Abstract
Mutational activation of the Ras oncogene products (H-Ras, K-Ras, and N-Ras) is frequently observed in human cancers, making them promising anticancer drug targets. Nonetheless, no effective strategy has been available for the development of Ras inhibitors, partly owing to the absence of well-defined surface pockets suitable for drug binding. Only recently, such pockets have been found in the crystal structures of a unique conformation of Ras⋅GTP. Here we report the successful development of small-molecule Ras inhibitors by an in silico screen targeting a pocket found in the crystal structure of M-Ras⋅GTP carrying an H-Ras–type substitution P40D. The selected compound Kobe0065 and its analog Kobe2602 exhibit inhibitory activity toward H-Ras⋅GTP-c-Raf-1 binding both in vivo and in vitro. They effectively inhibit both anchorage-dependent and -independent growth and induce apoptosis of H-rasG12V–transformed NIH 3T3 cells, which is accompanied by down-regulation of downstream molecules such as MEK/ERK, Akt, and RalA as well as an upstream molecule, Son of sevenless. Moreover, they exhibit antitumor activity on a xenograft of human colon carcinoma SW480 cells carrying the K-rasG12V gene by oral administration. The NMR structure of a complex of the compound with H-Ras⋅GTPT35S, exclusively adopting the unique conformation, confirms its insertion into one of the surface pockets and provides a molecular basis for binding inhibition toward multiple Ras⋅GTP-interacting molecules. This study proves the effectiveness of our strategy for structure-based drug design to target Ras⋅GTP, and the resulting Kobe0065-family compounds may serve as a scaffold for the development of Ras inhibitors with higher potency and specificity.
Keywords: molecular targeted therapy, small-molecule inhibitor
Ras oncoproteins belong to the Ras family of small GTPases and function as molecular switches by cycling between GTP-bound active and GDP-bound inactive forms in intracellular signaling pathways controlling cell growth, differentiation, and apoptosis (1). Interconversion between the two forms, which mainly involves the conformational changes of two flexible regions called switch I (residues 32–38) and switch II (residues 60–75), is reciprocally catalyzed by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (2). In particular, GEFs such as Son of sevenless (Sos) mediate upstream signals to enhance formation of the GTP-bound form. The oncogenic potential of Ras is activated by point mutations mainly involving the codons 12 and 61, which impair the intrinsic GTPase activity and, moreover, render Ras insensitive to the GAP action, leading to constitutive activation of downstream effectors such as Raf kinases including c-Raf-1 and B-Raf, PI3Ks, and Ral guanine nucleotide dissociation stimulator (RalGDS) family proteins (1). These mutations are observed in about 15–20% of human cancers, and specifically in about 60–90% and 30–50% of pancreatic and colorectal carcinomas, respectively (1, 3, 4). Cancer cells carrying the ras oncogene are known to exhibit a phenomenon called oncogene addiction, where their survival becomes dependent on the activated oncogene function (3). Consequently, inhibition of the activated Ras function has been shown to lead not only to reversal of the transformed phenotypes but also to cell death and tumor regression (4, 5). Despite their importance as an anticancer drug target, there is no effective molecular targeted therapy for Ras at present; the once highly anticipated farnesyl transferase inhibitors, which inhibit the posttranslational lipid modification, farnesylation, of Ras necessary for membrane targeting, have failed in clinical trials (1, 6). Although farnesylthiosalicylic acid has been reported to inhibit Ras by antagonizing its interaction with the Ras-escort proteins, its antitumor activity remains unclear (7).
Although recent success in drug discovery using structure-based drug design (SBDD) for AIDS and influenza has boosted hopes for the application of SBDD to anticancer drug development, Ras have been presumed refractory to this approach because they lack apparently “druggable” pockets on their surface, as seen from their crystal structures (1). Recently, by X-ray crystallography and NMR spectroscopy we solved the tertiary structures of H-Ras, its homolog M-Ras, and their mutants in complex with a nonhydrolyzable GTP analog, guanosine 5′-(β,γ-imido)triphosphate (GppNHp), all of which corresponded to a unique conformation (8–10) undergoing dynamic equilibrium with the previously known conformation. Intriguingly, the structures possessed surface pockets that seem suitable for drug binding. In this paper, we have applied SBDD to target Ras⋅GTP by using the structural information on these surface pockets. We report the successful discovery of a unique class of small-molecule compounds that have potent activity to block the interactions of Ras⋅GTP with their multiple effector molecules and, moreover, display antitumor activity on a xenograft of human colon carcinoma cells carrying the K-rasG12V gene.
Results
Discovery of Small-Molecule Compounds Inhibiting Ras–Raf Interaction by SBDD.
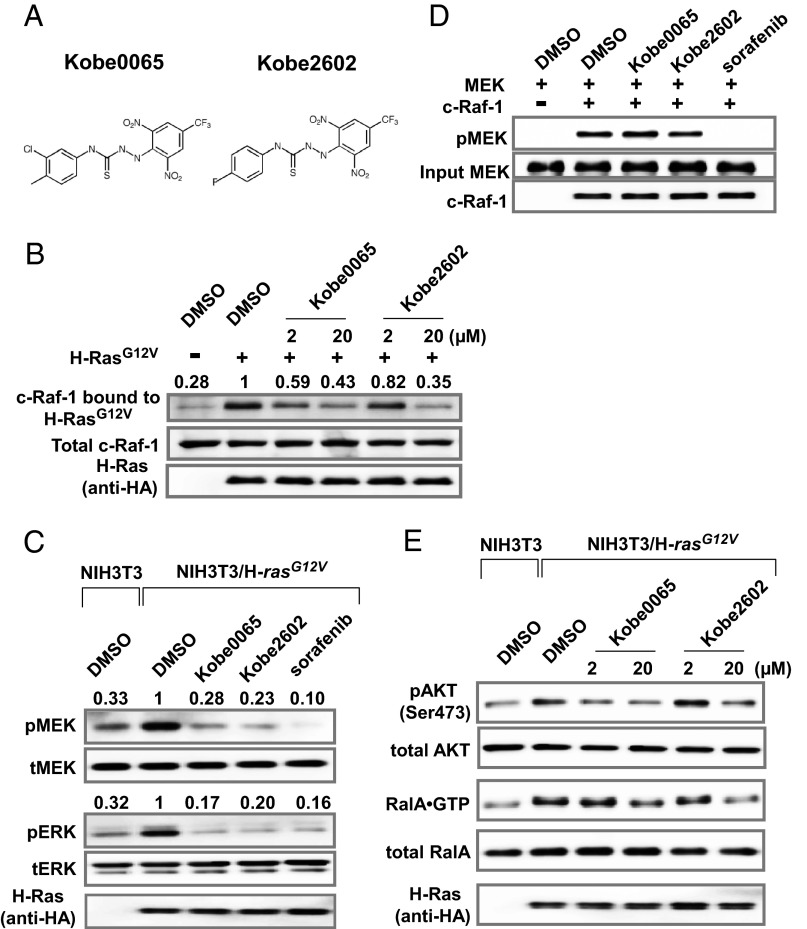
Aiming to discover small-molecule compounds fitting into the surface pockets of the unique conformation of Ras⋅GTP, we applied the molecular mechanics Poisson–Boltzman surface area (MMPB-SA) method with an Assisted Model Building and Energy Refinement (AMBER)96 force field to carry out a computer docking screen of a virtual library containing 40,882 compounds based on the high-resolution (1.35 Å) crystal structure of M-RasP40D⋅GppNHp (9). Ninety-seven candidates were selected and examined in vitro for their activity to inhibit the binding of M-RasP40D⋅GTP and H-Ras⋅GTP to the Ras-binding domain (RBD, amino acids 50–131) of c-Raf-1. Only one compound, named Kobe0065 (Fig. 1A), exhibited potent activity to competitively inhibit the binding of H-Ras⋅GTP to c-Raf-1 RBD with a Ki value of 46 ± 13 μM as estimated from the binding kinetics (Fig. S1). A subsequent computer-assisted similarity search of ∼160,000 compounds based on the Tanimoto coefficient selected 273 candidates, among which one positive, named Kobe2602 (Fig. 1A), with a Ki value of 149 ± 55 μM (Fig. S1), was identified. These two compounds, added to the culture medium at 2 and 20 μM, effectively reduced the amount of c-Raf-1 associated with H-RasG12V in NIH 3T3 cells in a dose-dependent manner, indicating the inhibition of the cellular activity of Ras (Fig. 1B). A rough estimate of the IC50 value for the cellular Ras–Raf-binding inhibition was around 10 μM (Fig. 1B), which was not much different from the Ki values for the in vitro Ras–Raf-binding inhibition considering the quite low cellular concentration of Raf. A similar inhibitory effect was also observed with NIH 3T3 cells overexpressing K-RasG12V (Fig. S2).
Fig. 1.
Inhibition of various downstream targets of Ras by the Kobe0065-family compounds. (A) Chemical structures of the compounds. (B) NIH 3T3 cells were transfected with pEF-BOS-HA-H-RasG12V or an empty vector and treated with the 2 and 20 μM compound or the vehicle (DMSO) in the presence of 2% FBS for 1 h. Cell lysate was subjected to detection of c-Raf-1 coimmunoprecipitated with an anti-H-Ras antibody (Top) and total c-Raf-1 (Middle) by Western blotting with an anti-c-Raf-1 antibody. Immunoprecipitated H-RasG12V were detected by an anti-HA antibody (Bottom). The numbers above the lanes show the values of H-Ras-bound/total c-Raf-1 relative to that of the vehicle-treated cells. (C) Lysate was prepared from cells treated with 20 μM Kobe0065, 20 μM Kobe2602, or 2 μM sorafenib as described in B and subjected to detection of phosphorylated MEK (pMEK) and ERK (pERK) by Western blotting with anti-pMEK and anti-pERK antibodies. Total amounts of MEK, ERK, and HA-tagged H-RasG12V were detected by anti-MEK, anti-ERK, and anti-HA antibodies, respectively. The numbers above the lanes show the values of pMEK/tMEK and pERK/tERK relative to those of the vehicle-treated cells. Four independent experiments yielded essentially equivalent results. (D) Recombinant c-Raf-1 was incubated with recombinant MEK in the presence of 20 μM Kobe0065, 20 μM Kobe2602, or 2 μM sorafenib, and pMEK formed was detected by Western blotting with an anti-pMEK antibody. (E) Lysate was prepared from cells treated with the indicated concentrations of the compound as described in B and subjected to the measurements of phosphorylated Akt (pAKT) by Western blotting with an anti-pAkt antibody and of RalA⋅GTP pulled down with GST-Sec5(1–99) immobilized on glutathione-sepharose resin by Western blotting with an anti-RalA antibody. Four independent experiments yielded essentially equivalent results.
Inhibitory Effects of the Kobe0065-Family Compounds on Various Ras Signaling Pathways.
We examined the effect of the compounds on the cellular activity of Raf, which is dependent on its interaction with Ras⋅GTP. Both Kobe0065 and Kobe2602 at 20 μM efficiently inhibited the phosphorylation of MEK and ERK, downstream kinases of Raf in NIH 3T3 cells transiently expressing H-RasG12V, although the effect was slightly weaker than that of 2 μM sorafenib (11), an inhibitor of multiple protein kinases including Raf (Fig. 1C). However, they failed to inhibit the kinase activity of c-Raf-1 measured in vitro (Fig. 1D), indicating the absence of direct inhibitory activity on Raf. Furthermore, the compound-treated cells showed substantial decreases of phosphorylated Akt and RalA⋅GTP, downstream molecules of PI3Ks and RalGDS, respectively, in a manner dependent on the compound concentrations (Fig. 1E), suggesting that the compounds exerted inhibitory effects toward multiple Ras effectors through inactivation of Ras.
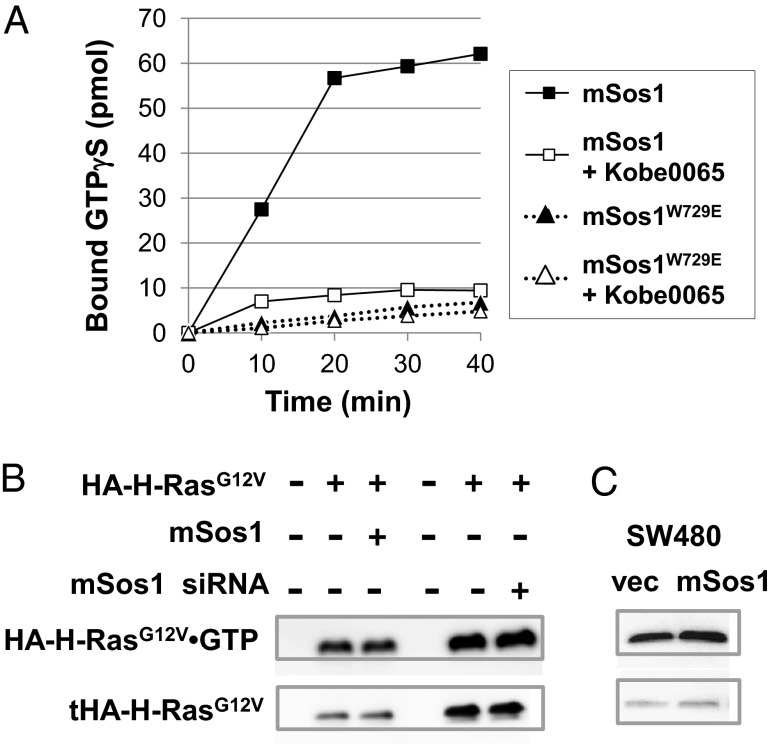
We next examined the effect of the compounds on Sos. Sos has two distinct Ras-binding sites: the GEF domain catalyzing the GDP–GTP exchange on Ras through interaction with Ras⋅GDP and the distal site allosterically accelerating the GEF catalytic activity through interaction with Ras⋅GTP (12), and thereby functions not only as a regulator but also as an effector of Ras. In vitro GDP–GTP exchange assays using mSos1 and mSos1W729E, carrying an inactivating mutation of the distal site (12), showed that Kobe0065 at 50 μM almost completely abolished the accelerating effect of the distal site without apparently affecting the catalytic activity of the GEF domain itself (Fig. 2A and Fig. S3A), suggesting that the compounds inhibited Ras⋅GTP but not Ras⋅GDP. The IC50 value of Kobe0065 was estimated to be around 20 μM (Fig. S3B). Kobe2602 also showed the same but weaker activity with an IC50 value of around 100 μM (Fig. S3C). This finding raised a possibility that the observed in vivo inhibition of H-RasG12V by the compounds (Fig. 1 C and E) might be accounted for by the decreased Ras⋅GTP level owing to Sos inhibition. However, we found that this was not the case, because the cellular RasG12V⋅GTP level was almost unaffected by either overexpression or siRNA-mediated knockdown of mSos1 in either NIH 3T3 cells transiently expressing H-RasG12V or human colon carcinoma SW480 cells carrying K-rasG12V (Fig. 2 C and D).
Fig. 2.
Inhibition of Sos by the Kobe0065-family compounds and effect of the Sos activity on the cellular RasG12V⋅GTP level. (A) GST-H-Ras(1-166)⋅GDP immobilized on glutathione-sepharose resin were incubated with [γ-35S]GTPγS and purified 6×His-tagged mSos1(563–1,049), wild-type, or a W729E mutant at 25 °C in the presence or absence of 50 μM Kobe0065. The radioactivity pulled down by glutathione-sepharose resin was measured. Three independent experiments yielded essentially equivalent results. (B) NIH 3T3 cells were transfected with pEF-BOS-HA-H-RasG12V in combination with pCMV-mSos1 or siRNA against mSos1. H-RasG12V⋅GTP pulled down by GST-c-Raf-1-RBD from the cell lysate was detected by an anti-HA antibody (Upper). Total amounts of HA-H-RasG12V in the lysates was also measured (Lower). (C) SW480 cells were transfected with pCMV-mSos1. K-RasG12V⋅GTP pulled down by GST-c-Raf-1-RBD from cell lysates was detected by an anti-K-Ras antibody (Upper). Total amounts of K-RasG12V in the lysates were also measured (Lower).
Inhibitory Effects of the Kobe0065-Family Compounds on the Growth of Cancer Cell Culture and Tumor Xenograft.
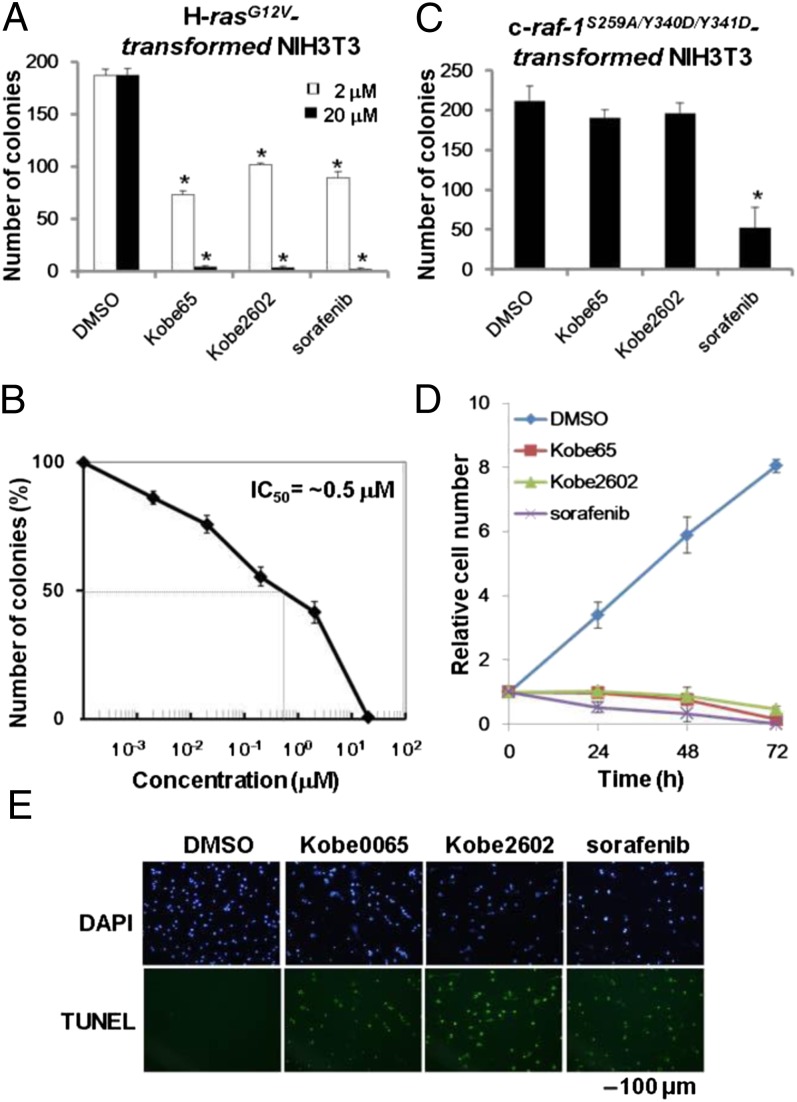
We next tested the effect of Kobe0065 and Kobe2602 on anchorage-independent proliferation of H-rasG12V–transformed NIH 3T3 cells. The compounds efficiently inhibited colony formation in soft agar in a dose-dependent manner (Fig. 3A and Fig. S3A). The IC50 values were around 0.5 and 1.4 μM for Kobe0065 (Fig. 3B) and Kobe2602 (Fig. S3B), respectively, which were comparable to the value of 2.1 μM observed for sorafenib (Fig. S3B). By contrast, the compounds failed to inhibit colony formation of NIH 3T3 cells transformed by the activated c-raf-1 gene carrying the S259A/Y340D/Y341D mutations (Fig. 3C and Fig. S4A), whereas sorafenib exhibited potent inhibitory activity.
Fig. 3.
Inhibition of proliferation of H-rasG12V–transformed cells by the Kobe0065-family compounds. (A) H-rasG12V–transformed NIH 3T3 cells (1 × 103 cells) were inoculated in 2 mL of DMEM containing 10% FBS, 0.33% SeaPlaque agarose, and the indicated concentrations of the compound. After incubation at 37 °C for 14 d, the number of colonies >200 μm in diameter was counted under a dissecting microscope. (B) The IC50 value for Kobe0065 was estimated from the dose–response curve. (C) Effects of the 20 μM compounds on soft agar colony formation of c-raf-1S259A/Y340D/Y341–transformed NIH 3T3 cells were measured similarly as described in A. (D) H-rasG12V–transformed NIH 3T3 cells were cultured under a low-serum condition (2% FBS) in the presence of the 20 μM compound. Each point represents the cell number relative to that of the 0-h treatment. The values are presented as the mean ± SEM (A, n = 4; B, n = 7; and C, n = 3). All experiments were performed in duplicate. One-way ANOVA with Dunnett’s test was used for the statistical analyses. *P < 0.001. (E) H-rasG12V–transformed NIH 3T3 cells cultured in the presence of the 20 μM compound for 24 h in 2% FBS were subjected to staining with DAPI (Upper) and the TUNEL assay for detection of apoptotic cells (Lower). A representative image is shown for each group.
We then assessed the ras specificity of inhibition by using several cancer cell lines carrying various oncogenes. The compounds effectively inhibited the colony formation of cancer cells carrying the activated ras oncogenes, such as SW480 and PANC-1 (K-rasG12V), EJ-1 (H-rasG12V), HT1080 (N-rasQ61L), and DLD-1 and HCT116 (H-rasG13D), but showed much weaker inhibition on those without the ras mutation, such as A375, T-47D, LNCap, BxPC-3, MCF-7, HepG2, and HeLa (Table S1). DLD-1 and HCT116 were sensitive to the compounds even though they carried additional activating mutations in PI3K, suggesting that the activated PI3K alone might be insufficient to sustain their anchorage independence. We next examined the effect of the compounds on anchorage-dependent proliferation (Fig. 3D). The compounds at 20 μM almost completely inhibited the proliferation of H-rasG12V–transformed NIH 3T3 cells in the presence of 2% FBS. The IC50 values were ∼1.5 and 2 μM for Kobe0065 and Kobe2602, respectively, which were a bit higher than that (0.8 μM) for sorafenib (Fig. S4C). The compound-treated cells exhibited frequent apoptosis (Fig. 3E), suggesting a contribution of the oncogene addiction mechanism to the antiproliferative effect.
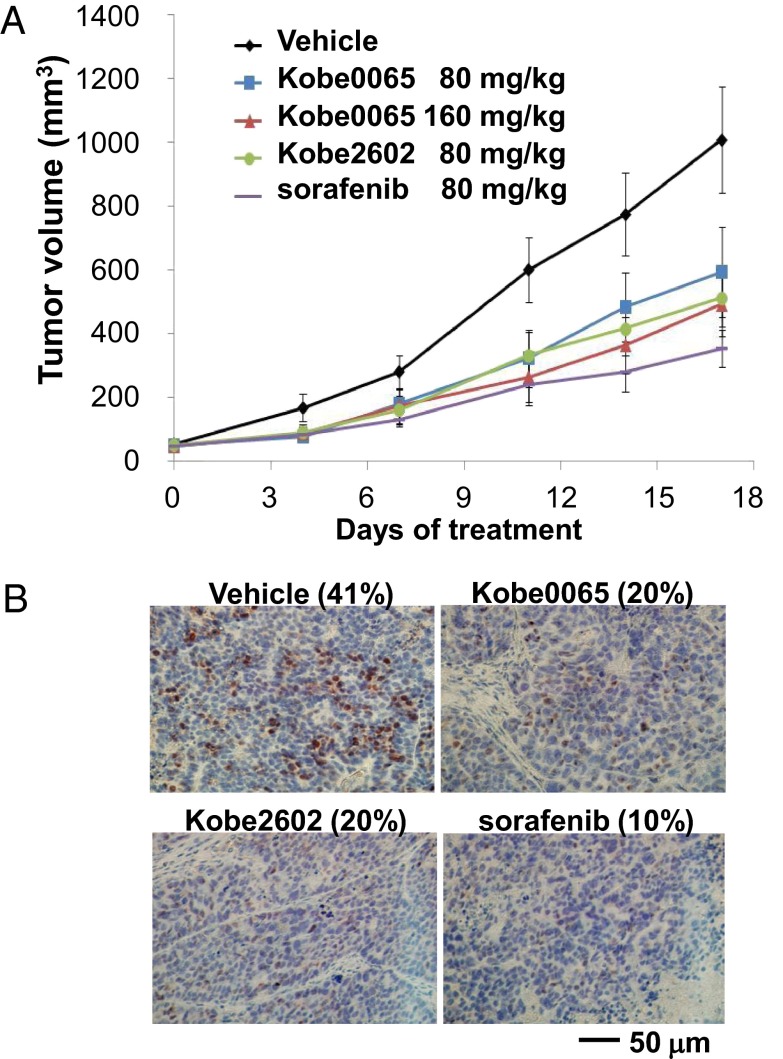
We next assessed the antitumor activity of the compounds by using a xenograft of SW480 cells in nude mice. Daily oral administration of the compounds at the dose of 80 mg/kg caused ∼40–50% inhibition of the tumor growth, which was weaker than the 65% inhibition by sorafenib (Fig. 4A). By doubling the dose to 160 mg/kg, the activity of Kobe0065 became more evident. During these compound treatments the mice did not exhibit any significant body weight loss (Fig. S5). Immunostaining of the tumor sections showed that the ERK activation was substantially compromised by the compound administration (Fig. 4B). Moreover, the compound-treated tumors showed a prominent increase of apoptotic cells (Fig. S6A), suggesting a contribution of the oncogene addiction mechanism to the antitumor effect. In contrast to the case of sorafenib, an antiangiogenesis effect was not observed (Fig. S6B).
Fig. 4.
Anti-proliferative activity of the Kobe0065-family compounds on a tumor xenograft. (A) Female athymic nude mice were implanted with SW480 cells (5 × 106 cells) in their right flanks. When the tumor sizes reached 52 ± 3 mm3, the compounds were administered orally for five consecutive days per week for 17–20 d at the indicated doses and the tumor volumes were continuously monitored. The values are presented as the mean ± SEM; n = 8–10 per group. P = 0.086 (t test) for 80 mg/kg Kobe0065, P < 0.05 for 160 mg/kg Kobe0065 and 80 mg/kg Kobe2602, and P < 0.01 for 80 mg/kg sorafenib at day 17. One-way ANOVA with Tukey’s test was used to analyze the significance of tumor size changes compared with the vehicle-treated group. (B) Phosphorylated ERK was detected by immunohistochemistry with an anti-pERK antibody in sections of tumors, which were treated daily with the 80 mg/kg compound for 17 d. The percentage of pERK-positive cells is shown on the top of each panel. A representative image is shown for each group.
Molecular Basis for Interaction of Ras⋅GTP with the Kobe0065-Family Compounds.
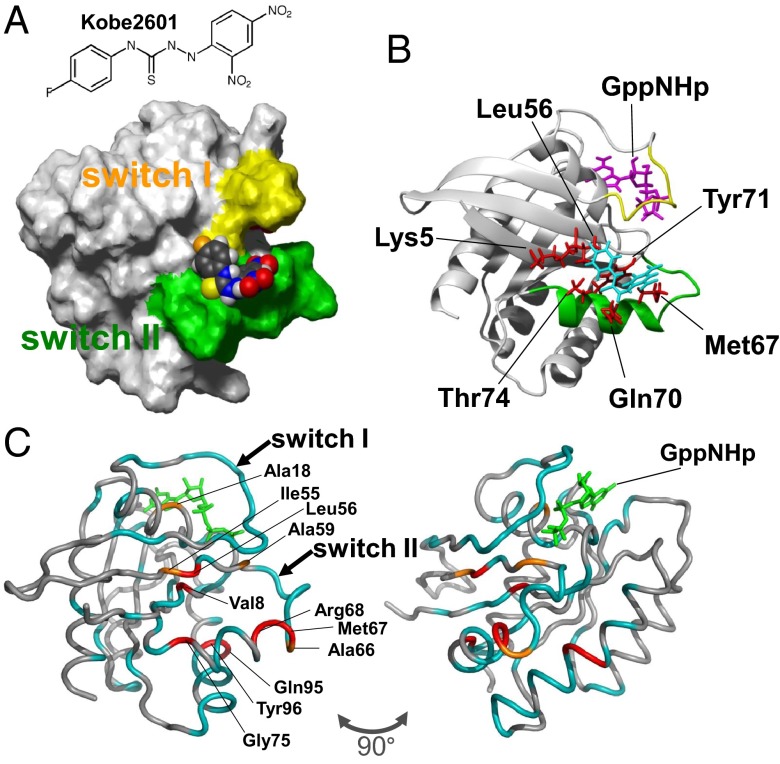
We used NMR spectroscopy to obtain structural information on the compound-binding interface on Ras⋅GTP. The NMR structure corresponding to the unique conformation of H-Ras⋅GppNHp was determined by using only its T35S mutant (13), because this mutation almost eliminated the slow conformational exchange process (14), which made NMR analysis of the wild-type protein impractical. Because of the low water solubility of Kobe0065 and Kobe2602, which made measurements of the NOEs impossible, we chose to use a water-soluble analog named Kobe2601 (Fig. 5A), which had also been identified by the similarity search of Kobe0065. Kobe2601 showed weak inhibitory activity toward in vitro Ras–Raf binding with a Ki value of 773 ± 49 μM (Fig. S1). NOEs between the benzene rings of Kobe2601 and the side chains of H-RasT35S⋅GppNHp were detected and the collected data were used for calculation of the tertiary structure of the H-RasT35S⋅GppNHp–Kobe2601 complex (Tables S2 and S3). The fluorobenzene moiety of Kobe2601 was located in close proximity to the side chains of Lys5, Leu56, Met67, Gln70, Tyr71, and Thr74 of H-Ras (Fig. 5 A and B). These six residues formed a hydrophobic surface pocket in the neighborhood of switch I (Fig. S7A), indicating that the fluorobenzene ring was inserted into the pocket through hydrophobic interaction. However, the dinitrobenzene moiety of Kobe2601 was located near switch II but not tightly fixed. Although direct assignment of the Kobe2601-interacting residues on wild-type H-Ras was difficult, measurement of the backbone amide 1H, 15N heteronuclear single quantum coherence (HSQC) spectra of H-Ras⋅GppNHp revealed that the resonances from Leu56, Met67, and their neighboring residues underwent significant chemical shift changes and line broadening by the addition of Kobe2601 (Fig. 5C and Fig. S8), suggesting sharing of a common binding pocket with H-RasT35S⋅GppNHp.
Fig. 5.
Molecular basis for the interaction of Ras⋅GTP with the Kobe0065-family compounds. (A) The lowest energy solution structure of the H-RasT35S⋅GppNHp–Kobe2601 complex. H-RasT35S⋅GppNHp is shown by a surface model (switch I, yellow; switch II, green) and Kobe2601 is shown by a space-filling model (C, black; O, red; N, blue; H, gray; S, yellow; and F, orange). (B) Spatial arrangements of the residues giving NOE contacts with Kobe2601. Stick representations of the residues giving intermolecular NOEs (red), Kobe2601 (cyan), and GppNHp (magenta) are shown on the backbone structure of H-RasT35S⋅GppNHp. (C) The residues that exhibited chemical shift perturbation and line broadening in the presence of Kobe2601 (Fig. S5) are shown on the crystal structure of H-Ras⋅GppNHp (PDB ID code 5P21). Moderately perturbed residues with 0.01 ≤ Δδ < 0.015 and I/I0 ≤ 0.8, orange; strongly perturbed residues with Δδ ≥ 0.015 and I/I0 ≤ 0.8, red; missing residues or residues exhibiting resonance overlaps, cyan. The models were generated using MOLMOL (27) and PyMOL (DeLano Scientific, LLC).
Superimposition of the NMR structure of the H-RasT35S–Kobe2601 complex with the crystal structures of various Ras–effector complexes (15–17) revealed that Kobe2601 overlapped with the effector-binding interfaces (Fig. S7 B–E). As for c-Raf-1 RBD (15), both the flurobenzene and nitrobenzene moieties of Kobe2601 were likely to cause steric hindrance with its surface residues (Fig. S7B), supporting our observation of the competitive inhibition by Kobe0065 and Kobe2602. Moreover, a major part of Kobe2601, including the thiosemicarbazide and nitrobenzene moieties, was predicted to interfere with PI3K (16) much more heavily than with c-Raf-1 RBD (Fig. S7C), which may account for the efficient inhibition of Akt phosphorylation (Fig. 1E). Likewise, Kobe2601 was predicted to interfere with the Ras-interacting domain of RalGDS (17) (Fig. S7D) and also more heavily with the distal site of hSos (11) (Fig. S7E), which was experimentally demonstrated in this work (Figs. 1E and 2A).
Because the residues forming the compound-binding pocket are well conserved among Ras family members (Fig. S9), the Kobe0065-family compounds were predicted to exhibit rather broad specificity. This was indeed the case when various small GTPases in their GppNHp-bound forms were tested for direct interaction with Kobe0065 and Kobe2602 by relaxation-edited 1D 1H NMR (18) (Fig. S10). The compounds bound efficiently to M-Ras, Rap2A, and RalA but weakly to Rap1A compared with H-Ras. As for Rho family small GTPases, both Kobe0065 and Kobe2602 showed very weak, if any, binding activity toward Cdc42 and Rac1, whereas Kobe0065, but not Kobe2602, seemed to have some binding activity toward RhoA. Also, we found that both Kobe0065 and Kobe2602 bound to H-Ras⋅GDP as well in the 1D 1H NMR analysis. This was rather unexpected considering no apparent inhibitory effect of the compounds on the intrinsic GEF catalytic activity of Sos (Fig. 2A and Fig. S3A). Interpretation of the significance of this result on the mode of action of the Kobe0065-family compounds will require further structural information on their actual binding site on H-Ras⋅GDP, which is totally lacking at present.
Discussion
Recently, Maurer et al. (19) reported discovery of small-molecule compounds that bound to K-Ras⋅GDP and inhibited the Sos-mediated nucleotide exchange both in vitro and in vivo. Their crystal structure analyses of the complexes of the compounds, benzimidazole (BZIM), benzamidine (BZDN), and 4,6-dichloro-2-methy-3-aminoethyl-indole (DCAI), with K-Ras⋅GDP, K-Ras in complex with guanosine-5′-[γ-thio]triphosphate (GTPγS), and K-Ras in complex with guanosine-5′-[(β,γ)-methyleno]triphosphate, respectively, provided a molecular basis for inhibition of the Ras⋅GDP–Sos interaction but not the K-Ras⋅GTP–effector interaction; the compounds apparently interfered with the binding of K-Ras to Sos but not any effectors. In a sharp contrast, our compounds exhibited a prominent inhibitory activity at both the biochemical and cellular levels toward H-Ras⋅GTP and K-Ras⋅GTP and effectively interfered with the Ras–effector interactions, although they also showed a sign of binding activity toward H-Ras⋅GDP in the 1D 1H NMR analysis (Fig. S10).
Although the residues whose interaction with BZDN and DCAI detected by the HSQC analysis of H-Ras⋅GTP (19) showed some overlap with those identified by our NOE analysis with Kobe2601, a considerable difference existed in the location of the binding pockets and the orientation of the compounds (Fig. S11), which seemed to account for the difference in their ability to interfere with the effector interaction (Fig. S7 shows the effector interaction sites). Namely, the binding pocket for BZDN and DCAI in K-Ras⋅GTP is located close to Asp54, whose side chain forms a direct hydrogen bond with the NH group of BZDN, whereas Kobe2601 is too far to establish any direct interactions with Asp54. Sun et al. (20) also reported discovery of small-molecule compounds inhibiting K-Ras⋅GDP, which showed only the inhibition toward the Sos-mediated nucleotide exchange in vitro and shared the binding pocket and the orientation of the compounds on Ras⋅GDP with BZIM, BZDN, and DCAI. At present, it is not clear whether Sos inhibition is an effective strategy for suppressing the constitutively activated Ras mutants, considering the great reduction of their GTPase activity and a vast excess of free GTP over GDP in cellular concentrations. In this regard, our results showing that the RasG12V⋅GTP level was almost unaffected by the mSos1 level (Fig. 2 B and C) indicated that H-RasG12V escaped from the regulation by Sos. However, Sos inhibition might be effective for some cancer types, considering that the function of wild-type Ras is required for the growth of tumors carrying the activated ras oncogene (21).
In conclusion, we found that the Kobe0065-family compounds bind to Ras⋅GTP and exhibit antiproliferative activity toward cancer cells carrying the activated ras oncogenes, by a strategy based on SBDD. The compounds efficiently inhibit the interaction of Ras⋅GTP with their multiple effectors including Raf, PI3K, and RalGDS and a regulator/effector Sos and show rather broad binding specificity toward various Ras family small GTPases, which may account for their higher potency at the cellular level compared with that of the in vitro binding inhibition. Although the inhibitory activity is not particularly potent at present with the order of 10−6 to 10−5 M, the Kobe0065-family compounds may serve as a lead scaffold for the development of Ras inhibitors with higher potency and specificity and low toxicity that are suitable for clinical application. For this purpose, we would propose two possible strategies for structural optimization: the addition of a functional group that gains a hydrogen-bonding or ionic interaction with the charged residues such as Asp54 to increase the avidity and the avoidance of the thiosemicarbazide structure, which is generally considered to lead to cellular toxicity.
Materials and Methods
In Silico Docking Screening.
Structure-based screening was targeted at a surface pocket of M-RasP40D⋅GppNHp (PDB ID code 3KKP) (9) surrounded by the two switch regions and the nucleotide. The MMPB-SA method was used with an AMBER96 force field, where the solvent effect on binding free energy was introduced upon simulation (22). A virtual library containing 40,882 compounds (Namiki Shoji Co., Ltd., www.namiki-s.co.jp) was filtered by application of “Lipinski’s rule of five” for the selection of drug-like compounds, yielding 40,307 compounds to be screened. Upon docking simulation, the targeting pocket was specified by amino acid residues located within a 6.5-Å distance from the probe points, which were generated by referring to the position of Asp67 (corresponding to Asp57 of H-Ras) in M-RasP40D⋅GppNHp. The initial 3D Ras-compound docking structures and electric charges of the molecules in the presence or absence of water molecules around a Mg2+ ion were calculated by using Sievegene in myPresto software (23, 24) and Tripos software, respectively. Candidates were selected based on the calculated docking free energy values and the Nihon Electric Company’s original scoring functions.
Computer-Assisted Similarity Search.
Approximately 160,000 compounds from three libraries (Maybridge, Labotest, and Bionet) were described in the binary fingerprint format with some fragment-type topological descriptor such as atom-pair descriptors (25). The Tanimoto coefficient (26) between Kobe0065 and each compound was calculated and 273 compounds with a coefficient ≥0.7 were selected as candidates.
Biochemical and Cellular Assays, Xenograft Assays, NMR Spectroscopy, and Structural Analyses.
SI Materials and Methods gives details.
Supplementary Material
Acknowledgments
We thank Takahiro Yasuda for his help in statistical analysis, Masahiro Neya and D. Sasahara for synthesis of the compounds, and Yoshikuni Ito, Yoshihiko Kitaura, Junko Suzuki, Hikaru Yabuuchi, Eiichi Muramatsu, and Yoichi Kurebayashi for helpful discussion. This work was supported by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (T. Kataoka), a Health and Labour Sciences Research grant (to T. Kataoka), Japan Society for the Promotion of Science Grants-in Aid for Scientific Research (KAKENHI) Grants 20590280 and 23590336 (to F.S.), Ministry of Education, Culture, Sports, Science, and Technology (MEXT) KAKENHI Grants 17014061 (to T. Kataoka) and 18057014 (to F.S.), and MEXT Global Center of Excellence Program A08.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.R.A. is a guest editor invited by the Editorial Board.
Data deposition: NMR, atomic coordinates, chemical shifts, and restraints have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2lwi).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217730110/-/DCSupplemental.
References
- 1.Karnoub AE, Weinberg RA. Ras oncogenes: Split personalities. Nat Rev Mol Cell Biol. 2008;9(7):517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294(5545):1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein IB. Cancer. Addiction to oncogenes—The Achilles heal of cancer. Science. 2002;297(5578):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 4.Chin L, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400(6743):468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 5.Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci USA. 2008;105(13):5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James GL, Goldstein JL, Brown MS. Polylysine and CVIM sequences of K-RasB dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro. J Biol Chem. 1995;270(11):6221–6226. doi: 10.1074/jbc.270.11.6221. [DOI] [PubMed] [Google Scholar]
- 7.Rotblat B, Ehrlich M, Haklai R, Kloog Y. The Ras inhibitor farnesylthiosalicylic acid (Salirasib) disrupts the spatiotemporal localization of active Ras: A potential treatment for cancer. Methods Enzymol. 2008;439:467–489. doi: 10.1016/S0076-6879(07)00432-6. [DOI] [PubMed] [Google Scholar]
- 8.Ye M, et al. Crystal structure of M-Ras reveals a GTP-bound “off” state conformation of Ras family small GTPases. J Biol Chem. 2005;280(35):31267–31275. doi: 10.1074/jbc.M505503200. [DOI] [PubMed] [Google Scholar]
- 9.Shima F, et al. Structural basis for conformational dynamics of GTP-bound Ras protein. J Biol Chem. 2010;285(29):22696–22705. doi: 10.1074/jbc.M110.125161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muraoka S, et al. Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants. FEBS Lett. 2012;586(12):1715–1718. doi: 10.1016/j.febslet.2012.04.058. [DOI] [PubMed] [Google Scholar]
- 11.Wilhelm S, et al. Discovery and development of sorafenib: A multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5(10):835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 12.Margarit SM, et al. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112(5):685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 13.Araki M, et al. Solution structure of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers. J Biol Chem. 2011;286(45):39644–39653. doi: 10.1074/jbc.M111.227074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito Y, et al. Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry. 1997;36(30):9109–9119. doi: 10.1021/bi970296u. [DOI] [PubMed] [Google Scholar]
- 15.Nassar N, et al. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375(6532):554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- 16.Pacold ME, et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103(6):931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 17.Huang L, Hofer F, Martin GS, Kim SH. Structural basis for the interaction of Ras with RalGDS. Nat Struct Biol. 1998;5(6):422–426. doi: 10.1038/nsb0698-422. [DOI] [PubMed] [Google Scholar]
- 18.Hajduk PJ, Olejniczak ET, Fesik SW. One-dimensional relaxation- and diffusion-edited NMR methods for screening compounds that bind to macromolecules. J Am Chem Soc. 1997;119:12257–12261. [Google Scholar]
- 19.Maurer T, et al. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci USA. 2012;109(14):5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51(25):6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim KH, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452(7187):646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem. 1994;98:1978–1988. [Google Scholar]
- 23.Fukunishi Y, Mikami Y, Nakamura H. The filing potential method: A method for estimating the free energy surface for protein-ligand docking. J Phys Chem B. 2003;107:13201–13210. [Google Scholar]
- 24.Fukunishi Y, Mikami Y, Nakamura H. Similarities among receptor pockets and among compounds: analysis and application to in silico ligand screening. J Mol Graph Model. 2005;24(1):34–45. doi: 10.1016/j.jmgm.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Carhart RE, Smith DH, Venkataraghavan R. Atom pairs as molecular features in structure-activity studies: Defenition and applications. J Chem Inf Comput Sci. 1985;25:64–73. [Google Scholar]
- 26.Willett P, Barnard JM, Downs GM. Chemical similarity searching. J Chem Inf Comput Sci. 1998;38:983–996. [Google Scholar]
- 27.Koradi R, Billeter M, Wuthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(1):51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.