Leaping drops of water help keep cicada wings clean
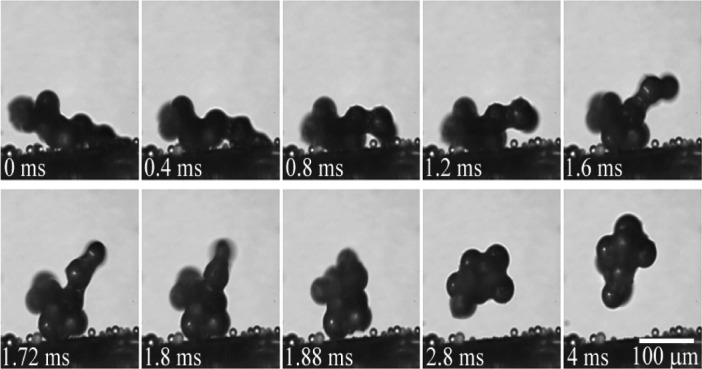
Particles aggregate and jump off a cicada wing during vapor condensation.
Like the leaves of the lotus plant, highly water-repellent surfaces known as superhydrophobic surfaces exhibit a remarkable ability to remove contaminating particles. Researchers commonly attribute the self-cleaning property, dubbed the lotus effect, to the action of water droplets rolling across the superhydrophobic surface under the influence of gravity. Katrina Wisdom et al. (pp. 7992–7997) report that water vapor can condense onto a superhydrophobic surface and produce self-propelled droplets of liquid condensate, which encloses contaminating particles and removes them via a jumping motion. Using the cicada wing as a model, the authors demonstrate that surface energy, released when a liquid condensate coalesces around a foreign particle, drives this jumping motion with sufficient force to dislodge particles similar in size to plant pollen and soil dust. Furthermore, the authors show that the process can spontaneously clean contaminants from superhydrophobic cicada wings that cannot be removed by gravity, wing vibration, or wind. The findings reveal a previously unreported mechanism for removing contaminants from superhydrophobic surfaces that may aid the development of self-cleaning materials, according to the authors. — T.J.
DNA repair observed in living bacteria
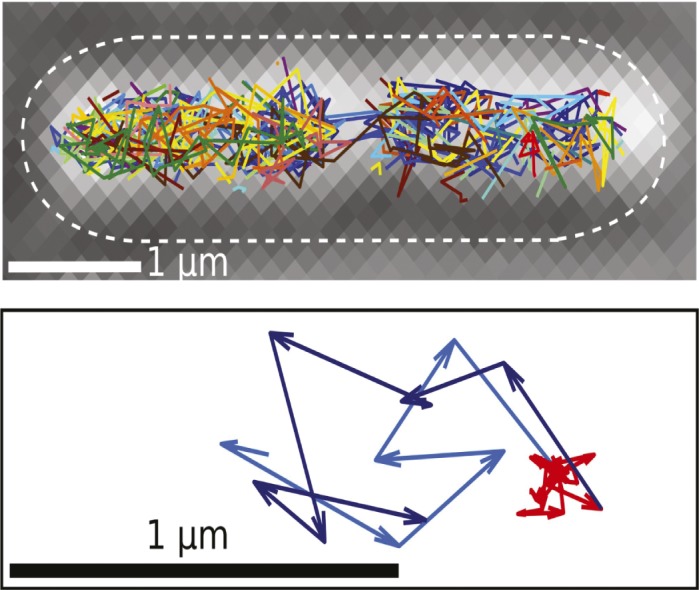
Tracks showing the movement of DNA polymerase I proteins in a live E. coli cell. Random search path of a single polymerase (in light blue) leads to a transient binding event for DNA repair (in red) and continued search (in dark blue).
Damage caused by mutagens to the nucleotide bases of DNA is reversed by a process called base excision repair, executed by a suite of enzymes. Two such enzymes—DNA polymerase I, which replaces missing nucleotides, and DNA ligase, which ligates DNA strands to help close nicks—perform the final steps of the repair process in a range of organisms from bacteria to humans, but the enzymes have not been directly observed in action. Stephan Uphoff et al. (pp. 8063–8068) used a technique called photoactivated localization microscopy to visualize the activity of single molecules of both enzymes in live Escherichia coli bacteria. Interpreting tracks of single enzyme molecules bound to DNA as evidence of DNA synthesis and ligation, the authors found that the DNA repair sites were spread throughout the cell. Only 2.7% of approximately 400 copies of polymerase and 3.8% of around 200 copies of ligase are active in replication and repair at any given time in a bacterial cell with undamaged DNA. The authors found that when the cell was subjected to DNA damage, both enzymes spent more than 80% of their lifespan searching for toxic intermediates to repair, thus minimizing the lifetimes of gaps and nicks. Direct measurements of DNA repair rates in living cells might lead to quantitative models that can help predict how organisms respond to DNA damage, according to the authors. — P.N.
Predicting forest expansion into alpine areas

Geomorphic processes limit temperature-driven, high-elevation forest encroachment into alpine areas in Kananaskis Country, in the Rocky Mountains of Alberta, Canada.
Numerous studies have predicted that as climate warming increases temperatures in mountain ecosystems, tree lines will shift to higher elevations. As a consequence, expanding forests will encroach into alpine areas and fragment alpine ecosystems, potentially degrading habitats and threatening certain species. Marc Macias-Fauria and Edward Johnson (pp. 8117–8122) devised a statistical model to investigate whether temperature alone can explain the distribution of high-elevation tree cover in a fine 10-m grid over roughly 100 km2. The authors examined a geologically diverse area in the Front Ranges of the Canadian Rocky Mountains, and by incorporating parameters to account for interrelated climatic and geomorphic processes, found that topographic characteristics influence how trees are distributed. The authors used the model to forecast tree cover in the late-21st century based on a moderate climate warming scenario, and determined that upward tree advance is likely to be severely limited by individual site characteristics such as slope, exposure, and soil properties. The findings suggest that at regional scales temperature alone does not adequately predict whether forests will encroach on alpine areas at high elevations, according to the authors. — T.J.
Bats harbor hepatitis C-related viruses

Colony of Egyptian fruit bats (Rousettus aegyptiacus) in a cave in Kenya.
Wildlife surveillance surveys have revealed surprising diversity among the viruses harbored by the estimated 1,150 bat species that exist worldwide. These surveys provide valuable tools for protecting bats and their habitats, and allow researchers to quickly identify bat viruses that emerge in humans. To better understand the diversity of bat viruses, Phenix-Lan Quan et al. (pp. 8194–8199) examined 415 serum samples from African and Central American bats. Using high-throughput sequencing analyses, the authors uncovered an extremely diverse and previously undocumented group of bat viruses that resemble known hepaciviruses and pegiviruses in the family Flaviridae, of which the human hepatitis C virus is a widely held example. The authors screened 1,673 specimens by consensus PCR from bats around the globe and found that the viruses were distributed in both North America and Asia. Their findings identified 83 previously undescribed bat viruses; evolutionary analyses revealed that the phylogenetic diversity of the viruses encompasses the entire spectrum of hepaciviruses and pegiviruses known to infect humans and other primates. According to the authors, the prevalence, worldwide distribution, and biodiversity of the viruses suggests that bats are a major and ancient reservoir for both hepaciviruses and pegiviruses. — A.G.


