Abstract
4-Methylideneimidazole-5-one (MIO)-containing aminomutases catalyze the conversion of l-α-amino acids to β-amino acids with either an (R) or an (S) configuration. l-Phenylalanine and l-tyrosine are the only two natural substrates identified to date. The enediyne chromophore of the chromoprotein antitumor antibiotic kedarcidin (KED) harbors an (R)-2-aza-3-chloro-β-tyrosine moiety reminiscent of the (S)-3-chloro-5-hydroxy-β-tyrosine moiety of the C-1027 enediyne chromophore, the biosynthesis of which uncovered the first known MIO-containing aminomutase, SgcC4. Comparative analysis of the KED and C-1027 biosynthetic gene clusters inspired the proposal for (R)-2-aza-3-chloro-β-tyrosine biosynthesis starting from 2-aza-l-tyrosine, featuring KedY4 as a putative MIO-containing aminomutase. Here we report the biochemical characterization of KedY4, confirming its proposed role in KED biosynthesis. KedY4 is an MIO-containing aminomutase that stereospecifically catalyzes the conversion of 2-aza-l-tyrosine to (R)-2-aza-β-tyrosine, exhibiting no detectable activity toward 2-aza-l-phenylalanine or l-tyrosine as an alternative substrate. In contrast, SgcC4, which stereospecifically catalyzes the conversion of l-tyrosine to (S)-β-tyrosine in C-1027 biosynthesis, exhibits minimal activity with 2-aza-l-tyrosine as an alternative substrate but generating (S)-2-aza-β-tyrosine, a product with the opposite stereochemistry of KedY4. This report of KedY4 broadens the scope of known substrates for the MIO-containing aminomutase family, and comparative studies of KedY4 and SgcC4 provide an outstanding opportunity to examine how MIO-containing aminomutases control substrate specificity and product enantioselectivity.
Keywords: amonia lyase, enzyme, natural product, Streptoalloteichus sp., Streptomyces globisporus
Aminomutases catalyze the interconversion of α-amino acids to β-amino acids, which are building blocks for the biosynthesis of various natural products of biological and medicinal significance. Aminomutases can be divided into two families based on their catalytic mechanisms. One of these families acts on lysine or leucine and uses either S-adenosyl methionine (SAM) and a [4Fe-4S] iron-sulfur cluster or adenosylcobalamin to generate the 5′-deoxyadenosyl radical that abstracts an unreactive hydrogen from the β-carbon of an α-amino acid to initiate the substrate radical isomerization with concomitant amino group migration facilitated by pyridoxal phosphate (1, 2). The other family of aminomutases, comprising tyrosine aminomutases (TAMs) and phenylalanine aminomutases (PAMs), features the 4-methylideneimidazol-5-one (MIO) prosthetic group that is formed posttranslationally from a highly conserved Ala-Ser-Gly amino acid motif. The amine of the glycine attacks the carbonyl of the alanine to generate a five-membered intermediate, which subsequently undergoes a two-step dehydration to afford the MIO prosthetic group as a potent electrophile (SI Appendix, Fig. S1A).
The mechanism of MIO-containing aminomutases begins with attack of the amino group onto the electrophilic MIO prosthetic group, followed by sequential or concerted deprotonation of the β-carbon and elimination of the MIO-bound amine to generate an α,β-unsaturated carboxylic acid intermediate. Subsequent addition of the MIO-bound amino group to the β-carbon of the α,β-unsaturated carboxylic acid intermediate affords a β-amino acid as the final product (SI Appendix, Fig. S1B) (3–5).
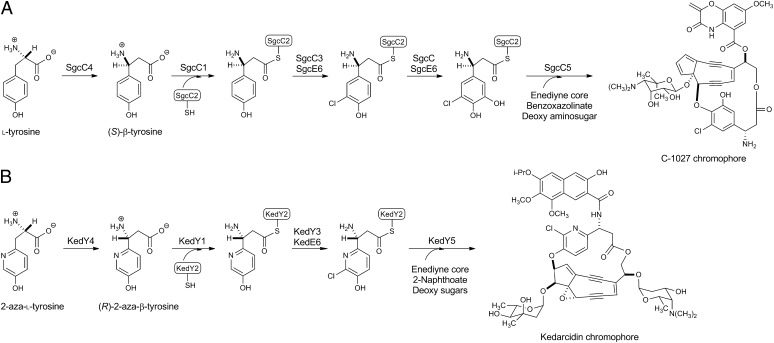
The first identified member of the MIO-containing aminomutase family, SgcC4, was discovered a decade ago during the course of our study of the biosynthesis of the enediyne antitumor antibiotic C-1027 in Streptomyces globisporus (Fig. 1A) (6, 7). Subsequent biochemical characterizations of SgcC4 have established that it (i) specifically catalyzes the conversion of l-tyrosine to (S)-β-tyrosine, shunting the primary metabolite into the C-1027 biosynthetic pathway, and (ii) releases 4-hydroxycinnamic acid as an intermediate, spuriously revealing weak activity as an ammonia lyase (7, 8). Crystal structures have been solved for SgcC4 either alone or with mechanism-based inhibitors, as well as for the active site mutant SgcC4 (Tyr70Phe) with bound l-tyrosine (9–12). These structures (i) unambiguously demonstrate the presence of the MIO prosthetic group, (ii) define the active site residues responsible for catalysis and substrate binding, and (iii) in comparison with structures of MIO-containing ammonia lyases, provide a method to bioinformatically differentiate MIO-containing aminomutases from ammonia lyases (3–5).
Fig. 1.
Comparison of biosynthetic pathways for the β-amino acid moieties of the C-1027 and KED chromophores featuring the MIO-containing aminomutases SgcC4 and KedY4. Shown are proposed biosynthetic pathways for the (S)-3-chloro-5-hydroxy-β-tyrosine moiety of C-1027 from ʟ-tyrosine (A) and the (R)-2-aza-3-chloro-β-tyrosine moiety of KED from 2-aza-ʟ-tyrosine (B).
The MIO-containing aminomutase family has grown steadily since the identification of SgcC4 (5). Current members, whose activity has been unambiguously confirmed experimentally, include (i) two PAMs, PaPAM from Pantoea agglomerans that yields (S)-β-phenylalanine (13–16) and TcPAM from Taxus canadensis that yields (R)-β-phenylalanine (17, 18), and (ii) five TAMs of SgcC4 from S. globisporus (6, 7), MdpC4 from Actinomadura madurae (19), MfTAM from Myxococcus fulvus (20), and MxTAM from Myxococcus sp. Mx-BO (20), all of which afford (S)-β-tyrosine, and CmdF from Chondromyces crocatus that yields (R)-β-tyrosine (21). Together these studies unveiled MIO-containing aminomutases that catalyze the conversion of l-phenylalanine and l-tyrosine to β-phenylalanine and β-tyrosine with either an (R) or an (S) configuration; however, l-phenylalanine and l-tyrosine are the only two natural substrates identified to date (3–5). Experiments designed to reveal the enzymatic mechanism of product stereochemistry failed to yield informative results (20). Attempts to convert MIO-containing aminomutases into MIO-containing ammonia lyases have not been successful (12, 20).
In our search for novel MIO-containing aminomutases to expand the catalytic landscape of this emerging family of enzymes and to exploit their utility for natural product structural diversity by combinatorial biosynthesis strategies, we were intrigued by the enediyne chromophore of the chromoprotein antitumor antibiotic kedarcidin (KED) (22–24). The KED chromophore harbors a (R)-2-aza-3-chloro-β-tyrosine moiety, which can be envisioned to derive from 2-aza-l-phenylalanine or 2-aza-l-tyrosine via an MIO-containing aminomutase in a mechanism analogous to that of the PAMs and TAMs. We recently completed the cloning and annotation of the KED biosynthetic gene cluster from Streptoalloteichus sp. ATCC 53650 (25) and proposed a convergent biosynthetic pathway for the KED chromophore based on a comparison of bioinformatics with the other characterized enediyne gene clusters, including C-1027 and maduropeptin (MDP) (6, 7, 19). Significantly, a subset of six genes that were absolutely conserved among the KED, C-1027, and MDP clusters—kedY1, kedY2, kedY3, kedY4, kedY5, and kedE6— inspired us to propose a pathway for (R)-2-aza-3-chloro-β-tyrosine biosynthesis starting from 2-aza-l-tyrosine (25). Thus, in a biosynthetic analogy to the (S)-3-chloro-5-hydroxy-β-tyrosine moiety of C-1027 (26-31), KedY4 acts as an aminomutase, converting 2-aza-l-tyrosine into (R)-2-aza-β-tyrosine. The latter is loaded onto the peptidyl carrier protein KedY2 by the adenylation enzyme KedY1 to generate a (R)-2-aza-β-tyrosyl-S-KedY2 intermediate, chlorinated by KedY3, an FAD-dependent halogenase requiring the KedE6 flavin reductase, and finally coupled to the enediyne core via an ester linkage catalyzed by the condensation enzyme KedY5 (Fig. 1) (25).
Here we report the biochemical characterization of KedY4, confirming its proposed role in KED biosynthesis. KedY4 is an MIO-containing aminomutase that stereospecifically catalyzes the conversion of 2-aza-l-tyrosine to (R)-2-aza-β-tyrosine, exhibiting no detectable activity toward 2-aza-l-phenylalanine or l-tyrosine as an alternative substrate. In contrast, SgcC4, the archetype MIO-containing aminomutase that stereospecifically catalyzes the conversion of l-tyrosine to (S)-β-tyrosine, displays minimal activity with 2-aza-l-tyrosine as an alternative substrate but generating (S)-2-aza-β-tyrosine, a product with the opposite stereochemistry of KedY4. Thus, KedY4 is a unique MIO-containing aminomutase that broadens the scope of known substrates for the MIO-containing aminomutase family.
Results
Bioinformatics Analysis of KedY4.
KedY4 shows high sequence homology, including the highly conserved Ala-Ser-Gly motif, to known MIO-containing aminomutases (SI Appendix, Fig. S1C). However, the overall sequence homology between KedY4 and TAMs is higher than that between KedY4 and PAMs; KedY4 shares 66% identity and 82% similarity with SgcC4, but only 31% identity and 49% similarity with TcPAM and 35% identity and 54% similarity with PaPAM. Close examination of the sequences further revealed the presence of the conserved polar residues in KedY4 (His93, Tyr415, Asn438, and Asn452, based on SgcC4 numbering) known to be responsible for binding the phenol moiety of l-tyrosine (SI Appendix, Fig. S1C) (5). These observations support the functional assignment of KedY4 as an MIO-containing aminomutase that uses 2-aza-l-tyrosine as the preferred substrate to generate (R)-2-aza-β-tyrosine (Fig. 1B).
In Vitro Characterization of KedY4 as an MIO-Containing Aminomutase.
KedY4 was overproduced in Escherichia coli BL21(DE3) as an N-terminal His6-tagged fusion protein, whose His6-tag can be removed by treatment with tobacco etch virus (TEV) protease (SI Appendix). The His6-tagged KedY4 was purified to homogeneity and subjected to SDS/PAGE analysis, which showed as a single band with an apparent molecular weight of 59 kDa (calculated 59,661 Da) (SI Appendix, Fig. S2A). UV-Vis spectroscopic analysis of the purified KedY4 revealed a characteristic UV absorption around 320 nm, indicative of the MIO prosthetic group (SI Appendix, Fig. S2B). Removal of the His6-tag on TEV treatment yielded a KedY4 preparation with reduced enzyme activity. Thus, we used the His6-tagged KedY4 in all experiments.
2-Aza-l-tyrosine was isolated from Streptomyces chibaensis SF-1346, and its structure and absolute stereochemistry were confirmed as reported previously (32) (SI Appendix, Figs. S3 and S4). This material was used as a substrate for in vitro characterization of KedY4. Both (R)- and (S)-2-aza-β-tyrosine were synthesized from 5-hydroxy-2-methylpyridine following a previously reported procedure (33) with slight modifications (SI Appendix, Figs. S5A–S23). They were used as authentic standards or as starting materials for preparing derivatives thereof, to establish the identity and stereochemistry of the products resulting from KedY4 catalysis, and as substrates to assay KedY4 and SgcC4 in the reverse direction.
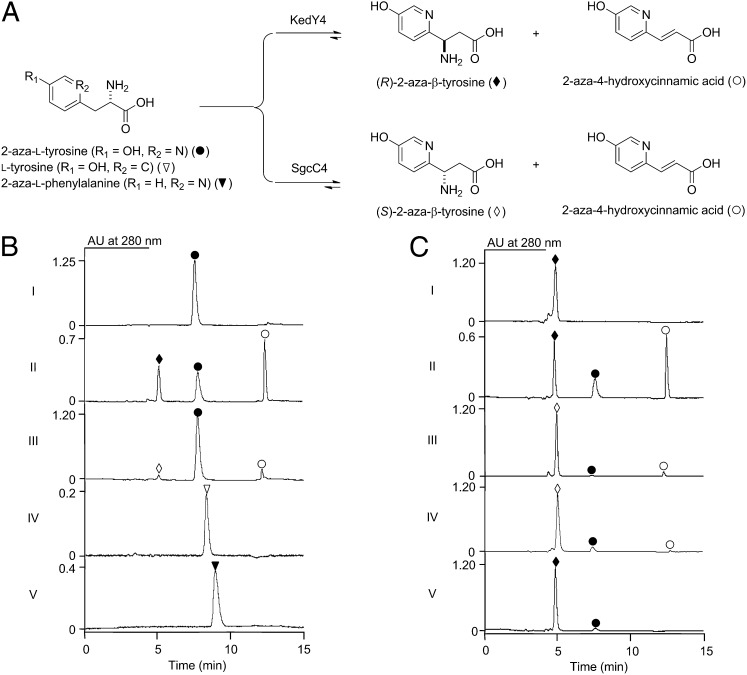
Enzyme assays of KedY4 with 2-aza-l-tyrosine as a substrate were carried out under standard condition [20 μM enzyme and 2 mM substrate in 50 mM KCl and 100 mM Bis-Tris propane (BTP) (pH 8.5), with incubation at 25 °C for 4 h], with SgcC4 as a control (Fig. 2A and SI Appendix). The assay reactions were followed by HPLC analysis, and 2-aza-l-tyrosine was eluted as single peak with a retention time of 7.8 min. Incubation of KedY4 with 2-aza-l-tyrosine resulted in the formation of two new products, one at a retention time of 5.0 min displaying a UV profile similar to that of 2-aza-l-tyrosine, suggesting 2-aza-β-tyrosine, and the other at a retention time of 12.5 min exhibiting a strong UV absorption from 250 to 330 nm, indicative of the deaminated intermediate 2-aza-4-hydroxycinnamic acid (Fig. 2B, II). In a control experiment with heat-denatured KedY4, 2-aza-l-tyrosine remained unchanged, and no new product was detected (Fig. 2B, I). The two new products were isolated and subjected to high-resolution electrospray ionization mass spectroscopy analysis. The product with a retention time of 5.0 min yielded an [M + H]+ ion at m/z 183.0762, consistent with 2-aza-β-tyrosine (C8H10N2O3 and calculated [M + H]+ ion at m/z 183.0764); this assignment was further supported by coelution with synthetic (R)- and (S)-2-aza-β-tyrosine standards. The product with a retention time of 12.5 min yielded an [M + H]+ ion at m/z 166.0496, consistent with 2-aza-4-hydroxycinnamic acid (C8H8NO3 and calculated [M + H]+ ion at m/z 166.0498).
Fig. 2.
Characterization of KedY4 as an MIO-containing aminomutase. (A) KedY4 catalyzed formation of (R)-2-aza-β-tyrosine and 2-aza-4-hydroxycinnamic acid from 2-aza-ʟ-tyrosine in comparison with SgcC4. (B) HPLC analysis of KedY4- and SgcC4-catalzyed forward reactions: (I) boiled KedY4 with 2-aza-ʟ-tyrosine, (II) KedY4 with 2-aza-ʟ-tyrosine, (III) SgcC4 with 2-aza-ʟ-tyrosine, (IV) KedY4 with ʟ-tyrosine, and (V) KedY4 with 2-aza-ʟ-phenylalanine. (C) HPLC analysis of KedY4- and SgcC4-catalzyed reverse reactions: (I) boiled KedY4 with (R)-2-aza-β-tyrosine, (II) KedY4 with (R)-2-aza-β-tyrosine, (III) KedY4 with (S)-2-aza-β-tyrosine, (IV) SgcC4 with (S)-2-aza-β-tyrosine, and (V) SgcC4 with (R)-2-aza-β-tyrosine. ●, 2-aza-ʟ-tyrosine; ▽, ʟ-tyrosine; ▼, 2-aza-ʟ-phenylalanine; ◆, (R)-2-aza-β-tyrosine; ◇, (S)-2-aza-β-tyrosine; ○, 2-aza-4-hydroxycinnamic acid. Note that the peaks for 2-aza-4-hydroxycinnamic acid reflect a minor product, because its extinction coefficient is much larger than those of 2-aza-ʟ-tyrosine and 2-aza-β-tyrosine at 280 nm.
Next, the enzymatic reactions, consisting of 0.625 μM KedY4 and 1 μM 2-aza-l-tyrosine in 50 mM KCl and 100 mM BTP (pH 6.5–10.0) or 100 mM N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) (pH 10.5–11.0), were incubated at 25 °C for 1 h to investigate the pH dependence of KedY4, and the initial rates of the Ked4-catalyzed formation of (R)-2-aza-β-tyrosine and 2-aza-4-hydroxycinnamic acid from 2-aza-l-tyrosine were determined by HPLC analysis (SI Appendix). KedY4 exhibited an optimal initial rate for the aminomutase activity at pH 8.5 and for the ammonia lyase activity at pH 9.5 (SI Appendix, Fig. S24); thus, 100 mM BTP (pH 8.5) was used in all experiments. Based of this pH profile, the pKa values controlling the KedY4 aminomutase activity were calculated as 7.44 ± 0.02 and 9.47 ± 0.02.
A large-scale enzymatic reaction under the optimized enzymatic reaction conditions was then carried out to produce the products for structural elucidation, and both products were purified by semipreparative HPLC (SI Appendix, Fig. S25A). Unexpectedly, 1H, 13C, and 2D NMR spectroscopic analyses revealed that both compounds were purified as complexes with the buffer component BTP. Nonetheless, the 1H and 13C NMR signals between both products and BTP were clearly resolved, allowing the structural determination of 2-aza-β-tyrosine and 2-aza-4-hydroxycinnamic acid, respectively (SI Appendix, Figs. S26–S32). Specifically, the β-amino functionality in 2-aza-β-tyrosine was unambiguously determined by 1H-1H COSY and HMBC NMR spectroscopic analysis (SI Appendix, Fig. S25B). The transdouble bond in 2-aza-4-hydroxycinnamic acid was supported by the characteristic chemical shifts of C-2′/H-2′ [δC124.4/δH 6.65, d (15.8 Hz)] and C-3′/H-3′ [δC141.2/δH 7.46, d (15.8 Hz)]. The 2-aza-β-tyrosine-BTP complex was finally converted to 2-aza-β-tyrosine methyl ester, which was isolated free from BTP (SI Appendix, Fig. S25A). The latter structure was confirmed by 1H and 13C NMR spectroscopic analyses (SI Appendix, Figs. S33 and S34) and was identical to synthetic (R)- and (S)-2-aza-β-tyrosine methyl ester standards (SI Appendix, Fig. S5A), excluding any ambiguity associated with the 2-aza-β-tyrosine structure deduced from its BTP complex.
Stereochemistry of KedY4-Catalyzed Conversion of 2-Aza-l-Tyrosine to (R)-2-Aza-β-Tyrosine.
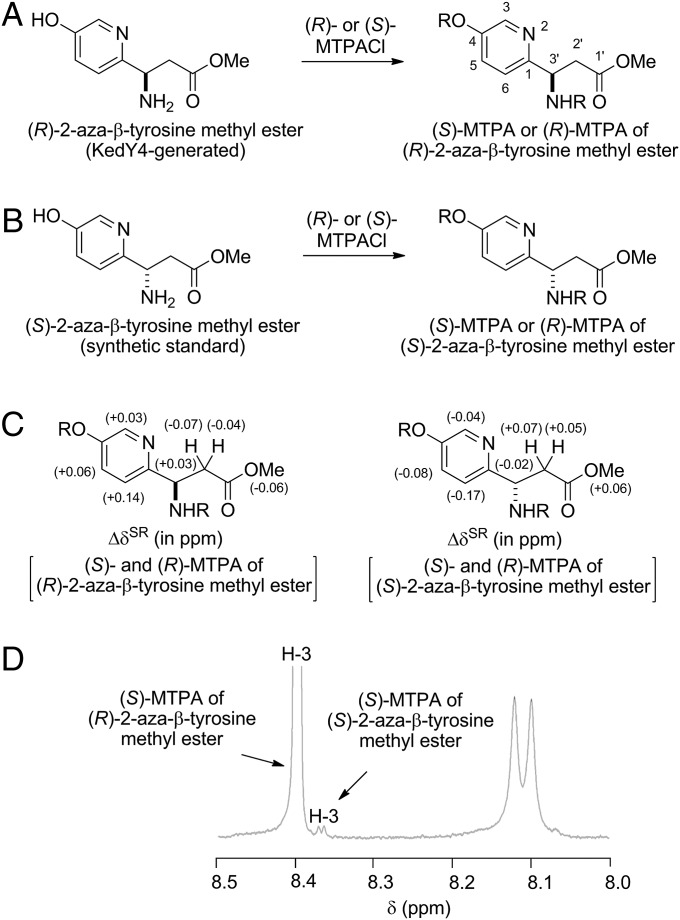
Attempts to determine the absolute configuration of the 2-aza-β-tyrosine product directly by chiral chromatography using synthetic (R)- and (S)-2-aza-β-tyrosine or their methyl esters as standards were unsuccessful, owing to poor resolution of these compounds under all conditions tested. The absolute configuration of the (R)-2-aza-β-tyrosine product was subsequently determined by the modified Mosher method (34, 35) (SI Appendix). The methyl ester of the (R)-2-aza-β-tyrosine product was converted to its (S)- and (R)-Mosher amides [α-methoxy-α-trifluoromethylphenylamide (MTPA)] (Fig. 3A and SI Appendix, Figs. S25A and S35–S42). Comparison of the 1H NMR chemical shift differences [ΔδSR (i.e., δS − δR), in ppm] between the resultant (S)- and (R)-MTPAs led to the assignment of the (R) configuration at C-3′ for 2-aza-β-tyrosine methyl ester (Fig. 3C). As controls, synthetic (R)- and (S)-2-aza-β-tyrosine methyl esters were similarly converted into the corresponding (S)- and (R)-MTPAs (Fig. 3B and SI Appendix, Fig. S5B). The 1H NMR chemical shift differences for (S)- and (R)-MTPAs derived from the enzymatic (R)-2-aza-β-tyrosine were identical to those derived from the synthetic (R)-2-aza-β-tyrosine. In contrast, the 1H NMR chemical shift differences for (S)- and (R)-MTPAs derived from synthetic (S)-2-aza-β-tyrosine were opposite to those derived from enzymatic or synthetic (R)-2-aza-β-tyrosine (Fig. 3C and SI Appendix, Figs. S43–S50). Taken together, these results establish KedY4 as an MIO-containing aminomutase that catalyzes the conversion of 2-aza-l-tyrosine to (R)-2-aza-β-tyrosine (Fig. 2A).
Fig. 3.
Determination of KedY4-catalyzed formation of (R)-2-aza-β-tyrosine from 2-aza-ʟ-tyrosine proceeding with high enantiospecificity. (A) Synthesis of (S)- and (R)-MTPAs from KedY4-generated (R)-2-aza-β-tyrosine methyl ester. (B) Synthesis of (S)- and (R)-MTPAs from synthetic (S)-2-aza-β-tyrosine methyl ester. (C) 1H NMR chemical shift differences ΔδSR (in ppm) for (S)- and (R)-MTPAs of (R)- and (S)-2-aza-β-tyrosine methyl esters establishing the absolute configuration at C-3′. (D) Expanded 1H NMR spectrum of (S)-MTPA of the KedY4-generated (R)-2-aza-β-tyrosine methyl ester showing H-3 at δH 8.40, with the doublet at δH 8.37, indicating the presence of (S)-MTPA of (S)-2-aza-β-tyrosine methyl ester as a minor product.
Attempts to resolve (S)- and (R)-MTPAs of (R)- or (S)-2-aza-β-tyrosine methyl ester by HPLC were unsuccessful, precluding direct determination by HPLC of the percent enantiomeric excess (% ee) value of the KedY4-generated (R)-2-aza-β-tyrosine product from 2-aza-l-tyrosine. Comparison of the 1H NMR spectra between (S)- and (R)-MTPAs of (R)- or (S)-2-aza-β-tyrosine methyl ester revealed clear differences, however, suggesting the 1H NMR could be used to determine the % ee value. The (S)-MTPAs of (R)- and (S)-2-aza-β-tyrosine methyl ester exhibited distinct doublets for the H-3 protons as doublets at δH 8.40 and 8.37, whereas the H-3 protons in the (R)-MTPAs of (R)- and (S)-2-aza-β-tyrosine methyl ester showed clear doublets at δH 8.44 and 8.40, respectively (Fig. 3 A and D). Thus, close examination of the H-3 proton in the 1H NMR spectrum of (S)-MTPA of (R)-2-aza-β-tyrosine methyl ester, prepared from KedY4-generated (R)-2-aza-β-tyrosine, indeed revealed the expected doublet at δH 8.40, with a minor doublet at δH 8.37, indicative of (S)-MTPA of (S)-2-aza-β-tyrosine methyl ester, the ratio of which indicated a >97.2% ee for the (R)-2-aza-β-tyrosine product (Fig. 3D). Taken together, these results establish KedY4 as an MIO-containing aminomutase that catalyzes the conversion of 2-aza-l-tyrosine to (R)-2-aza-β-tyrosine with high enantiospecificity (Fig. 2A).
Substrate Specificity of KedY4 and Comparison with SgcC4.
Both 2-aza-l-phenylalanine and l-tyrosine were examined as alternative substrates for KedY4 under the standard assay conditions, and SgcC4 was similarly investigated for comparison (Fig. 2A). No product was detected for KedY4 with either substrate even when the incubation time was extended from 4 h to 24 h (Fig. 2B, IV and V). In contrast, although no product was detected for SgcC4 with 2-aza-l-phenylalanine as a substrate, two new products, with the same retention time as 2-aza-β-tyrosine and 2-aza-4-hydroxycinnamic acid, were detected for SgcC4 with 2-aza-l-tyrosine, albeit at a fraction of the initial rate observed for KedY4 (Fig. 2B, III). The identities of these two products were confirmed by comparison with authentic standards.
KedY4 and SgcC4 generate β-amino acid products with opposite stereochemistry from their respective α-amino acid substrates, that is, (R)-2-aza-β-tyrosine from 2-aza-l-tyrosine for KedY4 and (S)-β-tyrosine from l-tyrosine for SgcC4 (Fig. 2A) (6, 7, 25). Thus, the stereochemistry of the (S)-2-aza-β-tyrosine product generated by SgcC4 from 2-aza-l-tyrosine had to be determined experimentally. The extremely weak activity of SgcC4 toward 2-aza-l-tyrosine precluded enzymatic synthesis of sufficient amounts of the product to establish its absolute stereochemistry by the modified Mosher method as was done for (R)-2-aza-β-tyrosine from 2-aza-l-tyrosine by KedY4.
Given that the MIO-containing aminomutase-catalyzed reactions are known to be reversible (3–5, 7, 8), the reverse reaction was exploited to investigate whether SgcC4 prefers (R)- or (S)-2-aza-β-tyrosine as a substrate (Fig. 2A). For this, synthetic (R)- and (S)-2-aza-β-tyrosine were incubated with SgcC4 or KedY4 under the standard assay conditions for 1 h, and the assays were followed by HPLC analysis. Although no change was observed with the boiled enzyme (Fig. 2C, I), (R)-2-aza-β-tyrosine was efficiently converted by KedY4 to 2-aza-l-tyrosine and 2-aza-4-hydroxycinnamic acid (Fig. 2C, II), and (S)-2-aza-β-tyrosine was a very poor substrate for KedY4, yielding barely detectable amounts of 2-aza-l-tyrosine and 2-aza-4-hydroxycinnamic acid (Fig. 2C, III). This finding is consistent with KedY4 catalyzing the forward reaction, yielding (R)-2-aza-β-tyrosine enantiospecifically from 2-aza-l-tyrosine. Similarly, whereas no change was observed for (S)-2-aza-β-tyrosine with the boiled SgcC4, (S)-2-aza-β-tyrosine was a significantly better substrate than (R)-2-aza-β-tyrosine for SgcC4, yielding 2-aza-l-tyrosine and 2-aza-4-hydroxycinnamic acid as products (Fig. 2C, IV and V). A careful comparison revealed that SgcC4-catalyzed formation of 2-aza-l-tyrosine from (S)-2-aza-β-tyrosine was least 1.8-fold faster than that from (R)-2-aza-β-tyrosine, a finding that supports SgcC4 catalyzing the forward reaction preferably yielding (S)-2-aza-β-tyrosine from 2-aza-l-tyrosine, similar to the reverse reactions with β-tyrosine (8).
Kinetic Properties of KedY4.
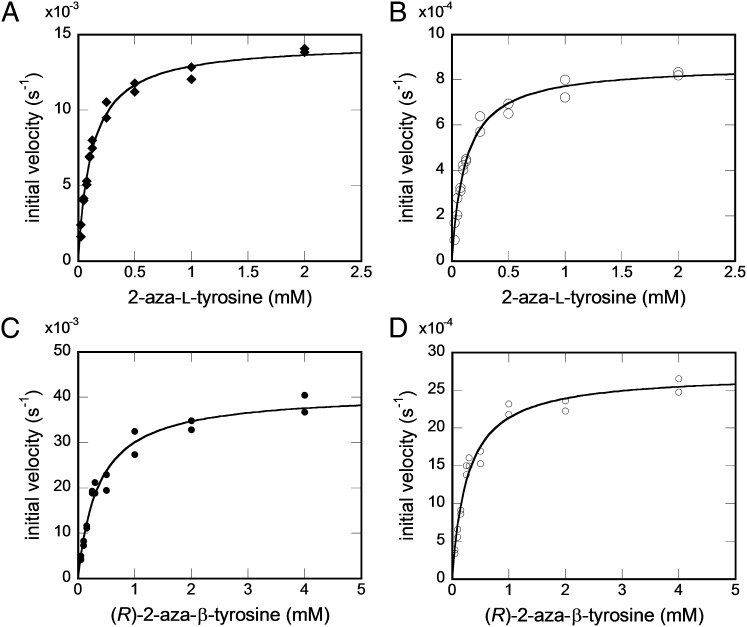
The HPLC assay was used to obtain Michaelis–Menten kinetic data for KedY4 with 2-aza-l-tyrosine (forward reaction) and (R)-2-aza-β-tyrosine (reverse reaction) as substrates (SI Appendix). Preliminary experiments established that the KedY4-catalyzed reaction was linear over a period of 60 min when KedY4 concentrations were kept below 10 μM. The aminomutase activity of KedY4 with 2-aza-l-tyrosine as a substrate had a Km of 121 ± 8 μM, a kcat of 14.5 ± 0.3 × 10−3 s−1, and a kcat/Km of 120 ± 8 M−1⋅s−1 (Fig. 4A), whereas ammonia lyase activity had a Km of 121 ± 9 μM, a kcat of 0.86 ± 0.02 × 10−3 s−1, and a kcat/Km of 7.1 ± 0.5 M−1⋅s−1 (Fig. 4B). The kinetic parameters for the reverse reaction with (R)-2-aza-β-tyrosine as a substrate show aminomutase activity with a Km of 366 ± 38 μM, a kcat of 41 ± 1 × 10−3 s−1, and a kcat/Km of 110 ± 10 M−1⋅s−1 (Fig. 4C) and ammonia lyase activity with a Km of 28 ± 3 μM, a kcat of 2.72 ± 0.08 × 10−3 s−1, and a kcat/Km of 10 ± 1 M−1⋅s−1 (Fig. 4D). These kinetic parameters for KedY4 are similar to those for SgcC4, with reported kcat/Km values for SgcC4 of 360 ± 44 M−1⋅s−1 for aminomutase activity and 50 ± 5 M−1⋅s−1 for ammonia lyase activity (8). The finding that KedY4 and SgcC4 share similar kinetic parameters supports the proposed role for KedY4 in catalyzing the conversion of 2-aza-l-tyrosine to (R)-2-aza-β-tyrosine in KED biosynthesis (Fig. 1).
Fig. 4.
Kinetic analysis of KedY4 with 2-aza-ʟ-tyrosine as a substrate for the forward reaction and (R)-2-aza-β-tyrosine as a substrate for the reverse reaction. (A and B) Formation of (R)-2-aza-β-tyrosine (◆; A) and 2-aza-4-hydroxycinnamic acid (○; B) with varying amounts of 2-aza-ʟ-tyrosine. (C and D) Formation of 2-aza-ʟ-tyrosine (●; C) and 2-aza-4-hydroxycinnamic acid (D) with varying amounts of (R)-2-aza-β-tyrosine.
Discussion
2-Aza-β-tyrosine is not known as a natural product, nor has it been identified as a component of any other natural products. Both 2-aza-l-tyrosine and of 2-aza-l-phenylalanine have been isolated, from S. chibaensis SF-1346 (32) and Streptomyces sp. SF2538 (36), respectively, but nothing is known about their biosynthesis. It was the comparative analysis of the KED, C-1027, and MDP clusters that led us to propose a pathway for (R)-2-aza-3-chloro-β-tyrosine biosynthesis starting from 2-aza-ʟ-tyrosine (Fig. 1) (25). Central to this proposal is KedY4, which was predicted to catalyze the formation of (R)-2-aza-β-tyrosine from 2-aza-ʟ-tyrosine, although the possibility of 2-aza-ʟ-phenylalanine as an alternative substrate could not be excluded. The results presented here establish that KedY4 is an MIO-containing aminomutase that efficiently and stereospecifically catalyzes the formation of (R)-2-aza-β-tyrosine from 2-aza-l-tyrosine. These findings support the proposed pathway for KED biosynthesis, and comparative studies of (R)-2-aza-3-chloro-β-tyrosine biosynthesis in KED and (S)-3-chloro-5-hydroxy-β-tyrosine biosynthesis in C-1027 provide outstanding opportunities to investigate distinct substrate (i) specificity, as exemplified by KedY1 for (R)-2-aza-β-tyrosine vs. SgcC1 for (S)-β-tyrosine, (ii) regiospecificity, as exemplified by KedY3 for C-6 chlorination of (R)-2-aza-β-tyrosyl-S-KedY2 vs. SgcC3 for C-3 chlorination of (S)-β-tyrosyl-S-SgcC2, and (iii) enantiospecificity, as exemplified by KedY4 affording (R)-2-aza-β-tyrosine vs. SgcC4 affording (S)-β-tyrosine, for this set of fascinating enzymes (Fig. 1).
The MIO-containing aminomutase family has grown steadily since the discovery of SgcC4 a decade ago (5–7). Mechanistic and structural characterizations of the MIO-containing aminomutases as a family have unveiled much unique chemistry, enzymology, and structural biology (5–20), and exploitation of these enzymes as biocatalysts has provided access to α- and β-amino acids, which are difficult to prepare by other means (4, 37). However, ʟ-phenylalanine and ʟ-tyrosine remain the only two known natural substrates (3–5). KedY4 shows high amino acid sequence homology to known MIO-containing aminomutases (SI Appendix, Fig. S1C), yet efficiently and specifically recognizes 2-aza-l-tyrosine as a substrate. Thus, the identification of KedY4 broadens the scope of known substrates for the MIO-containing aminomutase family, serving as an inspiration in the continuing search for new MIO-containing aminomutases and the engineering of new variants with designer properties.
The (R)-2-aza-β-tyrosine– and (S)-β-tyrosine–derived moieties in KED and C-1027 display opposite stereochemistries, defined by the activity of KedY4 and SgcC4, respectively. Whereas KedY4 catalyzes the conversion of 2-aza-l-tyrosine to (R)-2-aza-β-tyrosine but shows no detectable activity toward l-tyrosine, SgcC4 catalyzes the conversion of ʟ-tyrosine to (S)-β-tyrosine and is able to convert 2-aza-l-tyrosine to (S)-2-aza-β-tyrosine, albeit with very low activity. The current proposal for product stereochemistry indicates that (S)-β-amino acid-generating aminomutases reappend the amino group to the same face of the α,β-unsaturated carboxylic acid intermediate from which it was removed, whereas there is a concomitant rotation about the C1–Cα and Cβ–Cγ bonds of the intermediate, which presents the opposite face of the double bond to the amino group for (R)-β-amino acid-generating aminomutases (18). There is no defined structure–function relationship for how these aminomutases accomplish or preclude these rotations, however. Using the structure of SgcC4 and sequence alignments as a guide, we attempted to rationalize the difference between the stereochemical outcomes and product specificity for KedY4. The most significant difference in primary structure is the single amino acid insertion, Asn400, in KedY4 (SI Appendix, Fig. S1C). This insertion is very far away from the active site, however, and thus whether it contributes to product stereospecificity is far from certain. Although the active site residues lining the binding pocket of KedY4 are identical to the TAMs, there are many differences in the residues of the second sphere. Comparative studies of KedY4 and SgcC4 will provide an opportunity to decipher the determinants controlling substrate specificity and product enantioselectivity.
The MIO-containing aminomutases have been exploited as biocatalysts to generate novel α- and β-amino acids (8, 37). SgcC4 exhibits substrate promiscuity, including l-dopa and l-3-chlorotyrosine, albeit with significantly reduced activity (8). The PAMs have been demonstrated to accept a broad range of substrate analogs (37). Although KedY4 has shown no activity toward l-tyrosine and 2-aza-l-phenylalanine, the limited availability of other pyridine-containing l-α-amino acids has precluded testing of KedY4 substrate promiscuity; the similar kinetic parameters of KedY4 and SgcC4 would support such an exploration. The specificity of KedY4 and SgcC4 for 2-aza-l-tyrosine and l-tyrosine stems from a conserved set of polar residues lining the end of the substrate binding pocket that make key hydrogen bonds to the phenol group, as defined in the crystal structures of SgcC4 (9-12). The corresponding residues in the PAMs are hydrophobic in nature (15, 18). The preference of KedY4 for a hydroxypyridine-bearing amino acid promises to expand the number of α- and β-amino acids accessible by exploiting MIO-containing aminomutase biocatalysis. Furthermore, KedY4 and 2-aza-l-tyrosine (or the 2-aza-l-tyrosine biosynthetic machinery) can be envisioned to provide (R)-2-aza-β-tyrosine in vivo for engineered biosynthesis to further enrich natural product structural diversity.
Methods
Details on materials, methods, and experimental procedures are provided in SI Appendix. Included are posttranslational formation of the MIO prosthetic group from the conserved Ala-Ser-Gly motif, proposed mechanisms of MIO-containing ammonia lyases and aminomutases, bioinformatics analysis of KedY4 supporting its functional annotation as an MIO-containing aminomutase (SI Appendix, Fig. S1), expression of kedY4 and overproduction and purification of KedY4 (SI Appendix, Fig. S2), fermentation and isolation of 2-aza-l-tyrosine from S. chibaensis SF-1346 (SI Appendix, Figs. S3 and S4), chemical synthesis of (R)- and (S)-2-aza-β-tyrosine and their derivatives (SI Appendix, Figs. S5–S23), in vitro characterization of KedY4 as an MIO-containing aminomutase (SI Appendix, Figs. S24 and S25), and determination of the absolute stereochemistry of the (R)-2-aza-β-tyrosine product (SI Appendix, Figs. S26–S50).
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grant CA78747.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. JX679499).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304733110/-/DCSupplemental.
References
- 1.Frey PA. Radical mechanisms of enzymatic catalysis. Annu Rev Biochem. 2001;70:121–148. doi: 10.1146/annurev.biochem.70.1.121. [DOI] [PubMed] [Google Scholar]
- 2.Vey JL, Drennan CL. Structural insights into radical generation by the radical SAM superfamily. Chem Rev. 2011;111(4):2487–2506. doi: 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooke HA, Christianson CV, Bruner SD. Structure and chemistry of 4-methylideneimidazole-5-one–containing enzymes. Curr Opin Chem Biol. 2009;13(4):460–468. doi: 10.1016/j.cbpa.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Turner NJ. Ammonia lyases and aminomutases as biocatalysts for the synthesis of α-amino and β-amino acids. Curr Opin Chem Biol. 2011;15(2):234–240. doi: 10.1016/j.cbpa.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Lohman JR, Shen B. 4-methylideneimidazole-5-one–containing aminomutases in enediyne biosynthesis. Methods Enzymol. 2012;516:299–319. doi: 10.1016/B978-0-12-394291-3.00007-1. [DOI] [PubMed] [Google Scholar]
- 6.Liu W, Christenson SD, Standage S, Shen B. Biosynthesis of the enediyne antitumor antibiotic C-1027. Science. 2002;297(5584):1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 7.Christenson SD, Liu W, Toney MD, Shen B. A novel 4-methylideneimidazole-5-one–containing tyrosine aminomutase in enediyne antitumor antibiotic C-1027 biosynthesis. J Am Chem Soc. 2003;125(20):6062–6063. doi: 10.1021/ja034609m. [DOI] [PubMed] [Google Scholar]
- 8.Christenson SD, Wu W, Spies MA, Shen B, Toney MD. Kinetic analysis of the 4-methylideneimidazole-5-one–containing tyrosine aminomutase in enediyne antitumor antibiotic C-1027 biosynthesis. Biochemistry. 2003;42(43):12708–12718. doi: 10.1021/bi035223r. [DOI] [PubMed] [Google Scholar]
- 9.Christianson CV, Montavon TJ, Van Lanen SG, Shen B, Bruner SD. The structure of L-tyrosine 2,3-aminomutase from the C-1027 enediyne antitumor antibiotic biosynthetic pathway. Biochemistry. 2007;46(24):7205–7214. doi: 10.1021/bi7003685. [DOI] [PubMed] [Google Scholar]
- 10.Christianson CV, et al. The mechanism of MIO-based aminomutases in beta-amino acid biosynthesis. J Am Chem Soc. 2007;129(51):15744–15745. doi: 10.1021/ja0762689. [DOI] [PubMed] [Google Scholar]
- 11.Montavon TJ, Christianson CV, Festin GM, Shen B, Bruner SD. Design and characterization of mechanism-based inhibitors for the tyrosine aminomutase SgTAM. Bioorg Med Chem Lett. 2008;18(10):3099–3102. doi: 10.1016/j.bmcl.2007.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke HA, Bruner SD. Probing the active site of MIO-dependent aminomutases, key catalysts in the biosynthesis of beta-amino acids incorporated in secondary metabolites. Biopolymers. 2010;93(9):802–810. doi: 10.1002/bip.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin M, Fischbach MA, Clardy J. A biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J Am Chem Soc. 2006;128(33):10660–10661. doi: 10.1021/ja063194c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratnayake ND, Wanninayake U, Geiger JH, Walker KD. Stereochemistry and mechanism of a microbial phenylalanine aminomutase. J Am Chem Soc. 2011;133(22):8531–8533. doi: 10.1021/ja2030728. [DOI] [PubMed] [Google Scholar]
- 15.Strom S, Wanninayake U, Ratnayake ND, Walker KD, Geiger JH. Insights into the mechanistic pathway of the Pantoea agglomerans phenylalanine aminomutase. Angew Chem Int Ed Engl. 2012;51(12):2898–2902. doi: 10.1002/anie.201108525. [DOI] [PubMed] [Google Scholar]
- 16.Chesters C, Wilding M, Goodall M, Micklefield J. Thermal bifunctionality of bacterial phenylalanine aminomutase and ammonia lyase enzymes. Angew Chem Int Ed Engl. 2012;51(18):4344–4348. doi: 10.1002/anie.201200669. [DOI] [PubMed] [Google Scholar]
- 17.Walker KD, Klettke K, Akiyama T, Croteau R. Cloning, heterologous expression, and characterization of a phenylalanine aminomutase involved in Taxol biosynthesis. J Biol Chem. 2004;279(52):53947–53954. doi: 10.1074/jbc.M411215200. [DOI] [PubMed] [Google Scholar]
- 18.Feng L, Wanninayake U, Strom S, Geiger J, Walker KD. Mechanistic, mutational, and structural evaluation of a Taxus phenylalanine aminomutase. Biochemistry. 2011;50(14):2919–2930. doi: 10.1021/bi102067r. [DOI] [PubMed] [Google Scholar]
- 19.Van Lanen SG, Oh TJ, Liu W, Wendt-Pienkowski E, Shen B. Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J Am Chem Soc. 2007;129(43):13082–13094. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krug D, Müller R. Discovery of additional members of the tyrosine aminomutase enzyme family and the mutational analysis of CmdF. ChemBioChem. 2009;10(4):741–750. doi: 10.1002/cbic.200800748. [DOI] [PubMed] [Google Scholar]
- 21.Rachid S, Krug D, Weissman KJ, Müller R. Biosynthesis of (R)-β-tyrosine and its incorporation into the highly cytotoxic chondramides produced by Chondromyces crocatus. J Biol Chem. 2007;282(30):21810–21817. doi: 10.1074/jbc.M703439200. [DOI] [PubMed] [Google Scholar]
- 22.Leet JE, et al. Chemistry and structure elucidation of the kedarcidin chromophore. J Am Chem Soc. 1993;115(18):8432–8443. [Google Scholar]
- 23.Kawata S, Ashizawa S, Hirama M. Synthetic study of kedarcidin chromophore: Revised structure. J Am Chem Soc. 1997;119(49):12012–12013. [Google Scholar]
- 24.Ren F, Hogan PC, Anderson AJ, Myers AG. Kedarcidin chromophore: Synthesis of its proposed structure and evidence for a stereochemical revision. J Am Chem Soc. 2007;129(17):5381–5383. doi: 10.1021/ja071205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohman JR, et al. Cloning and sequencing of the kedarcidin biosynthetic gene cluster from Streptoalloteichus sp. ATCC 53650 revealing new insights into biosynthesis of the enediyne family of antitumor antibiotics. Mol Biosyst. 2013;9(3):478–491. doi: 10.1039/c3mb25523a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Lanen SG, et al. Biosynthesis of the β-amino acid moiety of the enediyne antitumor antibiotic C-1027 featuring β-amino acyl-S-carrier protein intermediates. J Am Chem Soc. 2005;127(33):11594–11595. doi: 10.1021/ja052871k. [DOI] [PubMed] [Google Scholar]
- 27.Van Lanen SG, Lin S, Dorrestein PC, Kelleher NL, Shen B. Substrate specificity of the adenylation enzyme SgcC1 involved in the biosynthesis of the enediyne antitumor antibiotic C-1027. J Biol Chem. 2006;281(40):29633–29640. doi: 10.1074/jbc.M605887200. [DOI] [PubMed] [Google Scholar]
- 28.Lin S, Van Lanen SG, Shen B. Regiospecific chlorination of (S)-β-tyrosyl-S-carrier protein catalyzed by SgcC3 in the biosynthesis of the enediyne antitumor antibiotic C-1027. J Am Chem Soc. 2007;129(41):12432–12438. doi: 10.1021/ja072311g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S, Van Lanen SG, Shen B. A free-standing condensation enzyme catalyzing ester bond formation in C-1027 biosynthesis. Proc Natl Acad Sci USA. 2009;106(11):4183–4188. doi: 10.1073/pnas.0808880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin S, et al. Specificity of the ester bond forming condensation enzyme SgcC5 in C-1027 biosynthesis. Org Lett. 2012;14(9):2300–2303. doi: 10.1021/ol300720s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin S, Huang T, Shen B. Tailoring enzymes acting on carrier protein-tethered substrates in natural product biosynthesis. Methods Enzymol. 2012;516:321–343. doi: 10.1016/B978-0-12-394291-3.00008-3. [DOI] [PubMed] [Google Scholar]
- 32.Inouye S, Shomura T, Tsuruoka T, Ogawa Y, Watanabe H. L-β-(5-hydroxy-2-pyridyl)-alanine and L-β-(3-hydroxyureido)-alanine from Streptomyces. Chem Pharm Bull (Tokyo) 1975;23(11):2669–2677. doi: 10.1248/cpb.23.2669. [DOI] [PubMed] [Google Scholar]
- 33.Adamczyk M, Reddy RE. Synthesis of (R)-(+)-methyl 3-amino-3-(5-hydroxy-2-pyridinyl)propanoate, an analog of ʟ-azatyrosine. Tetrahedron Asymmetry. 2001;12:1047–1054. [Google Scholar]
- 34.Dale JA, Mosher HS. Nuclear magnetic resonance enantiomer reagents: Configurational correlations via nuclear magnetic resonance chemical shifts of diastereomeric mandelate, O-methylmandelate, and α-methoxy-α-trifluoromethylphenylacetate (MTPA) esters. J Am Chem Soc. 1973;95(2):512–519. [Google Scholar]
- 35.Seco JM, Quiñoá E, Riguera R. A practical guide for the assignment of the absolute configuration of alcohols, amines and carboxylic acids by NMR. Tetrahedron Asymmetry. 2001;12:2915–2925. [Google Scholar]
- 36.Iwata M, et al. β-(2-Pyridyl)-l-α-alanine from Streptomyces. Sci Rep Meiji Seika Kaisha. 1988;27:63–66. [Google Scholar]
- 37.Wanninayake U, Deporre Y, Ondari M, Walker KD. (S)-Styryl-α-alanine used to probe the intermolecular mechanism of an intramolecular MIO-aminomutase. Biochemistry. 2011;50(46):10082–10090. doi: 10.1021/bi2012299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.