Abstract
Pediatric low-grade gliomas (PLGGs) are among the most common solid tumors in children but, apart from BRAF kinase mutations or duplications in specific subclasses, few genetic driver events are known. Diffuse PLGGs comprise a set of uncommon subtypes that exhibit invasive growth and are therefore especially challenging clinically. We performed high-resolution copy-number analysis on 44 formalin-fixed, paraffin-embedded diffuse PLGGs to identify recurrent alterations. Diffuse PLGGs exhibited fewer such alterations than adult low-grade gliomas, but we identified several significantly recurrent events. The most significant event, 8q13.1 gain, was observed in 28% of diffuse astrocytoma grade IIs and resulted in partial duplication of the transcription factor MYBL1 with truncation of its C-terminal negative-regulatory domain. A similar recurrent deletion-truncation breakpoint was identified in two angiocentric gliomas in the related gene v-myb avian myeloblastosis viral oncogene homolog (MYB) on 6q23.3. Whole-genome sequencing of a MYBL1-rearranged diffuse astrocytoma grade II demonstrated MYBL1 tandem duplication and few other events. Truncated MYBL1 transcripts identified in this tumor induced anchorage-independent growth in 3T3 cells and tumor formation in nude mice. Truncated transcripts were also expressed in two additional tumors with MYBL1 partial duplication. Our results define clinically relevant molecular subclasses of diffuse PLGGs and highlight a potential role for the MYB family in the biology of low-grade gliomas.
Keywords: cancer, aCGH, A-myb
Pediatric low-grade gliomas (PLGGs) are the most common brain tumors in children and, collectively with other CNS tumors, have surpassed leukemias as the leading cause of cancer-related deaths in children and young adults (1). PLGGs are generally categorized as “nondiffuse” or “diffuse” based on their extent of brain infiltration. Nondiffuse tumors exhibit minimal infiltration and are predominantly benign World Health Organization (WHO) grade I pilocytic astrocytomas (PAs), which are most often cured by surgery alone. In contrast, diffuse gliomas are associated with less favorable clinical outcomes, including recurrence after initial resection, by virtue of their extensive infiltration and invasion into the brain. These tumors are also more likely to progress to glioblastoma. PLGGs with diffuse growth patterns are further subclassified histologically as diffuse astrocytoma grade IIs (DA2s), gangliogliomas (GGs), angiocentric gliomas (AGs), pleomorphic xanthoastrocytomas (PXAs), and several other rare glioma types (2). However, these subclasses exhibit extensive heterogeneity and histologic overlap, often precluding categorical diagnosis. PLGGs that cannot be categorized are often referred to as low-grade gliomas, not otherwise specified (LGG-NOS) and represent nearly one-third of all PLGGs. Moreover, these histologic categories do not reliably predict biologic behavior and risk of malignant transformation.
Unifying genetic events have been identified in some PLGG subtypes, including v-raf murine sarcoma viral oncogene homolog B1 (BRAF) fusions in PAs and BRAF V600E mutations in PXAs and GGs, with substantial diagnostic, prognostic, and therapeutic implications (3–9). Identification of genetic alterations in diffuse PLGGs would increase biologic understanding of tumor behavior as well as define diagnostic molecular subclasses. However, unlike pilocytic astrocytomas, the rarity and diversity of diffuse PLGGs combined with the scarcity of frozen tissue available for genomic analyses has historically impeded identification of genetic alterations specific to these tumors. Prior studies have found that BRAF-KIAA1549 fusions are rare to nonexistent in diffuse PLGGs, particularly in DA2s (10). Diffuse PLGGs included in large cohorts of low- and high-grade gliomas were suggested to have increased expression of the proto-oncogene v-myb avian myeloblastosis viral oncogene homolog (MYB), including rare cases with genomic aberrations involving the gene (11), but no unifying recurrent genetic events have been identified.
Here we describe high-resolution copy-number profiles of 44 diffuse PLGGs, the largest collection ever to have been analyzed, and whole-genome sequencing of a diffuse PLGG. These studies reveal a recurrent rearrangement of the transcription factor v-myb avian myeloblastosis viral oncogene homolog-like 1 (MYBL1) that induces anchorage-independent growth of 3T3 cells as well as tumor growth in vivo. These findings indicate oncogenic events that define subclasses of diffuse PLGGs.
Results
Characteristics of the Diffuse PLGG Cohort.
To focus our studies on diffuse PLGGs, we collected a diverse set of carefully screened tumors through an international consortium of seven institutions (Table S1 and Fig. S1). Our cohort specifically excluded the more common, nondiffuse pilocytic astrocytomas, which are known to be driven primarily by BRAF alterations and are the focus of separate ongoing international collaborative sequencing efforts [International Cancer Genome Consortium (Germany)]. Our cohort included 18 DA2, three AG, three desmoplastic infantile ganglioglioma, nine GG, one subependymal giant cell astrocytoma, and 10 LGG-NOS tumors. Given the infiltrative growth and rarity of certain categories of diffuse PLGGs, the samples that we acquired were mostly archival formalin-fixed, paraffin-embedded (FFPE) tissue. We recently developed a method for reliable performance of array comparative genomic hybridization (aCGH) on FFPE archival samples (12) and used this technique to determine copy-number status at 1 million loci genome-wide. We also performed deep whole-genome or whole-exome sequencing to define and validate recurrent genetic events that drive tumorigenesis in these rare pediatric cancers.
Genomic Identification of Significant Targets in Cancer Analysis Identifies Significant Recurrent Events in Specific PLGG Subtypes.
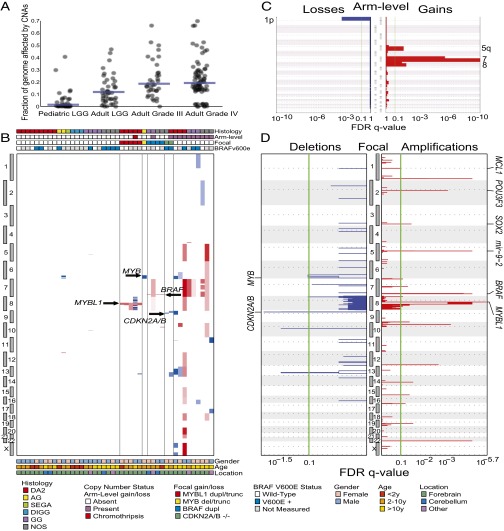
The percentage of the genome altered by copy-number alterations (CNAs) in diffuse PLGGs was significantly lower than among previously profiled adult low- and high-grade gliomas (P < 10−6, Mann–Whitney test, Fig. 1A) (13, 14). Few (12/44; 27%) of these tumors harbored alterations affecting more than 90% of the length of a chromosome arm (Fig. 1B), compared with an 83–97% rate among adult low- and high-grade tumors (15). One of the PLGG samples exhibited chromothripsis on chromosome 8 (chr8) (highlighted in Fig. 2A, PLGG27). The most significantly recurrent arm-level CNAs were gains of chromosomes 7 (11% of tumors), 8 (7%), and 5q (5%) and loss of 1p (2%) (Fig. 1C). These events have all been described in pediatric high-grade gliomas and adult gliomas with varying frequencies (15, 16).
Fig. 1.
CNAs among diffuse PLGGs. (A) Fraction of the genome altered by CNAs is lower among diffuse PLGGs compared with adult LGGs and high-grade gliomas (P < 10−6). Blue bars indicate medians. (B) Amplifications (red) and deletions (blue) among 44 diffuse PLGGs (x axis) across the genome (y axis) ordered by copy-number status. Significance (x axis) of (C) arm-level and (D) focal deletions (Left, blue) and amplifications (Right, red) across the genome (y axes). Putative gene targets within the peak regions are indicated where known.
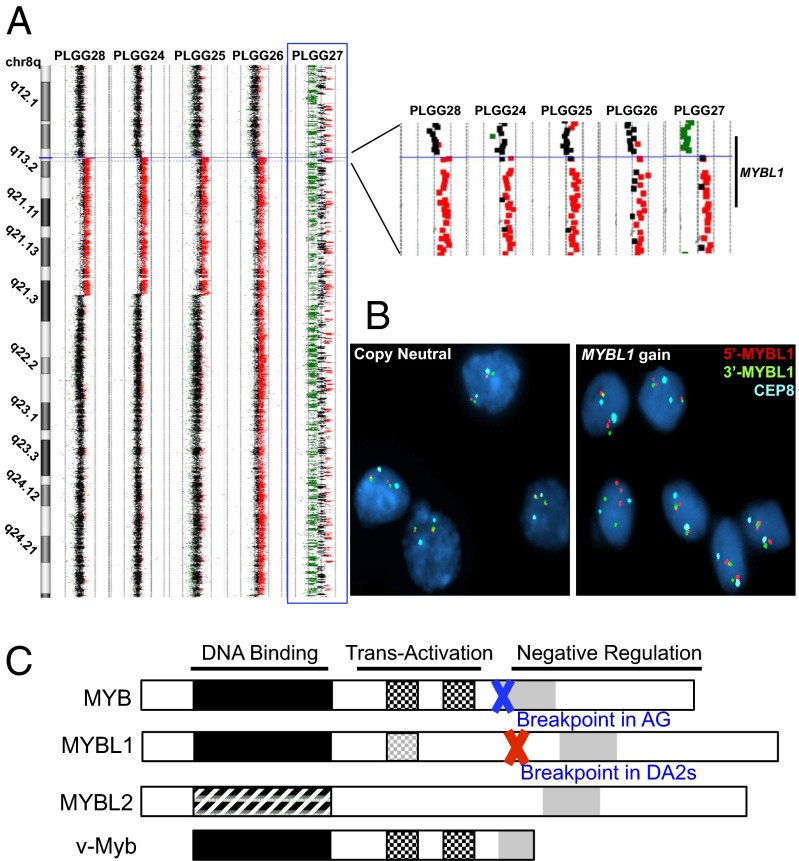
Fig. 2.
DA2 samples with focal 8q gains identified by aCGH share a common centromeric breakpoint within MYBL1. (A) aCGH data for the five DA2 samples with 8q gain, magnified on the right. Red: copy-number gain; green: loss. The highlighted sample (PLGG27, blue box) exhibits chromothripsis of chr8 but shows no other copy-number changes on other chromosomes. (B) FISH probes corresponding to sequences immediately distal (red) and proximal (green) of MYBL1 confirm that the single-copy gain identified by aCGH is a duplication involving one MYBL1 allele. The D8Z2 centromere enumeration probe (aqua) was used as a control. (C) Schematic representation of the proto-oncogene MYB family and breakpoints observed in our cohort in relation to the viral oncogene v-MYB.
We found 6 significantly recurrent regions of focal deletion and 17 significantly recurrent regions of focal amplification (Fig. 1D and Table S2). One deleted region on 9p21.3 contained cyclin-dependent kinase inhibitor 2A and 2B (CDKN2A and CDKN2B), known tumor suppressors that had previously been reported in diffuse PLGGs (17); a second region was immediately adjacent to this one. A third region (6q26) contained 252 genes, including the proto-oncogene MYB. Two regions (10q21.3 and 8p22) contained single genes with no known relation to cancer or neural development, catenin (cadherin-associated protein), alpha 3 (CTNNA3) and zeta sarcoglycan (SGCZ), respectively. The sixth region (13q31.3) contained 48 genes and was adjacent to the known tumor suppressor RB1. We did not identify any focal deletions of other known tumor suppressors involved in adult or pediatric brain tumors such as neurofibromin 1 (NF1), phosphatase and tensin homolog (PTEN), or cyclin-dependent kinase inhibitor 1C (CDKN1C).
One of the 17 focally gained regions contained BRAF. However, the canonical BRAF-KIAA1549 duplication-fusion was detected in only four samples: two GGs and two LGG-NOS. This is in contrast to pilocytic astrocytomas, among which >80% of tumors harbor a BRAF duplication (18) (P < 0.0001, Fisher’s exact test). We also determined BRAF V600E mutation status in 24 tumors with sufficient DNA for sequencing. We found mutations in 54% of the diffuse PLGGs (Fig. 1A and Table S1), consistent with previously published rates for diffuse PLGGs (8).
A second focally gained region (3q26.33) contained the stem cell and glial transcription factor sex determining region Y-box 2 (SOX2), which is amplified in adult glioblastomas (19). Two additional regions (2q12.1 and 5q14.3) contained factors that control telencephalic neural progenitor proliferation and differentiation: POU class 3 homeobox 3 (POU3F3) (also known as BRN1) and microRNA 9-2 (20, 21). A fifth region (1q21.3) contained myeloid cell leukemia sequence 1 (MCL1), a known oncogene amplified in several cancer types (22). Twelve regions either contained over 150 genes or did not contain genes with known roles in cancer or neural development. We did not observe any high-level amplification of receptor tyrosine kinases (e.g., EGFR, PDGFRA), which are observed frequently in both adult and pediatric high-grade gliomas (19, 23).
The most statistically significant recurrent focal aberration (q = 3.37 × 10−6) was a gain on chromosome 8q involving the transcription factor MYBL1. Although MYBL1 is not a known oncogene, it is closely related to the proto-oncogene MYB. In contrast to prior reports (11), no amplifications or gains of the proto-oncogene MYB were identified in our study set. All of the focal 8q gains occurred in DA2s (P = 0.0057, Fisher’s exact test), comprising 28% (5/18) of this histologic subtype. In contrast, MYBL1 was not in a significant amplification peak across 3,131 cancers comprising multiple other cancer types that we had previously analyzed (22) or, specifically, among adult low- or high-grade gliomas (15).
All five DA2 samples with 8q focal gains exhibited a common centromeric breakpoint within MYBL1 after exon 9, including the sample with chromothripsis of chr8 (Fig. 2A). To confirm the MYBL1 centromeric breakpoint, we performed fluorescence in situ hybridization (FISH) using a probe slightly telomeric to the breakpoint on all eight DA2 samples with sufficient tissue available (Fig. 2B and Fig. S2). All DA2 samples with 8q gain (3/3) demonstrated duplication of one allele in more than 60% of the nuclei in each tumor whereas none of the other DA2 samples showed duplication (0/5) (P = 0.018, Fisher’s exact test).
The tight clustering of these breakpoint sites, and particularly their location immediately preceding the C-terminal negative regulatory domains of MYBL1 and MYB, suggested a mechanism for rearrangement and creation of functional, truncated genes reminiscent of the viral oncogene v-MYB (Fig. 2C). Indeed, we also identified a homologous breakpoint between exons 10 and 11 of MYB on 6q in one angiocentric glioma with a focal 6q deletion (Fig. S3) similar to that previously reported in a single angiocentric glioma with a 6q deletion (11). An additional angiocentric glioma (PLGG45), not included in our initial diffuse PLGG cohort and genomic identification of significant targets in cancer (GISTIC) analysis, also exhibited a deletion in 6q at the same location in MYB as seen in PLGG29 (Fig. S3).
Whole-Genome Sequencing of a DA2 with 8q Focal Gain Defines a Tandem Duplication–Truncation of MYBL1.
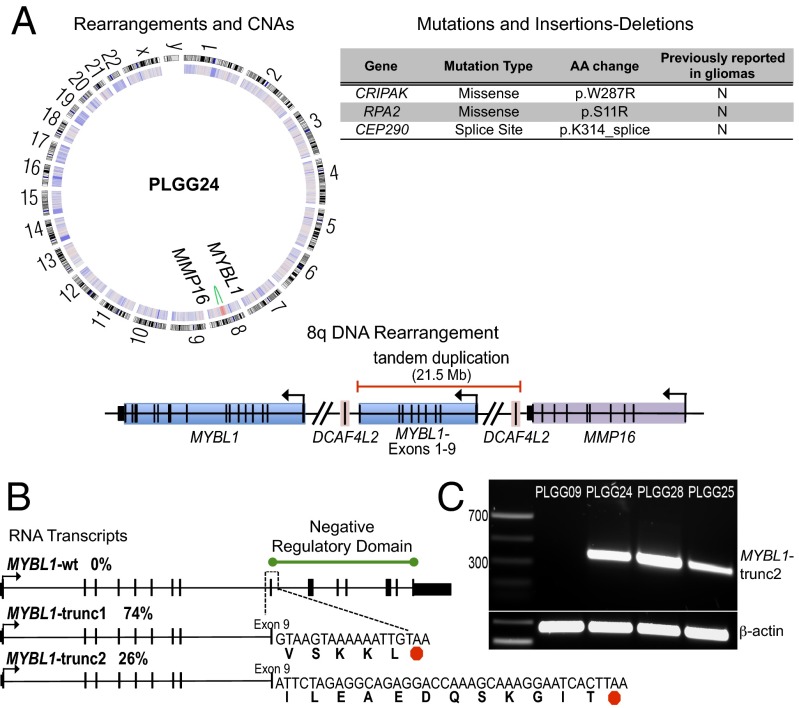
To further characterize the MYBL1 amplicon and its genetic context, we performed 90× whole-genome sequencing of a DA2 sample with MYBL1 gain but no other CNAs (PLGG24, Table S3). Whole-genome sequencing of PLGG24 determined the centromeric breakpoint of the 8q amplicon to single-base resolution between exons 9 and 10 of MYBL1 (Fig. 3A). The telomeric sequence was located in an intergenic region 38 kb from matrix metallopeptidase 16 (MMP16). We validated the breakpoint locations in this sample using PCR on native genomic DNA from the same tumor (Fig. S2 C and D). Taken together, our data define a tandem duplication–truncation of MYBL1.
Fig. 3.
Characterization of the MYBL1-truncating rearrangement in a DA2 sample (PLGG24). (A) Circos plot (Left) of 90× whole-genome sequence data showing a single-copy-number gain on 8q (red, inner heatmap ring) and corresponding intrachromosomal rearrangement (green) involving MYBL1 and an intergenic region outside MMP16. A schematic diagram represents the 8q tandem duplication (Lower Right). All nonsynonymous mutations and insertions and deletions are also listed (Upper Right). (B) Truncated MYBL1 transcripts (MYBL1-trunc1 and MYBL1-trunc2) identified by 3′-RACE. (C) RT-PCR demonstrating MYBL1-trunc2 expression in two additional PLGG samples with MYBL1 gain.
Apart from this event, whole-genome sequencing of PLGG24 revealed a sparsely altered genome. No other CNAs or fusion events were identified, and the BRAF V600E mutation was not present. Three nonsynonymous mutations in exons were identified (Table S4); none of these have been reported in association with cancer. The genome-wide mutation rate (1.48/Mb) and the number of nonsynonymous mutations in exons (three per genome) were low compared with pediatric and adult high-grade astrocytomas (mutation count means: 15 and 47.3 per genome, respectively) (16). The low rate of CNAs, mutations, and translocations in this sample highlight the potential biological impact of MYBL1 duplication–truncation.
Tandem Duplication–Truncation of MYBL1 Results in Expression of Oncogenic Transcripts.
To determine whether the MYBL1 duplication–truncation resulted in expression of fused transcripts, we performed 3′-rapid amplification of cDNA ends (3′-RACE) on cDNA generated from RNA of PLGG24. We identified two populations of MYBL1 transcripts, both of which contained MYBL1 exons 1–9 but also acquired short noncanonical sequences fused to the 3′ ends, leading to premature translation stops (Fig. 3B). The first transcript (MYBL1-trunc1) contained an additional 15 bp of intronic sequence, and the second transcript (MYBL1-trunc2) acquired 36 bp from the intergenic region near MMP16. MYBL1-trunc1 was retrieved more frequently (74%) than MYBL1-trunc2 (26%); the wild-type transcript was not observed. We then performed RT-PCR to detect MYBL1-trunc1 and MYBL1-trunc2 in the two other PLGGs with 8q focal amplification for which we had sufficient RNA (PLGG25 and PLGG28). We detected MYBL1-trunc2 in both of these samples (Fig. 3C), indicating that this transcript is recurrently expressed in DA2s.
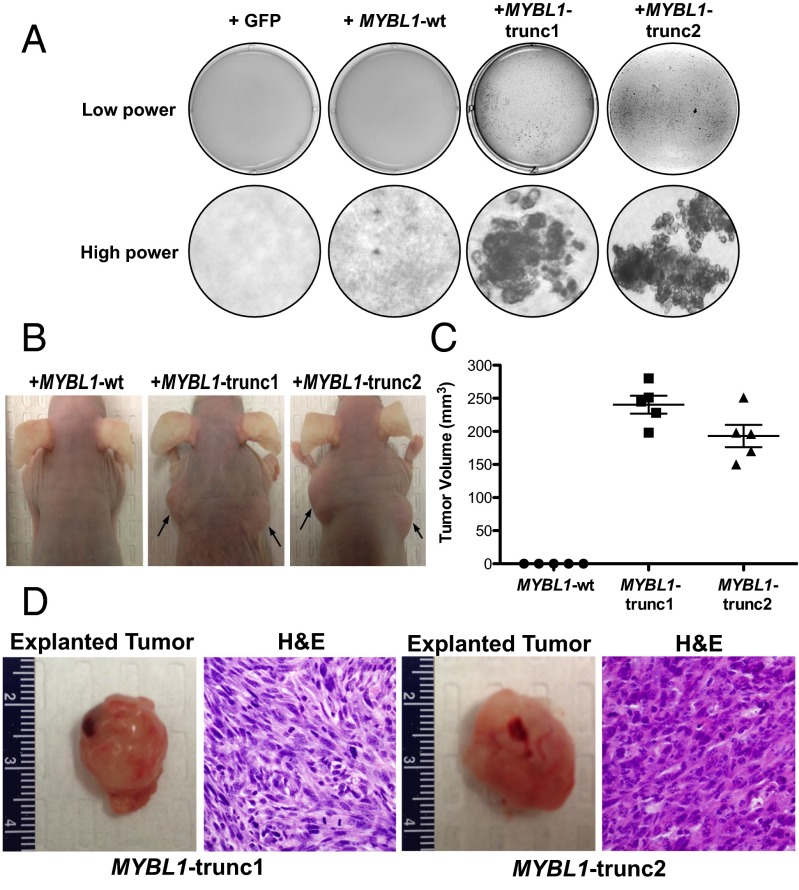
To assess the oncogenic potential of these transcripts, we transduced 3T3 cells with lentiviruses containing MYBL1-trunc1, MYBL1-trunc2, full-length MYBL1-wt, or GFP control constructs and plated them in soft agar. Both aberrantly truncated MYBL1 sequences produced soft agar colony growth indicative of transformation (Fig. 4A). No evidence of colony formation was noted with full-length MYBL1-wt or GFP control. Both MYBL1-trunc1– and MYBL1-trunc2–transformed 3T3 cells were also able to form tumors with malignant histology in Nude mice, whereas cells transduced with full-length MYBL1-wt constructs showed no evidence of tumor formation (Fig. 4 B–D). These results suggest that the truncation of MYBL1 is oncogenic.
Fig. 4.
(A) Anchorage-independent growth of 3T3 cells transduced with MYBL1-wt, MYBL1-trunc1, or MYBL1-trunc2 retroviruses, relative to GFP controls. (B) In vivo tumor formation after injection of 3T3 cells transduced with MYBL1-wt (n = 5), MYBL1-trunc1 (n = 5), or MYBL1-trunc2 (n = 5) (arrows) into the flanks of Nude mice. Tumor volume was calculated 6 wk post injection (C), and tumors were subjected to histologic analysis by H&E staining (D).
Discussion
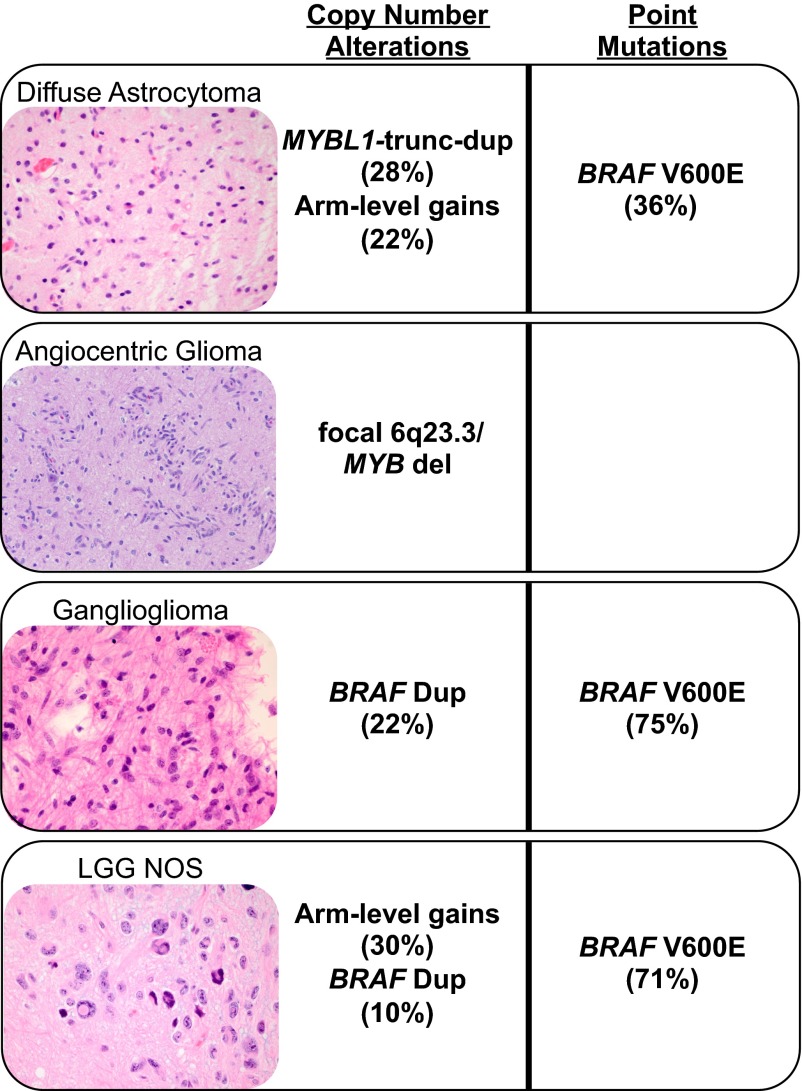
Our data identify several recurrent somatic genetic events in PLGG, some of which track with specific histologic types. The most significant of these was recurrent focal amplification of MYBL1, found exclusively in DA2 samples, none of which had other PLGG-associated lesions such as a BRAF duplication. Focal deletion of 6q involving the MYB locus was also observed in two angiocentric gliomas in our dataset, similar to deletions in two individual cases of angiocentric glioma noted in prior studies. Such aberrations may therefore be useful in identifying this tumor type (11, 24). In other cases, genetic events span histologic subtypes and tumor grades, such as duplications of BRAF in GG and NOS samples in our diffuse PLGG cohort as well as deletions involving CDKN2A/B (Fig. 5). Although previous work had identified high-level amplification of MYB and 6q deletions involving MYB in several DA2 tumors, we detected no such events in our DA2 cohort (11).
Fig. 5.
Schematic summarizing the highly characteristic genomic aberrations detected in our diffuse PLGG cohort.
These findings provide genetic and functional evidence of a role for MYBL1 and the MYB proto-oncogene family of transcription factors in low-grade gliomagenesis. MYB transcription factors are known to regulate cell-cycle progression in multiple cellular contexts, and MYB has been identified as an oncogene in T-ALL and adenoid cystic carcinomas (25). The MYB family members share extensive homology in their DNA-binding domains; however, they differ in their C-terminal domains. In vitro and in vivo studies have demonstrated roles for each transcription factor in cell-cycle regulation mediated by interactions with and modifications of the C terminus (26). Truncating deletions may affect the transcriptional activity of MYB transcription factors by loss of a conserved negative regulatory domain or may result in increased gene expression due to loss of a 3′ UTR targeted by microRNA 15a/16 and microRNA 150 (25). Our results suggest that truncations affecting the C terminus of MYB family transcription factors may be sufficient to drive oncogenesis in a discrete molecular subclass of diffuse PLGGs by acting as gain-of-function mutations.
Methods
Patients and Samples.
Institutional review board approval from all institutions (Boston Children's Hospital, Dana-Farber Cancer Institute, The University of Texas School of Medicine Southwestern, Children's Cancer Hospital-Egypt, The Johns Hopkins University School of Medicine, Children's National Medical Center, Hospital for Sick Children, and the Mayo Clinic) was obtained, and all samples were from patients who provided informed consent or were studied with waiver of the requirement for informed consent by the Dana-Farber Cancer Institutional review board. Samples of various histologic subtypes were identified, collected at multiple institutions, and central histopathologic review was performed by at least three board-certified neuropathologists using WHO criteria (K.L.L., S.S., S.H.R., or J.A.C.).
aCGH and Data Processing.
DNA extraction from archival FFPE samples and aCGH were performed as previously described (12). GC-normalized copy-number data for the samples were then cleaned of known germ-line copy-number variations. Circular Binary Segmentation was used to segment the copy-number data, using parameters (α = 0.001, undo.splits = sdundo, undo.SD = 1.5, minimum width = 5). Segmented data were analyzed with GISTIC 2.0 to determine statistically significant recurrent broad and focal CNAs using the following parameters: minimum segment size = 8, lesion amplitude threshold = 0.2, focal/broad cutoff = 0.9× chromosome arm length, q-value threshold = 0.10, and gene confidence level = 0.95. For comparison of diffuse PLGG data to previously published adult low-grade glioma (LGG) and high-grade glioma (HGG) data (13, 14), previously segmented copy-number data were subjected to the same GISTIC analysis parameters as above.
Whole-Genome Sequencing and Data Processing.
DNA from fresh-frozen tissue from PLGG24 and paired blood was extracted using the QIAGEN DNA Blood and Tissue kit. A target depth of 90× in blood and tumor was set for Illumina sequencing, using two different insert-size libraries (500 and 800 bp) to maximize detection of rearrangements (27). Sequencing quality control (QC) metrics are shown in Table S3.
Sequence data were aligned to the hg19 (b37) reference genome with the Burrows-Wheeler Aligner (28) with parameters [-q 5 -l 32 -k 2 -t 4 -o 1]. Aligned data were sorted, normalized, mate-fixed, duplicate-marked, and indexed with Samtools and Picard tools (29). Base-quality score recalibration and local realignment around insertions and deletions was achieved with the Genome Analysis Toolkit (30, 31).
Somatic mutations and small insertions-deletions were called with MuTect and Indelocator, filtered against a panel of normals, and annotated to genes with Oncotator (29, 32, 33). CNAs were called with SegSeq and standard parameters (34). Somatic rearrangements were identified with dRanger and BreakPointer algorithms (32, 33) with the cutoff SN parameter increased to 1,000 bp (reflecting the larger-than-normal insert sizes used for sequencing). Results are reported with high confidence if the dRanger score was ≥8 and the BreakPointer algorithm identified the exact breakpoints on both ends (equivalent to 8× high-confidence coverage of read pairs spanning the breakpoint).
Whole-Exome Sequencing.
DNA was extracted from tumors as above, and 250-bp libraries were prepared by Covaris sonication, followed by double-size selection (Agencourt AMPure XP beads) and ligation to specific barcoded adaptors (Illumina TruSeq) for multiplexed analysis. Exome hybrid capture was performed with the Agilent Human All Exon v2 (44 Mb) or Illumina TruSeq bait sets, and samples were sequenced as above. Tumors were manually reviewed for the presence of the BRAF V600E mutation.
FISH and PCR.
FISH was performed on 4-μm tissue sections using methods described previously (35) and Homebrew probes RP11-110J18 (5′ to MYBL1; directly labeled in SpectrumOrange) and RP11-707M3 (3′ to MYBL1; directly labeled in SpectrumGreen) that map to 8q13.1. MYBL1 status was assessed in 50 tumor nuclei per sample. PCR was performed on genomic DNA from PLGG0024 and control samples to confirm breakpoint sequences identified by dRanger. Primer sequences (5′-AATGCTATCCCTCCCCACTC-3′ and 5′-GAGGGAGCTTGGAAATTTGA-3′) targeting MYBL1 and intergenic sequences, respectively, amplified a 450-bp fragment. The band was gel-purified, cloned (TOPO TA Cloning; Invitrogen, and sequenced by Sanger sequencing to validate the fusion.
RT-PCR and 3′-RACE.
MYBL1-trunc1 and MYBL1-trunc2 were cloned from PLGG24 frozen tumor tissue using a 3′ RACE kit (Invitrogen) per manufacturer’s instructions. To amplify all MYBL1 specific transcripts, primers targeting the 5′ end of MYBL1 (5′-AAAACCCTGCAGGAGACTG-3′) were used in conjunction with a universal amplification primer (UAP). A second PCR was performed using a nested MYBL1 primer (5′-TGCGGTACTTGAAGGATGG-3′) along with the UAP. PCR products were subjected to Sanger sequencing, and results were aligned to the hg19 reference genome. For RT-PCR reactions, RNA was extracted from FFPE samples of PLGG09, PLGG25, and PLGG28 using RNeasy FFPE kit (Qiagen). cDNA was generated from 500 ng RNA using the iScript cDNA synthesis kit (Bio-Rad). PCR to detect the presence of MYBL1-trunc2 was performed using primers targeting the MYBL1 exon 7–8 junction (5′-ATTGTATAGAACATGTTCAGCCT-3′) and the MYBL1-trunc2 sequence (5′-GGTCCTCTGCCTCTAGAATAGATTC-3′).
Expression Constructs and Lentiviral Production.
Full-length MYBL1-wt (Open Biosystems), MYBL1-trunc1, and MYBL1-trunc2 cDNA sequences were subcloned into the pLenti7.3/V5 vector using the Gateway system (Invitrogen). For retroviral production, 293FT packaging cells were cotransfected with pBabe expression clones, Gag-pol, and vs.v-g. Viral supernatant was harvested 48 h after transfection, filtered through a 45-μm filter, and concentrated by ultracentrifugation.
Anchorage-Independent Growth Assay.
Wild-type 3T3 mouse embryonic fibroblasts (ATCC) were transduced by addition of viral supernatant to the growth medium. Twenty-four hours later, infection efficiency was evaluated based on GFP expression and determined to be >80%. Cells were harvested and mixed with growth medium containing 0.33% bactoagar, and 1 × 104 cells were plated in triplicate onto a bottom layer of medium with 0.5% agar in a six-well plate. Soft agar colonies were counted 2 wk later. Images were acquired using the AlphaInnotch FluorChem HD2 Imager.
Flank Tumor Growth Assay.
The Institutional Animal Care and Use Committee at Dana-Farber Cancer Institute preapproved all animal experiments. 3T3-MYBL1-wt, 3T3-MYBL1-trunc1, or 3T3-MYBL1-trunc2 cells (106) were suspended in 150 μL PBS, mixed with 150 μL Matrigel (BD Biosciences), and then injected into the flank of 6-wk-old male Nude (NU-Foxn1, Charles River) mice. Mice were then monitored for signs of distress or tumor growth. Six weeks post injection the mice were euthanized and analyzed for tumor growth. Tumors were subjected to standard histologic analysis.
Supplementary Material
Acknowledgments
The authors thank Denise Garcia and Heather Homer for technical help with FISH experiments; Yotam Drier and Cheng-Zhong Zhang for assistance with analytical algorithms; and Anna Pisarek-Horowitz and Sophie Horowitz for critical reading of the manuscript. This work was supported by the Pediatric Low Grade Astrocytoma Foundation; The Sontag Foundation (K.L.L., R.B.); Goldhirsh Foundation (K.L.L.); Boston Children’s Hospital Team Path to Cure (K.L.L.); American Brain Tumor Association Basic Research Fellowship in Honor of Joel A. Gingras, Jr. (to L.A.R.); the Eleanor and Miles Shore 50th Anniversary Fellowship Program for Scholars in Medicine (to A.H.L.); Nuovo-Soldati Foundation and Philippe Foundation, Inc. (to G.B.); and National Institutes of Health Grants R01 CA109467 and P30 HD018655 (to S.L.P.), U54CA143789 and K08CA122833 (to R.B.), and P01CA142536 (to K.L.L., W.C.H., S.S., C.D.S., L.E.M., and M.W.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The copy number data reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE45843). The sequence reported in this paper has been deposited in the database of Genotypes and Phenotypes (dbGaP), www.ncbi.nlm.nih.gov/gap (accession no. phs000614.v1.p1).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300252110/-/DCSupplemental.
References
- 1.Horner MJRL, et al. SEER Cancer Statistics Review 1975–2006. Bethesda, MD: National Cancer Institute; 2009. [Google Scholar]
- 2.Louis DN, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfister S, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118(5):1739–1749. doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dias-Santagata D, et al. BRAF V600E mutations are common in pleomorphic xanthoastrocytoma: Diagnostic and therapeutic implications. PLoS ONE. 2011;6(3):e17948. doi: 10.1371/journal.pone.0017948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sievert AJ, et al. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol. 2009;19(3):449–458. doi: 10.1111/j.1750-3639.2008.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones DT, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68(21):8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar EE, Lin A, Tihan T, Burger PC, Eberhart CG. Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. J Neuropathol Exp Neurol. 2008;67(9):878–887. doi: 10.1097/NEN.0b013e3181845622. [DOI] [PubMed] [Google Scholar]
- 8.Dougherty MJ, et al. Activating mutations in BRAF characterize a spectrum of pediatric low-grade gliomas. Neuro Oncol. 2010;12(7):621–630. doi: 10.1093/neuonc/noq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schindler G, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3):397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 10.Lin A, et al. BRAF alterations in primary glial and glioneuronal neoplasms of the central nervous system with identification of 2 novel KIAA1549:BRAF fusion variants. J Neuropathol Exp Neurol. 2012;71(1):66–72. doi: 10.1097/NEN.0b013e31823f2cb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatevossian RG, et al. MYB upregulation and genetic aberrations in a subset of pediatric low-grade gliomas. Acta Neuropathol. 2010;120(6):731–743. doi: 10.1007/s00401-010-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig JM, et al. DNA fragmentation simulation method (FSM) and fragment size matching improve aCGH performance of FFPE tissues. PLoS ONE. 2012;7(6):e38881. doi: 10.1371/journal.pone.0038881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotliarov Y, et al. High-resolution global genomic survey of 178 gliomas reveals novel regions of copy number alteration and allelic imbalances. Cancer Res. 2006;66(19):9428–9436. doi: 10.1158/0008-5472.CAN-06-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anonymous . REMBRANDT home page. Bethesda, MD: National Cancer Institute; 2005. [Google Scholar]
- 15.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104(50):20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 17.Schiffman JD, et al. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70(2):512–519. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatevossian RG, et al. MAPK pathway activation and the origins of pediatric low-grade astrocytomas. J Cell Physiol. 2010;222(3):509–514. doi: 10.1002/jcp.21978. [DOI] [PubMed] [Google Scholar]
- 19.Atlas TCG. Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295(5559):1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- 21.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31(9):3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paugh BS, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28(18):3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preusser M, et al. Angiocentric glioma: Report of clinico-pathologic and genetic findings in 8 cases. Am J Surg Pathol. 2007;31(11):1709–1718. doi: 10.1097/PAS.0b013e31804a7ebb. [DOI] [PubMed] [Google Scholar]
- 25.Persson M, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA. 2009;106(44):18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei W, Rushton JJ, Davis LM, Liu F, Ness SA. Positive and negative determinants of target gene specificity in myb transcription factors. J Biol Chem. 2004;279(28):29519–29527. doi: 10.1074/jbc.M403133200. [DOI] [PubMed] [Google Scholar]
- 27.Brastianos PK, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45(3):285–289. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Network TCGA. Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiang DY, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6(1):99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455(7212):547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.