Abstract
Viroids and most viral satellites have small, noncoding, and highly structured RNA genomes. How they cause disease symptoms without encoding proteins and why they have characteristic secondary structures are two longstanding questions. Recent studies have shown that both viroids and satellites are capable of inducing RNA silencing, suggesting a possible role of this mechanism in the pathology and evolution of these subviral RNAs. Here we show that preventing RNA silencing in tobacco, using a silencing suppressor, greatly reduces the symptoms caused by the Y satellite of cucumber mosaic virus. Furthermore, tomato plants expressing hairpin RNA, derived from potato spindle tuber viroid, developed symptoms similar to those of potato spindle tuber viroid infection. These results provide evidence suggesting that viroids and satellites cause disease symptoms by directing RNA silencing against physiologically important host genes. We also show that viroid and satellite RNAs are significantly resistant to RNA silencing-mediated degradation, suggesting that RNA silencing is an important selection pressure shaping the evolution of the secondary structures of these pathogens.
Viroids and most viral satellites, which are the smallest known infectious agents in plants, have single-stranded RNA genomes of 200-400 nt and do not encode proteins (1-3). Whereas viroids replicate autonomously by using host-encoded RNA polymerase, satellite RNAs multiply only in the presence of a helper virus that provides the appropriate RNA-dependent RNA polymerase (2, 4). Intriguingly, some viroids and satellites can induce unique, highly host species-specific disease symptoms despite their exceedingly small size and lack of mRNA activity. Previous studies have shown that one, or a few, nucleotide changes in their RNA genomes can dramatically alter the virulence of these subviral RNAs or the host-plant specificity of the disease symptoms (5-7). Despite intensive investigation, major questions remain as to how these minor sequence variations modulate viroid and satellite pathology and how host plants develop symptoms in response to specific sequences. A striking similarity among viroids and small satellites is that they tend to form characteristic secondary structures due to intramolecular base-pairing. These structures are clearly important, because the evolution of these small RNAs appears to be constrained by the need to preserve their distinct structural features. However, the host factor(s) that imposes this evolutionary pressure has yet to be identified.
RNA silencing is a sequence-specific RNA degradation process directed by double-stranded RNA (dsRNA) or self-complementary hairpin RNA (hpRNA). This dsRNA or hpRNA is cleaved by an RNase III-like enzyme known as Dicer to generate small (21- to 25-nt) RNAs, termed small interfering RNAs (siRNAs), which are used to guide siRNA-ribonuclease complexes [known as RNA-induced silencing complexes (RISC)] to degrade cognate single-stranded RNA (8). Recent studies have shown that plants infected with potato spindle tuber viroid (PSTVd) or cereal yellow dwarf virus RPV and its satellite (RPVSat) contain siRNAs derived from the pathogens' genomes (9-11). This finding has led to the hypothesis that RNA silencing might be involved in the processes of viroid and viral satellite pathogenicity (9, 12, 13). Until now, there has been no experimental evidence to support this hypothesis. Here, we provide evidence suggesting that viroid and satellite pathogenicities are mediated by RNA silencing and that these subviral RNAs have evolved secondary structures that minimize siRNA-mediated destruction.
Materials and Methods
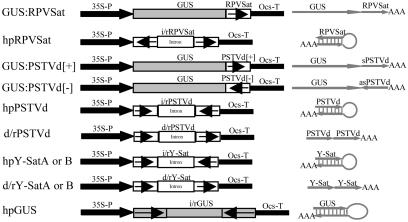
Plasmids. The various constructs used in this work are shown in Fig. 1. The hpRPVSat construct encoding hpRNA of the cereal yellow dwarf virus RPV satellite was prepared by cloning the same RPVSat sequence as in GUS:RPVSat (11) into pKannibal (14). To make the β-glucuronidase (GUS) fusion constructs, GUS:PSTVd[+] and GUS:PSTVd[-], the full-length sequence (with minor mutations; see Fig. 1) of the RG1 strain (15) of PSTVd, was assembled by PCR with overlapping oligonucleotides and then cloned into a 35S-GUS-Ocs cassette (11) in either sense (for GUS:PSTVd[+]) or antisense (for GUS:PSTVd[-]) orientation. For Agrobacterium-mediated transformation, the three constructs were all cloned into the binary vector pWBVec2a (16). The hpPSTVd construct was made by cloning into pKannibal a truncated PSTVd sequence (nucleotides 16-355) and the above-mentioned full-length PSTVd sequence in sense and antisense orientations, respectively. The hpY-Sat construct and the d/rY-Sat construct that encodes direct-repeat RNA of cucumber mosaic virus (CMV) Y satellite (Y-Sat) were prepared by cloning PCR-assembled (using several overlapping oligonucleotides) Y-Sat sequences (with minor mutations; see Fig. 1) into pKannibal. The PSTVd and Y-Sat constructs were cloned into pART27 (17) for Agrobacterium transformation. The hpGUS construct was prepared by cloning the hpGUS sequence (18) into pART7 (17). The resulting 35S-hpGUS-Ocs cassette was then inserted into pWBVec4 (16) for Agrobacterium transformation. The Agrobacterium tumefaciens strain AGL1 was used for plant transformation.
Fig. 1.
Schematic diagrams of transgene constructs used in tobacco and/or tomato transformation. (Right) The predicted structure of RNA transcripts from these transgenes. RPVSat is a full-length (322-nt) sequence of the cereal yellow dwarf virus RPV satellite. The PSTVd sequence in GUS:PSTVd[+] and GUS:PSTVd[-] contains several minor sequence mutations (arising from cloning) compared with the 359-nt wild-type PSTVd-RG1 sequence (deletions of G168 and C318G319 and single-nucleotide substitutions of C185→T and C231→G), but these mutations do not cause significant alteration to the predicted rod-like structure of the viroid RNA. The sense PSTVd sequence in hpPSTVd starts at nucleotide 16 and ends at nucleotide 355 of the RG1 strain, with one nucleotide substitution of G281→A, and the antisense sequence is the same as that in the GUS:PSTVd fusion constructs. There are two versions (A and B) for the hpY-Sat and d/rY-Sat constructs; the Y-SatA sequence has the size of a wild-type Y-Sat RNA (369 nt) but with two single-nucleotide substitutions of A293→G and A308→G, and Y-SatB has a 5-nt deletion (nucleotides 192-196), which constitutes part of the yellow symptom domain, plus four single-nucleotide substitutions of G80→A, G128→ C, G204→T, and A214→T. The hpGUS sequence is the same as that described in ref. 18. 35S-P, cauliflower mosaic virus 35S promoter; Ocs-T, 3′ region of Agrobacterium octopine synthase gene; i/r, inverted-repeat sequences.
Plant Transformation. Tobacco was transformed as described (11) by using 20 mg/liter hygromycin (for pWBVec2-based plasmids), 50 mg/liter kanamycin (for pART27-based plasmids), or 15 mg/liter phosphinothricin (for hpGUS) as the selective agent. The Y-Sat constructs were transformed into Nicotiana tobacum Samsun, and all of the others were transformed into N. tobacum Wisconsin 38. Transformation of tomato (Lycopersicon esculentum Money Maker) with hpPSTVd or d/rPSTVd was performed as described (19).
Analysis of Transgenic Plants. GUS activity in transgenic tobacco was measured at 37°C by the fluorometric 4-methylumbelliferryl-β-glucuronide assay (20) by using 5 μg of leaf protein extract. For Northern blot hybridization analysis, total RNA was prepared by using the TRIzol reagent (Invitrogen), separated in formaldehyde-agarose gels (for normal Northern analysis) or in 15% polyacrylamide gels (for siRNA detection), blotted to Hybond-N filter, and hybridized with T7 or SP6 polymerase-synthesized, α-32P-labeled riboprobes (11).
Virus/Satellite RNA Infection. The Y-Sat culture was initiated by mechanically inoculating CMV-infected young tobacco with transcript synthesized from an infectious Y-Sat clone (21). Extracts of the CMV/Y-Sat-infected tobacco leaves were then used for subsequent inoculation. Infection by potato leaf roll virus, which was required to support RPVSat replication, was performed as described (11).
Results
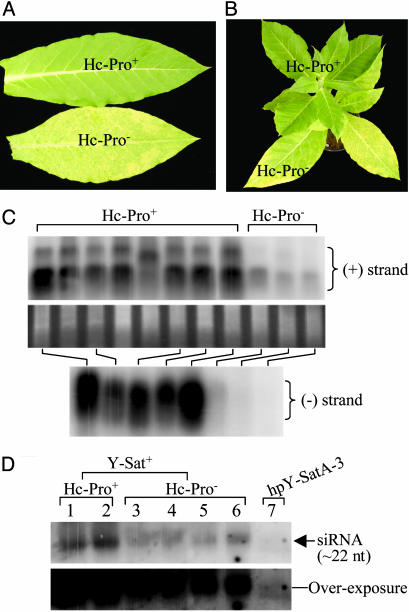
Evidence That RNA Silencing Mediates Viroid and Satellite Pathogenicity. Expressing an RNA silencing suppressor in plants dramatically reduces satellite symptoms. To test whether RNA silencing is involved in the pathology of subviral RNAs, we examined the symptom development of CMV Y-Sat in transgenic tobacco expressing P1/Hc-Pro from tobacco etch virus (22). Hc-Pro is a potent suppressor of RNA silencing induced by either transgenes or viruses (23). We infected 21 plants expressing P1/Hc-Pro as a transgene (Hc-Pro+) and 7 plants without the transgene (Hc-Pro-) with CMV plus Y-Sat and monitored the development of the bright yellow mosaic symptom. It should be noted that this yellowing symptom is unique to Y-Sat infection (24) and that the helper virus CMV alone induces only light green mosaic symptoms. All seven Hc-Pro- plants developed the characteristic yellow mosaic symptom ≈17 days postinoculation (dpi), which turned to severe systemic chlorosis (Fig. 2A) from 25 dpi. In contrast, none of the 21 Hc-Pro+ plants showed such severe systemic yellowing. Some young leaves of the Hc-Pro+ plants initially showed yellow vein clearing, usually near the midribs (data not shown), but the symptoms disappeared in expanded leaves (Fig. 2A). Reciprocal grafting between the Y-Sat-infected Hc-Pro+ and Hc-Pro- plants gave the predicted result: leaves of Hc-Pro- scions or stocks developed severe chlorosis, and those of the Hc-Pro+ scions or stocks showed little or no yellowing (see Fig. 2B). These results suggest that a fully functional RNA silencing mechanism is required for satellite symptom development, making it likely that this mechanism is the mediator of those symptoms. An alternative explanation, i.e., the reduction in symptoms was due to reduced levels of Y-Sat RNA replication, can be ruled out because the levels of both the plus (+) and minus (-) strands of Y-Sat RNA were much higher in the Hc-Pro+ plants than in the Hc-Pro- plants (Fig. 2C).
Fig. 2.
Expression of Hc-Pro in tobacco dramatically reduces the yellow mosaic symptom of Y-Sat. (A) Y-Sat induces severe chlorosis in Hc-Pro- tissue but not in Hc-Pro+ tissue. The Hc-Pro+ and Hc-Pro- plants were siblings of the same F1 plant that was obtained by pollinating N. tobacum Wisconsin 38 with pollen from the previously described Hc-Pro plant (22). The presence or absence of the Hc-Pro transgene was confirmed by PCR analysis. (B) An Hc-Pro- scion in a grafted plant showed severe Y-Sat symptom, whereas the shoot from the Hc-Pro+ stock exhibited very little yellowing. (C) Detection of plus (+) strand (Top) or minus (-) strand (Bottom) of Y-Sat RNA in Hc-Pro+ and Hc-Pro- tobacco infected with Y-Sat. (Middle) RNA loading control. Twenty micrograms of total RNA was hybridized with either antisense (Top) or sense (Bottom) Y-Sat RNA probe. (D) Detection of siRNA from Hc-Pro+ (lanes 1 and 2) or Hc-Pro- (lanes 3-6) tobacco infected with Y-Sat, or uninfected tobacco transformed with the hpY-Sat construct (lane 7). Total RNA (40 μg) was hybridized with the full-length antisense Y-Sat sequence. (Lower) An overexposure of Upper. Note that the infecting Y-Sat produced at least 100-fold more siRNA than the hpY-Sat transgene; hpY-SatA-3 is the same as that in Fig. 4A, where small RNA enriched from 120 μg of total RNA was loaded.
Northern blot analysis revealed extremely high levels of Y-Sat-specific siRNAs in the satellite-infected plants (Fig. 2D, lanes 1-6), which were at least 100-fold more abundant than Y-Sat-specific siRNAs derived from the hpY-Sat transgene (Fig. 2D, lane 7). The Y-Sat siRNA levels were higher in the Hc-Pro+ plants than in the Hc-Pro- plants (Fig. 2D, lanes 1 and 2 versus lanes 3-6), indicating that Hc-Pro does not block siRNA production from dsRNA, consistent with previous findings (25).
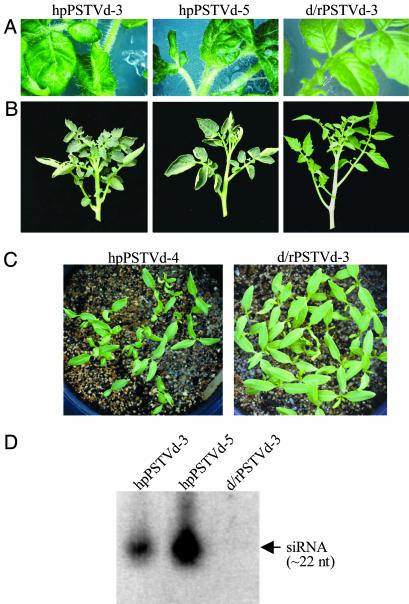
Plants expressing hpRNA of a viroid sequence develop viroid-like symptoms. We reasoned that if RNA silencing mediates viroid and satellite symptoms, then the production of dsRNA or siRNAs, corresponding to viroid or satellite sequences, from a nonreplicating hpRNA should give symptoms that mimic those of viroid or satellite infection. To test this, we transformed tomato plants with a transgene designed to express an hpRNA that contains sequences corresponding to a virulent strain (RG1) of PSTVd (15). A total of five transgenic lines containing the hpPSTVd construct (Fig. 1) and eight lines containing a nonhairpin (direct repeat) PSTVd control construct (d/rPSTVd) (Fig. 1) were examined. In tissue culture, the leaves of the hpPSTVd lines showed rugosity and abnormal shapes with dark green color (Fig. 3A). In contrast, the lines with the control construct showed normal leaf phenotypes. Under glasshouse conditions, the five hpPSTVd lines displayed a range of abnormalities compared with the control lines. The two extreme lines, hpPSTVd-3 and hpPSTVd-5, had shorter internodes and stunted, epinastic leaves (Fig. 3B), which resemble the symptoms caused by infection with PSTVd (26). Fruit development of most hpPSTVd lines, including hpPSTVd-3 and hpPSTVd-5, was impaired. One line, hpPSTVd-4, which showed a milder abnormality than hpPSTVd-3 and hpPSTVd-5, yielded a small number of seed. As shown in Fig. 3C, the T1 seedlings of hpPSTVd-4 displayed significant developmental defects compared with the T1 progeny of the healthy control line (d/rPSTVd-3). PSTVd-specific siRNA was detectable in the hpPSTVd lines but not in the control line (Fig. 3D). Northern blot hybridization showed that no replicating PSTVd RNA arose from the hpPSTVd transgene (data not shown), which was expected, because both the sense and antisense sequences in hpPSTVd contain small sequence mutations outside the virulence modulating region (Fig. 1). This ruled out the possibility that the symptoms are due to the generation of infectious PSTVd. The symptoms in hpPSTVd plants did not appear to be as strong as those described for plants infected with the natural RG1 strain (6, 15). This is probably due to lower levels of siRNA being produced from the hpPSTVd transgenes than from replicating PSTVd. Overall, these results suggest that PSTVd-like symptoms are induced by siRNAs corresponding to PSTVd sequences, thus supporting a direct role of RNA silencing in mediating viroid symptoms.
Fig. 3.
Expression of PSTVd hpRNA in tomato induces symptoms reminiscent of PSTVd infection. (A) Phenotypes of regenerated tomato shoots in tissue culture that were transformed with hpPSTVd or d/rPSTVd. (B) Phenotypes of hpPSTVd or d/rPSTVd transgenic tomato in the glasshouse. (C) T1 seedlings of hpPSTVd-4 and d/rPSTVd-3. (D) Analysis of hpPSTVd and d/rPSTVd transgenic tomato for the presence of siRNAs. Total RNA (30 μg) was hybridized with full-length antisense PSTVd RNA.
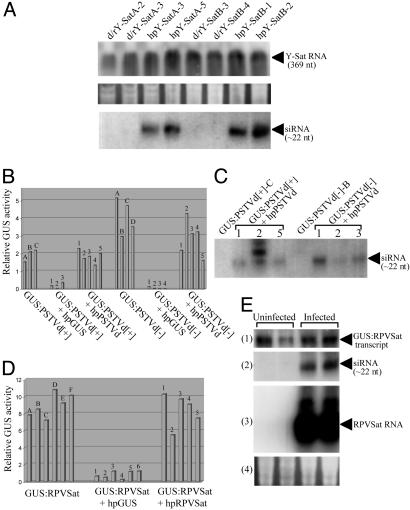
Evidence That RNA Silencing Plays a Role in the Evolution of Viroid and Satellite RNA. Replicating satellite RNAs are resistant to hpRNA-induced silencing. hpRNA transgenes are highly effective at inducing the silencing of a wide range of plant genes and in conferring resistance to RNA viruses (14, 27, 28). Therefore, we tested whether satellite RNAs are similarly vulnerable to hpRNA-induced degradation. Tobacco plants were transformed with a transgene encoding an hpRNA containing sequences from either Y-Sat (a linear satellite RNA) (24) or a circular satellite RNA of RPVSat (29). Plants transformed with the Y-Sat hairpin construct (hpY-Sat) or a control nonhairpin (direct repeat) Y-Sat construct (d/rY-Sat, Fig. 1), together with nine untransformed plants, were infected with CMV plus Y-Sat and monitored for symptom development. At ≈18 dpi, T1 plants of all 14 hpY-Sat lines tested had developed the characteristic bright yellow mosaic symptom (data not shown). There was no observable difference in symptom severity or Y-Sat RNA accumulation between the hpY-Sat plants and untransformed plants or between the hpY-Sat plants and the d/rY-Sat plants that did not produce siRNA (Fig. 4A). Infection of the hpY-Sat and d/rY-Sat plants with the helper CMV alone did not induce the yellowing symptom (data not shown), indicating that no infectious Y-Sat RNA arose from the Y-Sat transgenes, which would continuously provide Y-Sat inocula. Similarly, the hpRPVSat tobacco plants showed no resistance to RPVSat in tobacco, and Northern blot hybridization analysis revealed similar levels of RPVSat RNA in hpRPVSat plants and the untransformed plants at 28 dpi (data not shown). These results suggest that satellite RNAs evade or are protected from host-mediated RNA silencing.
Fig. 4.
(A) Replicating Y-Sat RNA is resistant to hpRNA-induced RNA silencing. (Top) Northern blot hybridization analysis of (+) strand Y-Sat RNA in Y-Sat-infected d/rY-Sat or hpY-Sat tobacco. Total RNA (20 μg) isolated from leaf tissue collected from a pool of approximately two to five Y-Sat-infected T1 plants of each line was hybridized with antisense Y-Sat RNA probe. (Bottom) Detection of Y-Sat siRNA in uninfected d/rY-Sat or hpY-Sat tobacco. High-molecular weight RNA was removed from 120 μg of total RNA (isolated from a pool of several uninfected T1 plants of each line) by precipitation with 5% polyethylenglycol 8000 and 0.5 M NaCl, and the small RNAs were pelleted from the supernatant with 0.3 M NaOAc and 3 volumes of ethanol and used for the analysis. Hybridization was performed by using full-length antisense Y-Sat RNA as probe. (Middle) RNA loading control for Top. (B-E) Nonreplicating subviral RNA sequences are also resistant to RNA silencing. (B and D) Relative GUS activity of 5 μg of protein from tobacco transformed with GUS:PSTVd[+], GUS:PSTVd[-], or GUS:RPVSat or their supertransformants with hpGUS, hpPSTVd or hpRPVSat. The uppercase letter above each bar represents plants regenerated from the same primary transformant (for GUS:PSTVd[+] and GUS:PSTVd[+]) or T1 plant (for GUS:RPVSat), and the number above each bar indicates an independent supertransformant. (C) Detection of siRNA from a subset of plants in B by Northern blot hybridization using full-length sense PSTVd RNA as probe. (E) Northern blot hybridization analysis of duplicate GUS:RPVSat regenerants uninfected or infected with potato leaf roll virus that is required to support RPVSat replication. The blots, from 1 to 4, show total RNA hybridized with antisense GUS RNA (11); siRNA hybridized with antisense RPVSat sequence (11); total RNA hybridized with antisense RPVSat RNA, where the strong signals indicate the presence of replicating RPVSat RNA; and RNA loading control for blots 1 and 3.
Nonreplicating satellite and viroid sequences are resistant to RNA silencing. The ability of Y-Sat or RPVSat to escape hpRNA-induced RNA silencing could be due to the high rates of replication and spread of the satellites outcompeting the capacity of the RNA silencing machinery. Therefore, we examined the effect of RNA silencing on nonreplicating, nonspreading viroid (PSTVd) or satellite (RPVSat) sequences present as part of a transgene-derived GUS-fusion transcript (GUS:PSTVd[+], GUS:PSTVd[-] or GUS:RPVSat; Fig. 1). Plants expressing the GUS:PSTVd[+] or GUS:PSTVd[-] transgene were supertransformed with hpPSTVd (Fig. 1). The doubly transformed plants showed no dramatic reduction in either GUS activity (Fig. 4B) or levels of the full-length fusion transcripts (data not shown) despite the presence of PSTVd siRNA (Fig. 4C). Similarly, GUS:RPVSat plants supertransformed with hpRPVSat showed little reduction in GUS activity (Fig. 4D). Furthermore, the GUS:RPVSat plants, infected with RPVSat, showed no significant reduction in the level of GUS:RPVSat fusion transcript, despite the production of abundant RPVSat siRNAs by the replicating RPVSat RNA (Fig. 4E). In contrast, GUS:PSTVd[+], GUS:PSTVd[-], and GUS:RPVSat plants supertransformed with the hpGUS construct, which targets silencing to the GUS sequence of the fusion mRNAs, showed high levels of GUS silencing (Fig. 4 B and D). These results suggest that resistance to RNA silencing, while not a feature of most messenger or viral RNAs, is a common feature of viroid and satellite genomic sequences and is intrinsic to their sequences rather than to their replication rates.
Discussion
Does RNA Silencing Mediate the Pathogenicity of Viroids and Viral Satellites? One of the intriguing features of viroids and viral satellites is their ability to induce symptoms in their host plants without encoding proteins. Various models involving the primary sequence or secondary structural features of their RNA genomes have been proposed to account for the pathogenicity of these small RNAs (7, 30). Here, we present two lines of evidence supporting an alternative pathogenicity model based on RNA silencing (refs. 9, 12, and 13 and Fig. 5). We show that (i) tobacco plants expressing the strong silencing suppressor Hc-Pro no longer support systemic induction of the bright yellow symptoms of CMV Y-Sat, despite the increased accumulation of satellite RNA in these plants, and (ii) tomato plants expressing the noninfectious hpRNA of PSTVd develop the corresponding viroid-like symptoms. These findings strongly argue against a direct involvement of unprocessed genomic RNAs, or secondary structures, of viroids and satellites in their pathogenicity.
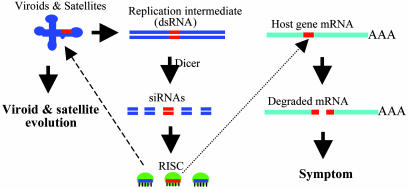
Fig. 5.
A model for the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellite. Replication of the subviral RNAs generates dsRNA intermediates, which are processed by Dicer into 21- to 25-nt siRNAs, and these siRNAs are then incorporated into siRNA-ribonuclease complexes (RISC). If significant sequence identity exists between a region in the subviral RNA genome and a region in host gene mRNA (shown in red), RISC will target the host gene for degradation leading to symptom development. RISC can also target the subviral genome for degradation, forcing the subviral RNA to evolve and to adopt and maintain RNA silencing-resistant secondary structure.
siRNA-directed degradation requires a minimum sequence identity of ≈19 nt between the siRNA and the cognate target RNA (8, 31). Previous studies have shown that the pathogenicity of viroids and viral satellites is generally determined by the nucleotide sequences within particular small (≈20-nt) regions of their RNA genomes (3, 7, 32), such as the previously defined virulence modulating region of PSTVd (3). Interestingly, a blast search (33) with the full-length sequence of PSTVd-RG1 revealed numerous sequences from several plant species that have 19- to 20-nt identities with the PSTVd sequence. Almost all of these 19- to 20-nt sequences correspond to the A+G-rich virulence modulating region (nucleotides 45-68) of PSTVd. The identified plant sequences have not been annotated, but at least two ESTs (GenBank accession nos. BJ473247 and BI969092) appear to encode putative transcription factors, and another (GenBank accession no. BI265876) encodes a putative chromodomain helicase DNA-binding protein. Although no potato or tomato sequences were found, possibly because of insufficient sequence entries in the databases for the two species, the blast search result raises the possibility that siRNAs derived from the virulence modulating region of PSTVd may target the silencing of host regulatory genes.
A seeming inconsistency with the RNA silencing-mediated pathogenicity model is that symptom induction by certain pathogenic CMV satellites appears to be helper virus-dependent (7). However, it is known that RNA silencing is dose-dependent, especially when the target sequence is relatively small (31, 34). Therefore, it is possible that the variation in symptoms induced by CMV satellites reflects their replication efficiency by different CMV strains. Indeed, strong symptom induction by the pathogenic CMV satellites appears to require helper virus strains that support high levels of double-stranded satellite RNA (and hence siRNA) accumulation (35-37). Our results show that replicating Y-Sat RNA produced an extremely high level of siRNAs (Fig. 2D). It is possible that such a high level of siRNA is essential for the induction of the yellow mosaic symptom in tobacco. This could explain why no chlorotic symptoms were observed in the transgenic tobacco containing the hpY-Sat transgene that yields a much lower abundance of siRNAs.
In addition to targeting RNA for degradation, siRNAs generated by viroids and satellites could also act like micro-RNAs (38) to form mismatched dsRNA complexes with cognate sequences of host gene mRNAs and thereby inhibit their translation and induce symptoms. Translational inhibition by micro-RNAs, a class of small ≈21-nt RNAs generated by Dicer cleavage of endogenous hpRNA precursors (38), occurs naturally in plants (39). Another alternative to symptom induction by means of the RNA degradation-based mechanism is the RNA-directed DNA methylation model proposed in ref. 40. Both viroids and satellites induce heavy de novo cytosine methylation of homologous nuclear DNA (11, 41), which could lead to transcriptional silencing of cognate endogenous genes. However, this model would not account for the prevention of Y-Sat symptom development by Hc-Pro, which does not block RNA-directed methylation (22, 25).
Does RNA Silencing Mediate the Evolution of Viroids and Viral Satellites? A major role attributed to RNA silencing in plants is defense against viral infection (42). The siRNAs produced from the pathogen's dsRNA replication intermediates target degradation of the pathogen's genome. To evade or block this host defense mechanism, viruses encode silencing suppressor proteins (43). Viroids and small viral satellites, on the other hand, do not encode any functional proteins and yet are capable of accumulating to high levels in plants. Our results suggest that viroid and satellite RNAs have developed a nuclease resistance strategy to protect themselves against degradation by RNA silencing. Satellites are encapsidated by helper virus coat proteins, which may provide some protection against RNA silencing-mediated degradation. However, the level of such protection is clearly limited, because the helper viruses themselves are, in general, highly susceptible to hpRNA-induced silencing (14, 27, 28). Most viroids, including PSTVd, replicate in the nucleus, and this subcellular localization may allow viroid RNA to avoid contact with RISC, which is believed to act mainly in the cytoplasm (44). However, a recent report shows that nucleolar RNAs are also susceptible to RNA silencing (45). Furthermore, viroids must traverse the cytoplasm during cell-to-cell movement in plants, thereby potentially being exposed to cytoplasmic RISC degradation.
A likely explanation for the resistance of viroids and viral satellites to RNA silencing is that their extensive intramolecular base-pairing renders them less accessible to the RISC complex for degradation. Additionally, the presence of mismatches in the duplex regions restricts perfectly paired regions of the predicted secondary structures of subviral RNAs (1) to no longer than 14 bp (data not shown). This would be sufficient to protect them against cleavage by Dicer, which requires a minimum of ≈19 bp of dsRNA (46). Several recent observations support a direct role of secondary RNA structures in conferring resistance to RNA silencing: (i) regions of a plant mRNA that have the potential to form duplex structure have been shown to accumulate in cells where the mRNA is silenced (47), (ii) a short defective interfering viral RNA, with the potential to form a stable secondary structure, is significantly more resistant to RNA silencing than is its helper virus (48), and (iii) PSTVd or viroid-like RNAs are highly resistant to Dicer cleavage in an in vitro system (49).
The resistance of viroids and satellites to RNA silencing-mediated degradation implies that RNA silencing may have directed the evolution of plant subviral RNAs (Fig. 5). This view is supported by the observation that viroid and satellite sequence variants retain their secondary structures. For instance, a recently discovered natural variant of citrus exocortis viroid retains the rod-like secondary structure of the wild-type strain, despite the insertion of an additional 96-nt sequence (50). The RNA silencing-mediated model is also consistent with the lack of significant sequence identity between viral satellites and their corresponding helper viruses. A viral satellite with significant sequence identity (e.g., >19 nt) to its helper virus would induce degradation of the helper virus genome, thereby compromising its own evolutionary survival.
In conclusion, our results suggest that RNA silencing in plants plays a central role in both the pathogenicity of viroids and viral satellites and in the evolution of their secondary structures (Fig. 5). Like viruses, the evolutionary pathway that viroid and satellite RNAs appear to have adopted allows them not only to use the host functions for their replication but also to evade host defenses and to elicit pathogenic reactions. Whereas viruses achieve these functions by means of an array of encoded proteins, viroids and viral satellites appear to ensure their evolutionary survival using an exclusively sequence and structure-based strategy.
Acknowledgments
We thank Paul Chu for providing the CMV strain; Peter Palukaitis for sharing information on CMV satellites; Geoff Ellacott for maintaining potato leaf roll virus culture; Judith Gaudron and Jasmina Dedic for PCR primers; Carl Davies for photography; Fei Zhang for advice on blast searches; Tony Arioli, Rogerio Margis, and Jean Finnegan for helpful discussions; Jim Peacock, Bill Taylor, and James Hutchinson for support; and three referees for their critical reviews of the manuscript.
Abbreviations: CMV, cucumber mosaic virus; hpRNA, hairpin RNA; PSTVd, potato spindle tuber viroid; Y-Sat, Y satellite; dsRNA, double-stranded RNA; siRNA, small interfering RNA; RISC, RNA-induced silencing complex; RPVSat, cereal yellow dwarf virus RPV satellite; GUS, β-glucuronidase; dpi, days postinoculation.
References
- 1.Pelchat, M., Rocheleau, L., Perreault, J. & Perreault, J. P. (2003) Nucleic Acids Res. 31, 444-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diener, T. O. (1991) FASEB J. 5, 2808-2813. [DOI] [PubMed] [Google Scholar]
- 3.Steger, G. & Riesner, D. (2003) in Viroids, eds. Hadidi, A., Flores, R., Randles, J. W. & Semancik, J. S. (CSIRO Publishing, Collingwood, Australia), pp. 15-29.
- 4.Symons, R. H. (1997) Nucleic Acids Res. 25, 2683-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickson, E., Robertson, H. D., Niblett, C. L., Horst, R. K. & Zaitlin, M. (1979) Nature 277, 60-62. [Google Scholar]
- 6.Qi, Y. & Ding, B. (2003) Plant Cell 15, 1360-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Arenal, F. & Palukaitis, P. (1999) Curr. Top. Microbiol. Immunol. 239, 37-63. [DOI] [PubMed] [Google Scholar]
- 8.Zamore, P. D. (2001) Nat. Struct. Biol. 8, 746-750. [DOI] [PubMed] [Google Scholar]
- 9.Papaefthimiou, I., Hamilton, A. J., Denti, M. A., Baulcombe, D. C., Tsagris, M. & Tabler, M. (2001) Nucleic Acids Res. 29, 2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itaya, A., Folimonov, A., Matsuda, Y., Nelson, R. S. & Ding, B. (2001) Mol. Plant-Microbe Interact. 14, 1332-1334. [DOI] [PubMed] [Google Scholar]
- 11.Wang, M.-B., Wesley, S. V., Finnegan, E. J., Smith, N. A. & Waterhouse, P. M. (2001) RNA 7, 16-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conejero, V. (2003) in Viroids, eds. Hadidi, A., Flores, R., Randles, J. W. & Semancik, J. S. (CSIRO Publishing, Collingwood, Australia), pp. 67-70.
- 13.Wang, M.-B., Finnegan, E. J.& Waterhouse, P. M. (2004) in Viruses and Virus Diseases of Poaceae, eds. Hervé Lapierre, H. & Signoret, P. (Institut National de la Recherche Agronomique, Versailles, France).
- 14.Wesley, S. V., Helliwell, C. A., Smith, N. A., Wang, M.-B., Rouse, D. T., Liu, Q., Gooding, P. S., Singh, S. P., Abbott, D., Stoutjesdijk, P. A., et al. (2001) Plant J. 27, 581-590. [DOI] [PubMed] [Google Scholar]
- 15.Gruner, R., Fels, A., Qu, F., Zimmat, R., Steger, G. & Riesner, D. (1995) Virology 209, 60-69. [DOI] [PubMed] [Google Scholar]
- 16.Wang, M.-B., Li, Z.-Y., Matthews, P. R., Upadhyaya, N. M. & Waterhouse, P. M. (1998) Acta Hortic. 461, 401-407. [Google Scholar]
- 17.Gleave, A. P. (1992) Plant Mol. Biol. 20, 1203-1207. [DOI] [PubMed] [Google Scholar]
- 18.Wang, M.-B. & Waterhouse, P. M. (2000) Plant Mol. Biol. 43, 67-82. [DOI] [PubMed] [Google Scholar]
- 19.Frary, A. & Earle, E. D. (1996) Plant Cell Rep. 16, 235-240. [DOI] [PubMed] [Google Scholar]
- 20.Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. (1987) EMBO J. 6, 3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuwata, S., Masuta, C. & Takanami, Y. (1988) Ann. Phytopath. Soc. Jpn. 54, 510-515. [Google Scholar]
- 22.Mallory, A. K., Ely, L., Smith, T. H., Marathe, R., Anandalakshmi, R., Fagard, M., Vaucheret, H., Pruss, G., Bowman, L. & Vance, V. B. (2001) Plant Cell 13, 571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anandalakshmi, R., Pruss, G. J., Marathe, R., Mallory, A. C., Smith, T. H. & Vance, V. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuta, C. & Takanami, Y. (1989) Plant Cell 1, 1165-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mette, M. F., Matzke, A. J. & Matzke, M. A. (2001) Curr. Biol. 11, 1119-1123. [DOI] [PubMed] [Google Scholar]
- 26.Branch, A. D., Benenfeld, B. J., Franck, E. R., Shaw, J. F., Varban, M. L., Willis, K. K., Rosen, D. L. & Robertson, H. D. (1988) Virology 163, 538-546. [DOI] [PubMed] [Google Scholar]
- 27.Smith, N. A., Singh, S. P., Wang, M.-B., Stoutjesdijk, P. A., Green, A. G. & Waterhouse, P. M. (2000) Nature 407, 319-320. [DOI] [PubMed] [Google Scholar]
- 28.Wang, M.-B., Abbott, D.-C. & Waterhouse, P. M. (2000) Mol. Plant Pathol. 1, 347-356. [DOI] [PubMed] [Google Scholar]
- 29.Miller, W. A., Hercus, T, Waterhouse, P. M. & Gerlach, W. L. (1991) Virology 183, 711-720. [DOI] [PubMed] [Google Scholar]
- 30.Semancik, J. S. (2003) in Viroids, eds. Hadidi, A., Flores, R., Randles, J. W. & Semancik, J. S. (CSIRO Publishing, Collingwood, Australia), pp. 61-66.
- 31.Vanitharani, R., Chellappan, P. & Fauquet, C. M. (2003) Proc. Natl. Acad. Sci. USA 100, 9632-9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owens, R. A., Steger, G., Hu, Y., Fels, A., Hammond, R. W. & Riesner, D. (1996) Virology 222, 144-158. [DOI] [PubMed] [Google Scholar]
- 33.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, D., Lu, H. & Erickson, J. W. (2000) Curr. Biol. 10, 1191-1200. [DOI] [PubMed] [Google Scholar]
- 35.Sleat, D. E. & Palukaitis, P. (1990) Virology 176, 292-295. [DOI] [PubMed] [Google Scholar]
- 36.Taliansky, M. E., Ryabov, E. V., Robinson, D. J. & Palukaitis, P. (1998) Mol. Plant-Microbe Interact. 11, 1214-1222. [Google Scholar]
- 37.Xu, P. & Roossinck, M. J. (2000) Plant Cell 12, 1079-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ambros, V., Lee, R. C., Lavanway, A., William, P. T. & Jewell, D. (2003) Curr. Biol. 13, 807-818. [DOI] [PubMed] [Google Scholar]
- 39.Aukerman, M. J. & Sakai, H. (2003) Plant Cell 15, 2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sänger, H. L., Schiebel, L., Riedel, T., Pélissier, T. & Wassenegger, M. (1996) in Biology of Plant-Microbe Interactions, eds. Stacey, G. Mullin, B. & Gresshoff, P. M. (Int. Soc. for Molecular Plant-Microbe Interactions, St. Paul), pp. 533-540.
- 41.Wassenegger, M., Heimes, S., Riedel, L. & Sänger, L. C. (1994) Cell 76, 567-576. [DOI] [PubMed] [Google Scholar]
- 42.Vance, V. & Vaucheret, H. (2001) Science 292, 2277-2280. [DOI] [PubMed] [Google Scholar]
- 43.Li, W. X. & Ding, S. W. (2001) Curr. Opin. Biotechnol. 12, 150-154. [DOI] [PubMed] [Google Scholar]
- 44.Matzke, M. A., Matzke, A. J. M., Pruss, G. J. & Vance, V. B. (2001) Curr. Opin. Genet. Dev. 11, 221-227. [DOI] [PubMed] [Google Scholar]
- 45.Liang, X.-H., Liu, Q. & Michaeli, S. (2003) Proc. Natl. Acad. Sci. USA 100, 7521-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, J. Y., DeRuiter, S. L. & Turner, D. L. (2002) Proc. Natl. Acad. Sci. USA 99, 6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metzlaff, M., O'Dell, M., Cluster, P. D. & Flavell, R. B. (1997) Cell 88, 845-854. [DOI] [PubMed] [Google Scholar]
- 48.Szittya, G., Molnár, A., Silhavy, D., Hornyik, C. & Burgyán, J. (2002) Plant Cell 14, 359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang, J., Provost, P. & Taylor, J. M. (2003) J. Virol. 77, 11919-11917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fadda, Z., Daròs, J. A., Flores, R. & Duran-Vila, N. (2003) Virus Res. 97, 145-149. [DOI] [PubMed] [Google Scholar]