Abstract
During plant defense against bacterial pathogens, the hypersensitive response (HR) functions to restrict pathogen growth and spread. The mechanisms driving this growth restriction are poorly understood. We used a water stress-responsive transcriptional fusion to quantify the water potential sensed by individual Pseudomonas syringae pv. tomato DC3000 cells during infection of Arabidopsis thaliana leaves. A nonpathogenic DC3000 hrcC mutant defective in type III secretion, as well as the saprophyte Pseudomonas fluorescens A506, sensed water potentials of -0.3 to -0.4 MPa at 48 h postinfiltration (hpi). During pathogenesis, DC3000 sensed lower water potentials (-0.4 to -0.9 MPa), demonstrating that it can modify the intercellular environment, and these water potentials were associated with optimal DC3000 growth in culture. During the HR, DC3000 cells sensed water potentials (-1.6 to -2.2 MPa) that were low enough to prevent cell division in the majority of cells in culture. This water potential decrease occurred within only 4 hpi and was influenced by avirulence gene expression, with avrRpm1 expression associated with lower water potentials than avrRpt2 or avrB expression at 48 hpi. The population sizes of the DC3000 variants tested were significantly correlated with the apoplastic water potential at 48 hpi, with a decrease of -0.9 MPa associated with a 10-fold decrease in cells per gram of leaf. These results suggest that the apoplastic water potential is a determinant of endophytic bacterial population size, and water stress, resulting from high osmolarity or tissue desiccation, is at least one factor restricting bacterial growth during the HR.
Bacterial foliar pathogens such as Pseudomonas syringae have been the subject of intense study aimed at identifying the molecular and physiological basis of plant-bacterial interactions. In susceptible host plants, P. syringae can establish large populations in the intercellular spaces of leaves and produce necrotic lesions. In nonhost plants and in host plants with R-gene-mediated resistance, P. syringae can induce a localized defense response that involves rapid, programmed plant cell death. This hypersensitive response (HR) is associated with cessation of the growth and spread of the bacteria. The actual mechanisms by which bacterial growth is promoted during pathogenesis and restricted during the HR are poorly understood. One of the best-studied model systems for bacterial-plant interactions is that of P. syringae pv. tomato strain DC3000 and Arabidopsis thaliana. Pathogenic DC3000 can be converted to an avirulent form by introducing one of several known avirulence genes. Genetic analyses of DC3000 have led to the identification of genes essential to pathogenesis and HR induction. Functions of these genes include the production of a type III secretion system that promotes the delivery of various effector proteins into plant cells (1, 2). At least one role of effector proteins is in suppressing, interfering with, or modulating host defense signaling pathways (3-5). Genetic analyses of A. thaliana have profiled genes induced in response to DC3000 and its avirulent derivatives and have demonstrated that the plant response is rapid, dynamic, and distinct for susceptibility versus R-gene-specific resistance (6, 7).
During pathogenesis, P. syringae pv. tomato often attains populations as high as 107 to 108 colony-forming units (CFU) per cm2 in A. thaliana leaves (8, 9). P. syringae genes that are induced during infection include hrp/hrc genes, which are involved in producing the type III secretion system, and genes encoding Hop (Hrp-dependent outer protein) and Avr (avirulence) effector proteins, a putative plant cell wall-degrading enzyme, phytotoxin biosynthetic enzymes, and adaptation factors for growth in planta (10). Whereas the collective effect of these expressed genes is enhanced bacterial growth in the apoplast, the nature of the changes that occur in the apoplastic environment has not been well characterized. Foliar bacterial pathogens can induce a host plasma membrane K+ efflux and H+ influx (11), which has been associated with an increase in apoplastic sucrose levels (12) and thus an improvement in the nutritional environment in the apoplast. Numerous recent studies provide evidence that some of the bacterial effectors, such as AvrPtoB, HopPtoD2, and AvrPto in DC3000 (3-5, 13), suppress defense responses in susceptible plants, thus reducing plant inhibition of bacterial growth.
During the HR, P. syringae pv. tomato populations attain concentrations that are typically 10- to 200-fold smaller than those attained during pathogenesis (8, 9). This potential restriction of pathogen growth could result from the production of antimicrobial compounds such as phytoalexins and reactive oxygen species (ROS), or, as will be explored in this work, from restrictive physical conditions in the apoplast. Any or all of these phenomena may occur concurrently. Indolic compounds, including the phytoalexin camalexin, are the predominant secondary metabolites that accumulate in A. thaliana Columbia in response to DC3000 (14). Although these metabolites accumulate more rapidly and to higher concentrations in response to avirulent than virulent DC3000, their role in restricting the growth of the avirulent pathogen appears to be limited based on the finding that they accumulate after the populations of avirulent and virulent bacteria have diverged in size (14). Furthermore, avirulent DC3000 established similarly sized populations in camalexin-deficient mutants and wild-type plants, indicating that camalexin does not have a significant role in growth restriction during the HR (15). ROS, including hydrogen peroxide and superoxide anions, are also produced in response to bacterial pathogens. Hydrogen peroxide accumulates to detectable levels within hours after recognition of an avirulent pathogen, with accumulation sites being primarily in the plant cell walls (16). Interestingly, A. thaliana mutants that were deficient in the production of a probable NADPH oxidase and thus produced reduced levels of hydrogen peroxide were similar to wild-type A. thaliana in their support of avirulent DC3000 growth (17), suggesting that hydrogen peroxide accumulation does not have a significant role in growth restriction during the HR. In laboratory studies, we observed that bacterial exposure to an osmotic upshift often restricts further cell division rather than causing cell death, as results from exposure to hydrogen peroxide or phytoalexins. This observation led us to hypothesize that bacterial growth restriction during the HR may result from physiological changes in leaves, such as increased apoplastic osmolarity or tissue desiccation, that limit water availability to the bacteria.
Previously, we reported on the effectiveness of a water stress-responsive transcriptional fusion as a tool for quantifying the water potential sensed by individual bacterial cells (18). This fusion, which involves the Escherichia coli proU promoter, responds to both the solute and matric components of the total water potential, and responds specifically to low water potential and not to ion toxicity or oxidative stress (18). Here, we have used this fusion to measure the water potentials that avirulent and virulent DC3000 encounter during their interactions with A. thaliana. We report that in contrast to nonpathogens and virulent DC3000, which encounter favorable water potentials for growth in the leaf apoplast, avirulent DC3000 encounters water potentials that severely restrict its growth. These results indicate that exposure to water stress is likely one of the mechanisms by which bacterial growth is restricted during the HR.
Materials and Methods
Strains and Plasmids. P. syringae pv. tomato strain DC3000 (19), DC3000 hrcC (previously designated hrpH) (20), and Pseudomonas fluorescens A506 (21) were used in this study. E. coli DH5α was used for cloning. Strains were grown in LB (22) or, for induction studies, K medium (23) amended with NaCl. Antibiotics were used at the following concentrations (μg/ml): rifampin, 50; kanamycin, 50; streptomycin, 20; spectinomycin, 20, and tetracycline, 20. pPProIce was constructed by inserting a 612-bp EcoRI-BamHI fragment with the proU promoter from pOSEX4 (24) in front of inaZ in pPROBE-KI (25). pPNptIce was constructed by PCR amplifying the nptII promoter from Tn5 while introducing flanking SalI sites, and inserting a 365-bp SalI fragment with the promoter in front of inaZ in pPROBE-KI (25). Plasmids pLH12 (8), pLH12-Ω (8), pK48-8 (26), and pPGS002 (27) contained avrRpt2, avrRpt2:: Ω, avrRpm1, and avrB, respectively. Plasmids were mobilized by using pRK2073 (28).
Plant Growth and Inoculation. A. thaliana seeds, ecotype Columbia (Col-O) and an rps2-201 mutant (29, 30), were grown in an 11-hr light/13-hr dark cycle at 25°C and 20-25% relative humidity. Bacterial cells grown on K agar for 2 d at 24°C were suspended in 10 mM MgSO4. Bacterial suspensions (106 or 108 cells per ml) were syringe-infiltrated into the abaxial side of the leaves of 5- to 6-week-old plants (26).
Measurement of Ice Nucleation Activity (INA). The effect of NaCl concentration on proU-inaZ expression was assessed by growing cells at 24°C on solid medium containing NaCl at concentrations from 0 to 500 mM NaCl (31), suspending cells to 103 cells per ml in K broth, and measuring their INA at -5°C, unless otherwise stated, by a droplet freezing assay (32). The INA of cells recovered from plants was estimated by excising ≈1 g of leaf tissue, weighing it, homogenizing it, and subjecting dilutions made with K broth to the droplet freezing assay.
Characterization of Water Potential Effects on DC3000 Growth in Culture. Cells were grown on solid K medium amended with 0, 25, 50, 100, 200, 300, 400, and 500 mM NaCl; colonies did not develop at higher NaCl concentrations. The time until the initial appearance of colonies was determined for each NaCl concentration by examining plates every 3 h; this was done without the aid of a microscope. Colony diameters (mm) were used to estimate growth rate after confirming that colony size and number of cells per colony were directly related. The culturability of cells at each NaCl concentration was evaluated by comparing the number of colonies on plates with and without NaCl amendment. Cells were also grown at 28°C in K broth amended with 0, 50, 100, 200, 300, 400, 500, 600, 800, and 1000 mM NaCl, and the density of the cultures was monitored by viable plate counts and optical density at 600 nm.
Results
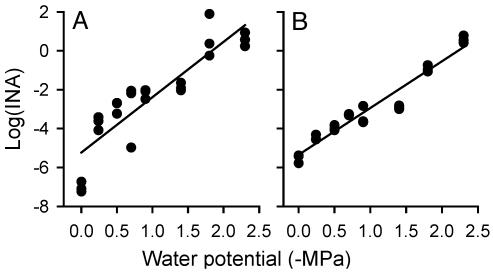
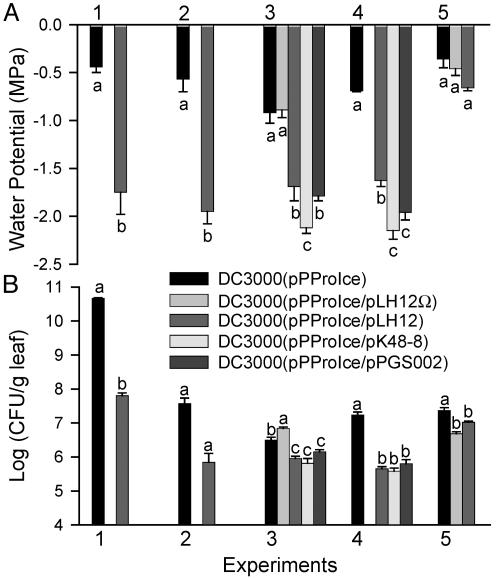
DC3000 Is Exposed to Significantly Lower Water Potentials During the HR than During Pathogenesis. To evaluate the water potential sensed by bacteria in both compatible and incompatible interactions with A. thaliana, a plasmid pPProIce containing a proU-inaZ fusion was introduced into DC3000 and DC3000 derivatives containing the cloned avirulence genes avrRpt2, avrRpm1, or avrB. The log10(INA) of these strains was directly related to the water potential to which the bacteria were exposed (Fig. 1). We previously reported that a proU-gfp fusion was induced by distinct permeating solutes, including Na2SO4, KCl, and the nonpermeating solute polyethylene glycol 8000, demonstrating that the proU promoter responds to low water potentials rather than specifically to NaCl (18). Forty-eight hours after bacterial cells were infiltrated into A. thaliana (Col-O) leaves, DC3000(pPProIce) exhibited a significantly higher INA when it contained the avrRpt2-encoding plasmid pLH12 than when it did not. This result was observed in seven independent experiments. By using the relations between INA and water potential on solid media (Fig. 1), we estimated that DC3000(pPProIce) cells were exposed to water potentials of -0.4 to -0.9 MPa in these experiments (Fig. 2A, experiments 1-4), whereas DC3000(pPProIce/pLH12) cells were exposed to -1.6 to -1.9 MPa. The disruption of avrRpt2 in DC3000(pPProIce/pLH12Ω) resulted in the bacteria sensing water potentials similar to those sensed by DC3000(pPProIce) on Col-O (Fig. 2A, experiment 3). Similarly, when DC3000(pPProIce/pLH12) was introduced into an A. thaliana rps2-201 mutant, the cells sensed water potentials similar to those sensed by DC3000(pPProIce) on Col-O and the rps2 mutant (Fig. 2A, experiment 5).
Fig. 1.
Correlation between the INA of cells and the water potential of the solid K medium on which they were grown. (A) DC3000(pPProIce), y = -2.85x - 5.23 (R2 = 0.79). (B) DC3000(pPProIce/pLH12), y = -2.40x - 5.35 (R2 = 0.94).
Fig. 2.
Water potentials sensed by bacteria (A) and bacterial population sizes (B) in A. thaliana leaves 48 h postinfiltration (hpi) with DC3000(pPProIce) or a derivative containing an avirulence gene. Bacteria were inoculated into A. thaliana Col-O (experiments 1-4), or into an A. thaliana rps2-201 mutant (experiment 5). Bacterial inocula contained ≈108 cells per ml for experiment 1 and 106 cells per ml for experiments 2-5. Bacteria were inoculated into a larger leaf area for experiment 1 than for experiments 2-5. The ice nucleation assay temperature was -9°C for experiment 3 and -5°C for all of the other experiments. Values represent the mean ± SE (n = 3). Values indicated by the same letter within an experiment do not differ significantly by Student's t test (experiments 1-2) or by Fisher's least significant difference test (experiments 3-5) (P < 0.05). Values shown for experiments 1, 3, and 4 are each representative of two replicate experiments. Values shown for experiment 5 are representative of three replicate experiments.
On Col-O, DC3000(pPProIce) consistently sensed lower water potentials when it expressed any of the three avirulence genes avrRpt2, avrRpm1, or avrB than when it did not (Fig. 2A, experiments 3 and 4). In three of four experiments, DC3000(pPProIce/pK48-8) was exposed to significantly lower water potentials than either DC3000(pPProIce/pLH12) or DC3000(pPProIce/pPGS002), suggesting a greater impact of avrRpm1 than of avrRpt2 or avrB on changes in apoplastic osmolarity.
The control plasmid pPNptIce, which conferred constitutive inaZ expression, was used to determine whether nontarget conditions influenced the activity of the InaZ protein. The log(INA) of DC3000(pPNptIce) did not change significantly when cultured cells were exposed to low water potentials (data not shown). Similarly, the log(INA) of DC3000(pPNptIce) during pathogenesis did not differ significantly from that of DC3000(pPNptIce/pLH12) during the HR, indicating that conditions in the apoplast did not affect InaZ protein activity (data not shown). The plasmids pPProIce and pPNptIce were stable under low- and high-osmolarity conditions for at least 30 generations (data not shown).
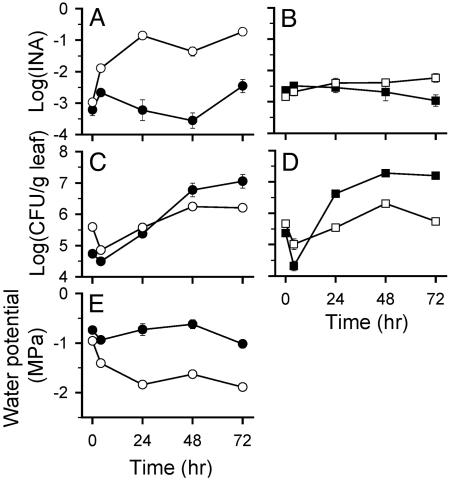
The Apoplastic Water Potential Decreases Rapidly During an HR. The water potential sensed by bacteria during the HR decreased significantly in the 4 h after inoculation, with further decreases thereafter (Fig. 3E). In the first 4 h postinfiltration (hpi), DC3000(pPProIce) experienced a significant decrease of 0.2 MPa, whereas DC3000(pPProIce/pLH12) experienced a significantly greater decrease of 0.45 MPa. By 24 hpi, DC3000(pPProIce) cells were exposed to a water potential of -0.73 ± 0.12 MPa, whereas DC3000(pPProIce/pLH12) cells were exposed to a water potential of -1.84 ± 0.02 MPa. The control strain DC3000(pPNptIce) did not exhibit a significant change in INA during the 48 hpi, whereas DC3000(pPNptIce/pLH12) experienced only a small increase.
Fig. 3.
Dynamics of the in planta INA (A and B), population sizes (C and D), and water potentials sensed (E) by DC3000(pPProIce) (•), DC3000(pPProIce/pLH12) (○), DC3000(pPNptIce) (▪), and DC3000(pPNptIce/pLH12) (□) after infiltration into A. thaliana Col-O. Values represent the mean ± SE (n = 3).
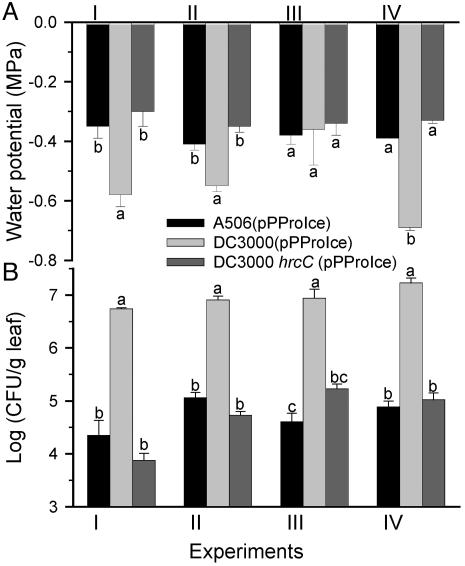
Nonpathogens Encountered Higher Water Potentials in Planta than Did DC3000. To measure the water potential sensed by nonpathogens within the leaf apoplast, we infiltrated P. fluorescens A506 into A. thaliana Col-O leaves. Like DC3000, A506 does not exhibit natural ice nucleation activity. In three of four independent studies, A506(pPProIce) encountered significantly higher water potentials than did DC3000(pPProIce) (Fig. 4A), with A506(pPProIce) sensing water potentials of -0.35 to -0.4 MPa, whereas DC3000(pPProIce) typically sensed water potentials of -0.5 to -0.7 MPa. The nonpathogenic hrcC mutant of DC3000, which was defective in pathogenicity and HR induction on A. thaliana, was similar to A506(pPProIce) in encountering significantly higher water potentials than did DC3000(pPProIce) (Fig. 4A).
Fig. 4.
Water potentials sensed by bacteria (A) and bacterial population sizes (B) in A. thaliana Col-O leaves 48 hpi with A506(pPProIce), DC3000 hrcC (pPProIce), or DC3000(pPProIce). Water potential estimates were derived from the equations y = 0.04x - 5.34 (R2 = 0.87) for A506(pPProIce), y = 0.03x - 3.76 (R2 = 0.89) for DC3000 hrcC (pPProIce), or as shown in Fig. 1 for DC3000(pPProIce), where x = -MPa and y = log(INA). Values represent the mean ± SE (n = 3). Values indicated by the same letter do not differ within each experiment by Fisher's least significant difference test (P < 0.05). The results of four independent experiments are shown.
Lower Water Potentials Were Strongly Correlated with Smaller Bacterial Population Sizes in Planta. Bacterial population sizes were significantly smaller during the HR than during pathogenesis (Fig. 2B), as has been observed in many previous studies. In general, the population sizes of the DC3000 variants during a compatible interaction were 8- to 33-fold larger than those during an incompatible interaction at 48 hpi. Similar differences between compatible and incompatible interactions were observed with DC3000 variants containing pPProIce and pPNptIce (Figs. 2B, 3C, and 3D). Populations of DC3000(pPProIce) were considerably larger than populations of A506(pPProIce) and DC3000 hrcC (pPProIce) at 48 hpi (Fig. 4B); specifically, they were ≈80- to 200-fold greater than those of A506(pPProIce) and 50- to 1,000-fold greater than those of DC3000 hrcC(pPProIce).
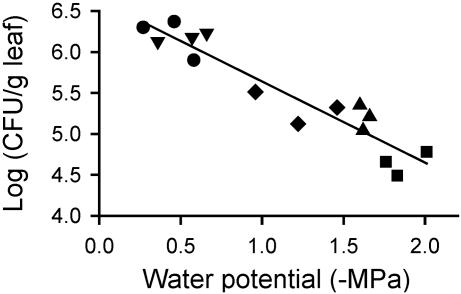
In the experiments with Col-O described in Fig. 1, there was a significant linear correlation between the log(CFU/g of leaf) values and the water potential sensed by the bacteria (P < 0.01). Although the large population sizes in the plants involved in compatible interactions contributed to this correlation, the correlation was significant even when these populations were removed from the analyses. On average, a decrease in water potential of -0.9 MPa correlated with a 10-fold decrease in CFU/g of leaf (Fig. 5).
Fig. 5.
Correlation between bacterial population sizes in planta 48 hpi and the water potential sensed by those bacteria. DC3000(pPProIce) (•), DC3000(pPProIce/pLH12Ω) (▾), DC3000(pPProIce/pK48-8) (♦), DC3000- (pPProIce/pPGS002) (▴), and DC3000(pPProIce/pLH12) (▪). Results are shown for a replicate of experiment 3 described in Fig. 2, and are representative of the results in the other experiments with Col-O described in Fig. 2. Regression analysis yielded the following equation: y = -0.99x + 6.63, R2 = 0.89.
Physiological Response of DC3000 to Low Water Potentials. We evaluated the influence of low water potential, conferred by NaCl amendment, on a range of DC3000 growth characteristics. Experiments performed with the osmolyte KCl and DC3000(pPProIce) yielded results similar to those with NaCl and DC3000.
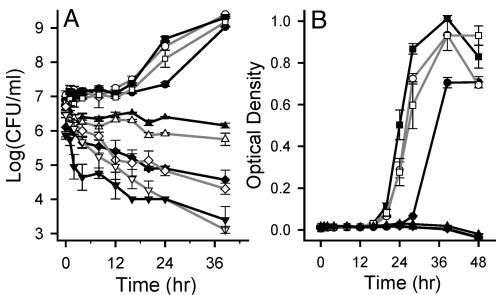
DC3000 growth was hindered by water potentials of -1.4 to -2.3 MPa (300 to 500 mM NaCl), which were the levels sensed by bacteria during the HR. DC3000 cells did not increase in number over a 40-h period in broth with water potentials of -1.4 MPa and lower (Fig. 6), but did eventually develop visible colonies (0.6-0.7 mm) on agar plates with water potentials of -1.4 to -2.3 MPa. The period preceding the detection of these colonies, and thus the time required for osmoadaptation, increased significantly with decreasing water potential; it increased from ≈30 h at 0 and -0.5 MPa to 36, 40, 49, and 80 h at -0.9, -1.4, -1.8, and -2.3 MPa, respectively.
Fig. 6.
DC3000 growth and survival in K broth containing NaCl to confer various water potentials. Cell density was measured by viable plate count (A) and optical density at 600 nm (B). The water potentials examined were 0 (•), -0.2 (○), -0.5 (▪), -0.9 (□), -1.4 (▴), -1.8 (▵), -2.3 (♦2), -2.7 (⋄), -3.7 (▾), and -4.6 (▿) MPa. These results are representative of two independent experiments. Values represent the mean ± SE (n = 3).
Water potentials of -1.8 MPa (400 mM NaCl) and lower showed evidence of toxicity toward DC3000. After a DC3000 culture was plated onto K agar at various water potentials, a significantly smaller number of colonies developed at -1.8 and -2.3 MPa than at higher water potentials (Table 1); thus only a portion (<5%) of the population appeared to divide. Water potentials of -2.7 MPa (600 mM NaCl) and lower inhibited cell division in the entire population based on the absence of colonies on agar plates, even after 265 h of incubation. Decreases in the population sizes in broth at these lower water potentials (Fig. 6) suggest that these levels cause cell death, or at least loss of culturability.
Table 1. Percentage of cells that grew on K agar amended with NaCl relative to on unamended K agar.
| Cells that grew, %
|
||
|---|---|---|
| Water potential, MPa | DC3000 | DC3000 (pPProlce) |
| 0 | 100 ± 0a | 100 ± 0a |
| −0.1 | 108 ± 10a | 176 ± 77a |
| −0.2 | 114 ± 14a | 146 ± 27a |
| −0.5 | 80 ± 25a | 172 ± 36a |
| −0.9 | 93 ± 9a | 141 ± 52a |
| −1.4 | 82 ± 17a | 136 ± 47a |
| −1.8 | 5 ± 1b | 11 ± 2b |
| −2.3 | 3 ± 0.3b | 6 ± 3b |
Values indicated by the same letter within a strain do not differ by Fisher's least significant difference test performed on colony numbers on the plates (P < 0.05).
Several growth parameters suggest that DC3000 exhibits optimal growth at water potentials of -0.2 to -0.9 MPa (50 to 200 mM), which were in the range sensed by DC3000 during pathogenesis. The lag phase was significantly shorter for cells grown in broth at -0.2 to -0.9 MPa, with the shortest lag phase occurring at -0.5 MPa (Fig. 6). Based on the results shown in Fig. 6B, the growth rate was also significantly faster at -0.2 and -0.5 MPa (μ = 0.08 to 0.09) than at 0 MPa (μ = 0.06). Last, there was a consistent trend toward recovering a larger number of colonies on plates with a moderately low water potential than on unamended plates (Table 1); a similar phenomenon was observed in the 4 h after inoculation into broth.
Discussion
In this report, we demonstrate that DC3000 cells encounter significantly lower water potentials in an incompatible than in a compatible plant-pathogen interaction. We determined this by measuring the expression level of the proU-inaZ fusion in DC3000(pPProIce) after infiltration into A. thaliana leaves and using this expression level to estimate the water potential sensed by the bacteria. We examined incompatible interactions by using DC3000 containing avrRpt2, which interacts with the A. thaliana gene RPS2, and avrRpm1 and avrB, which both interact with the A. thaliana gene RPM1. We examined compatible interactions by using either A. thaliana Col-O with DC3000(pPProIce), which itself does not contain an avirulence gene that interacts with the A. thaliana Col-O resistance genes; A. thaliana Col-O with DC3000(pPProIce/pLH12Ω), which contains a disrupted avrRpt2 gene; or an A. thaliana rps2 mutant with DC3000(pPProIce/pLH12), which encodes avrRpt2. In all cases, the results were consistent: cells of the P. syringae pv. tomato pathogen encountered significantly lower water potentials during incompatible interactions, i.e., the HR, than during compatible interactions, i.e., pathogenesis.
During an HR, DC3000 encountered water potentials of -1.6 to -2.3 MPa. Based on the growth characteristics of DC3000 in culture, these water potentials are sufficiently low to prevent cell division in the majority of cells in a population, as well as to dramatically delay cell division in those cells that do divide. The reduced water potentials during the HR thus may be a major factor restricting the size of the DC3000 populations that develop during the HR. Alternatively, low water potentials may arise simultaneous with or after other unknown factors that restrict bacterial growth. The strong correlation between apoplastic water potential and apoplastic bacterial populations further suggests that water availability within leaves is an important determinant of bacterial population size.
Distinct avirulence genes in DC3000 differentially influenced the extent to which DC3000 cells were exposed to low water potentials in the apoplast. DC3000 cells expressing the avrRpm1 gene were exposed to significantly lower water potentials (-2.1 to -2.2 MPa) than cells expressing avrRpt2 or avrB (-1.6 to -2 MPa). Previous studies have demonstrated differences in the rate at which AvrRpm1, AvrRpt2, and AvrB induce changes in A. thaliana leaves (16, 33, 34). For example, AvrRpm1 induced visible leaf collapse 5 h after infiltration (34), whereas AvrRpt2 and AvrB required 9-14 h and 8-10 h, respectively (33, 35). Similarly, AvrRpm1 induced a sustained increase in cytosolic Ca2+ concentration and accumulation of H2O2 more rapidly than did AvrB (16). Our findings are consistent with these differential responses to the avirulence gene products.
We believe that during the HR, the proU-inaZ fusion was responding to high osmolarity rather than to desiccation. Although the proU promoter responds specifically to the water status of a cell and not to ion toxicity (18), hydrogen peroxide, ethanol, or the superoxide-generating compound paraquat (unpublished data), it responds to osmotic (solute) stress as well as to matric stress, in which the bacteria are dehydrated by the loss of water from their surroundings (18, 36). The well-characterized K+ efflux that occurs across the host plasma membrane during HR would drive an increase in apoplastic osmolarity. This efflux, as evaluated based on the associated H+ influx, or alkalinization, begins within only 1 h after inoculation of an incompatible pathogen into A. thaliana cell cultures (37), within 2 h in tobacco cell cultures (38), and within 4 h in soybean leaves (39). It is possible, however, that proU-inaZ induction resulted from bacterial exposure to desiccation. Such desiccation could result from continued transpiration while restricting xylem flow to the infection site, as was observed in Nicotiana edwardsonii in response to the tobacco mosaic virus (40), and maintaining open stomata, as was suggested in the response of Cucumis sativus to P. syringae pv. syringae (41) and Nicotiana sylvestris to Erwinia amylovora harpin (42). Given that these changes generally were not observed until 10-11 hpi, however, it seems unlikely that sufficient tissue desiccation to cause a measurable water potential reduction occurs within only 4 hpi.
In a compatible interaction, DC3000 encountered water potentials of -0.4 to -0.9 MPa, which were significantly lower than the water potentials encountered by the saprophyte P. fluorescens A506 in A. thaliana. The DC3000-mediated reductions in water potential likely resulted, at least in part, from active modification of the intercellular environment. For example, P. syringae can promote increases in apoplastic sucrose levels by increasing the apoplastic pH and thus decreasing the activity of symporters directing sucrose into the phloem (12, 43). Interestingly, although P. syringae pv. syringae induced an H+ influx in a compatible interaction with Phaseolus vulgaris leaves, albeit one that was slower and weaker than that in an incompatible interaction (11), DC3000 did not induce a detectable H+ influx in a compatible interaction with A. thaliana, at least not in ecotype Fi-3 cell cultures (37). DC3000 may also alter the intercellular environment by secreting proteins that bind to lipid bilayers and form pores, as has been shown for the P. syringae pv. phaseolicola HrpZ harpin (44) and for several P. syringae pv. syringae phytotoxins (45). Although DC3000 has not been shown to produce pore-forming phytotoxins, it does produce harpins that may perform this function (46). Water potentials of -0.4 to -0.9 MPa are equivalent to those conferred by 90-200 mM KCl or 150-300 mM sucrose (31); these solute concentrations are significantly higher than the concentrations in the apoplast of uninfected leaves (43). Although these sucrose concentrations are higher than those required to support abundant growth in culture, the apoplastic water potentials during the compatible interaction were themselves in the range that supported optimal DC3000 growth in culture.
The DC3000 hrcC mutant, which is deficient in the production of the type III secretion apparatus and thus the secretion of effectors into plant cells (20), appeared to be identical to the saprophyte P. fluorescens A506 in its effect on the plant. Both strains encountered water potentials of -0.3 to -0.4 MPa in A. thaliana at 48 hpi. These levels may not indicate the water status of the apoplast of uninfected leaves because these bacteria can elicit plant responses involved in innate immunity (47). The most well-studied elicitor of plant innate immunity, a highly conserved domain of bacterial flagellin, is present in a wide range of eubacteria, including P. syringae pv. tomato and P. fluorescens (48). Flagellin induces plant responses, including alkalinization of the medium supporting cultured A. thaliana cells, a concomitant efflux of K+, and production of ROS (48). These responses occur within minutes after exposure of A. thaliana leaf tissue to a purified flagellin elicitor (49) and thus may have influenced the water potentials sensed by bacteria 48 hpi. Based on the model that host defenses are suppressed by pathogenic strains but activated by nonpathogenic and avirulent strains, with the latter being unable to attenuate those responses via effectors (13), we predicted that the nonpathogenic and avirulent strains would elicit lower water potentials than DC3000. However, the nonpathogenic strains elicited less severe water potentials than DC3000. This result remains unexplained at present.
The dynamics of DC3000 populations during HR and pathogenesis may be partly explained by water potential changes in the apoplast. The DC3000 population showed no net increase by 24 h during the HR, which was consistent with low water potential restricting cell division. The population decline in the first 4 hpi was similar in magnitude to the decline in cultured cell number in the 4 h after equivalent water potential reductions. The subsequent increase may have reflected growth of a subpopulation of cells that tolerated water stress, as we observed for cells grown on solid but not liquid media, or the eventual expression of adaptation mechanisms that were specific to the apoplast, such as the uptake of plant-derived compatible solutes. During pathogenesis, DC3000 showed a 10- to 20-fold net population increase, which was consistent with the water potentials favoring growth. The population decrease within 4 hpi may reflect a transient loss of culturability or, alternatively, cell death and subsequent division of the survivors. Although the mechanisms driving the population decreases in the first 4 hpi are not clear, they may result from the rapid host oxidative burst that typically occurs within the first 2 hpi during both pathogenesis and HR (50).
In summary, these studies demonstrate that DC3000 cells rapidly encounter much lower water potentials during the HR than during pathogenesis. Furthermore, the effects of low water potentials on DC3000 growth in culture indicate that water potential reduction during the HR may be at least one factor restricting the growth of P. syringae pv. tomato DC3000. ROS and phytoalexins have been the primary factors considered to have this role. Although they may have an impact on bacterial growth in planta, their role in growth restriction during the HR appears not to be significant based on the dynamics of bacterial growth in various A. thaliana mutants deficient in ROS and phytoalexin production (15, 17). Future studies exploring water stress and bacterial growth restriction in planta may benefit from A. thaliana mutants altered in their water relations.
Acknowledgments
We thank Sheng Yang He for providing strains DC3000 and DC3000 hrcC, Andrew Bent for providing the cloned avirulence genes and Col-O and rps2-201 seeds, Steven Lindow for providing strain A506, and Lindsay Schulz, Daniel Schlangen, Lindsay Carpp, and Zeb McMillan for technical assistance. We are also grateful to Sheng Yang He and Andrew Bent for their helpful comments on the manuscript. This work was supported by the United States Department of Agriculture National Research Initiative Grant 2001-35319-10929.
Abbreviations: hpi, hours postinfiltration; HR, hypersensitive response; CFU, colony-forming units; ROS, reactive oxygen species; INA, ice nucleation activity.
References
- 1.Alfano, J. R. & Collmer, A. (1997) J. Bacteriol. 179, 5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collmer, A., Badel, J. L., Charkowski, A. O., Deng, W.-L., Fouts, D. E., Ramos, A. R., Rehm, A. H., Anderson, D. M., Schneewind, O., van Dijk, K. & Alfano, J. R. (2000) Proc. Natl. Acad. Sci. USA 97, 8770-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abramovitch, R. B., Kim, Y.-J., Chen, S., Dickman, M. B. & Martin, G. B. (2003) EMBO J. 22, 60-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinosa, A., Guo, M., Tam, V. C., Fu, Z. Q. & Alfano, J. R. (2003) Mol. Microbiol. 49, 377-387. [DOI] [PubMed] [Google Scholar]
- 5.Hauck, P., Thilmony, R. & He, S. Y. (2003) Proc. Natl. Acad. Sci. USA 100, 8577-8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Torres, M., Sanchez, P., Fernandez-Delmond, I. & Grant, M. (2003) Plant J. 33, 665-676. [DOI] [PubMed] [Google Scholar]
- 7.Quirino, B. F. & Bent, A. F. (2003) Mol. Plant Pathol. 4, 517-530. [DOI] [PubMed] [Google Scholar]
- 8.Whalen, M. C., Innes, R. W., Bent, A. F. & Staskawicz, B. J. (1991) Plant Cell 3, 49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jambunathan, J., Siani, J. M. & McNellis, T. W. (2001) Plant Cell 13, 2225-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boch, J., Joardar, V., Gao, L., Robertson, T. L., Lim, M. & Kunkel, B. N. (2002) Mol. Microbiol. 44, 73-88. [DOI] [PubMed] [Google Scholar]
- 11.Atkinson, M. M. & Baker, C. J. (1987) Phytopathology 77, 1273-1279. [Google Scholar]
- 12.Atkinson, M. M. & Baker, C. J. (1987) Phytopathology 77, 1573-1578. [Google Scholar]
- 13.Brown, I., Mansfield, J. & Bonas, U. (1995) Mol. Plant-Microbe Interact. 8, 825-836. [Google Scholar]
- 14.Hagemeier, J., Schneider, B., Oldham, N. J. & Hahlbrock, K. (2001) Proc. Natl. Acad. Sci. USA 98, 753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glazebrook, J. & Ausubel, F. M. (1994) Proc. Natl. Acad. Sci. USA 91, 8955-8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A. & Mansfield, J. (2000) Plant J. 23, 441-450. [DOI] [PubMed] [Google Scholar]
- 17.Torres, M. A., Dangl, J. L. & Jones, J. D. G. (2002) Proc. Natl. Acad. Sci. USA 99, 517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Axtell, C. A. & Beattie, G. A. (2002) Appl. Environ. Microbiol. 68, 4604-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore, R. A., Starratt, A. N., Ma, S. W., Morris, V. L. & Cuppels, D. A. (1989) Can. J. Microbiol. 35, 910-917. [Google Scholar]
- 20.Yuan, J. & He, S. Y. (1996) J. Bacteriol. 178, 6399-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson, M. & Lindow, S. E. (1993) Phytopathology 83, 117-123. [Google Scholar]
- 22.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 23.Kennedy, E. P. (1982) Proc. Natl. Acad. Sci. USA 79, 1092-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbst, B., Kneip, S. & Bremer, E. (1994) Gene 151, 137-142. [DOI] [PubMed] [Google Scholar]
- 25.Miller, W. G., Leveau, J. H. J. & Lindow, S. E. (2000) Mol. Plant-Microbe Interact. 13, 1243-1250. [DOI] [PubMed] [Google Scholar]
- 26.Debener, T., Lehnackers, H., Arnold, M. & Dangl, J. L. (1991) Plant J. 1, 289-302. [DOI] [PubMed] [Google Scholar]
- 27.Staskawicz, B., Dahlbeck, D., Keen, N. & Napoli, C. (1987) J. Bacteriol. 169, 5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Better, M. & Helinski, D. R. (1983) J. Bacteriol. 155, 311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee, D., Zhang, X. & Bent, A. F. (2001) Genetics 158, 439-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunkel, B. N., Bent, A. F., Dahlbeck, D., Innes, R. W. & Staskawicz, B. J. (1993) Plant Cell 5, 865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris, R. F. (1981) in Water Potential Relations in Soil Microbiology, eds. Parr, J. F., Gradner, W. R. & Elliot, L. F. (Soil Science Society of America, Madison, WI), pp. 23-96.
- 32.Lindow, S. E., Arny, D. C. & Upper, C. D. (1978) Phytopathology 68, 523-527. [Google Scholar]
- 33.Bent, A. F., Kunkel, B. N., Dahlbeck, D., Brown, K. L., Schmidt, R., Giraudat, J., Leung, J. & Staskawicz, B. J. (1994) Science 265, 1856-1860. [DOI] [PubMed] [Google Scholar]
- 34.Grant, M. R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R. W. & Dang, J. L. (1995) Science 269, 843-846. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro, A. D. & Zhang, C. (2001) Plant Physiol. 127, 1089-1101. [PMC free article] [PubMed] [Google Scholar]
- 36.Overdier, D. G., Fletcher, S. & Csonka, L. N. (1992) in Water and Life, eds. Somero, G. N., Osmond, C. B. & Bolis, L. (Springer, Berlin), pp. 61-69.
- 37.Davis, K. R., Schott, E. & Ausubel, F. M. (1991) Mol. Plant-Microbe Interact. 4, 477-488. [Google Scholar]
- 38.Atkinson, M., Bina, J. & Sequeira, L. (1993) Mol. Plant-Microbe Interact. 6, 253-260. [Google Scholar]
- 39.Orlandi, E. W., Hutcheson, S. W. & Baker, C. J. (1992) Physiol. Mol. Plant Pathol. 40, 173-180. [Google Scholar]
- 40.Wright, K. M., Duncan, G. H., Pradel, K. S., Carr, F., Wood, S., Oparka, K. J. & Cruz, S. S. (2000) Plant Physiol. 123, 1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pike, S. M. & Novacky, A. (1988) in Current Topics in Plant Biochemistry and Physiology, eds. Randall, D. D., Belvins, D. G., Campbell, W. H., Long, J. M. & Rapp, B. J. (Interdisciplinary Plant Biochemistry and Physiology Program, University of Missouri, Columbia), Vol. 7, pp. 233. [Google Scholar]
- 42.Boccara, M., Boué, C., Garmier, M., De Paepe, R. & Boccara, A.-C. (2001) Plant J. 28, 663-670. [DOI] [PubMed] [Google Scholar]
- 43.Lohaus, G., Pennewiss, K., Sattelmacher, B., Hussmann, M. & Muehling, K. H. (2001) Physiol. Plant. 111, 457-465. [DOI] [PubMed] [Google Scholar]
- 44.Lee, J., Klüsener, B., Tsiamis, G., Stevens, C., Neyt, C., Tampakaki, A. P., Panopoulos, N. J., Nöller, J., Weiler, E. W., Cornelis, G. R., Mansfield, J. W. & Nürnberger, T. (2001) Proc. Natl. Acad. Sci. USA 98, 289-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutchison, M. L. & Gross, D. C. (1997) Mol. Plant-Microbe Interact. 10, 347-354. [DOI] [PubMed] [Google Scholar]
- 46.Hoyos, M. E., Stanley, C. M., He, S. Y., Pike, S., Pu, X. A. & Novacky, A. (1996) Mol. Plant-Microbe Interact. 9, 608-616. [Google Scholar]
- 47.Gómez-Gómez, L. & Boller, T. (2002) Trends Plant Sci. 7, 251-256. [DOI] [PubMed] [Google Scholar]
- 48.Felix, G., Duran, J. D., Volko, S. & Boller, T. (1999) Plant J. 18, 265-276. [DOI] [PubMed] [Google Scholar]
- 49.Nuhse, T. S., Peck, S. C., Hirt, H. & Boller, T. (2000) J. Biol. Chem. 275, 7521-7526. [DOI] [PubMed] [Google Scholar]
- 50.Baker, C. J. & Orlandi, E. W. (1995) Annu. Rev. Phytopathol. 33, 299-321. [DOI] [PubMed] [Google Scholar]