 2009;10(5):862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9(7):537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl. 2006;45(48):8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 21.Yang L, et al. Aptamer-conjugated nanomaterials and their applications. Adv Drug Deliv Rev. 2011;63(14–15):1361–1370. doi: 10.1016/j.addr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 23.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 24.Shangguan D, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103(32):11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sefah K, Shangguan D, Xiong X, O’Donoghue MB, Tan W. Development of DNA aptamers using Cell-SELEX. Nat Protoc. 2010;5(6):1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 26.Shi H, et al. In vivo fluorescence imaging of tumors using molecular aptamers generated by cell-SELEX. Chem Asian J. 2010;5(10):2209–2213. doi: 10.1002/asia.201000242. [DOI] [PubMed] [Google Scholar]
- 27.Meng L, et al. Targeted delivery of chemotherapy agents using a liver cancer-specific aptamer. PLoS One. 2012;7(4):e33434. doi: 10.1371/journal.pone.0033434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Y-L, Shiao Y-S, Huang Y-F. Release of photoactivatable drugs from plasmonic nanoparticles for targeted cancer therapy. ACS Nano. 2011;5(10):7796–7804. doi: 10.1021/nn201592s. [DOI] [PubMed] [Google Scholar]
- 29.Ruggiero A, et al. Paradoxical glomerular filtration of carbon nanotubes. Proc Natl Acad Sci USA. 2010;107(27):12369–12374. doi: 10.1073/pnas.0913667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkataraman S, Dirks RM, Ueda CT, Pierce NA. Selective cell death mediated by small conditional RNAs. Proc Natl Acad Sci USA. 2010;107(39):16777–16782. doi: 10.1073/pnas.1006377107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Choi HMT, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol. 2010;28(11):1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Elbaz J, Orbach R, Magen N, Willner I. Amplified analysis of DNA by the autonomous assembly of polymers consisting of DNAzyme wires. J Am Chem Soc. 2011;133(43):17149–17151. doi: 10.1021/ja2076789. [DOI] [PubMed] [Google Scholar]
- 33.Shangguan D, et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J Proteome Res. 2008;7(5):2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther. 2006;5(12):2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 35.Xiao Z, Shangguan D, Cao Z, Fang X, Tan W. Cell-specific internalization study of an aptamer from whole cell selection. Chemistry. 2008;14(6):1769–1775. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 36.Zhu G, et al. Self-assembled aptamer-based drug carriers for bispecific cytotoxicity to cancer cells. Chem Asian J. 2012;7(7):1630–1636. doi: 10.1002/asia.201101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geng Y, et al. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat Nanotechnol. 2007;2(4):249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.